The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Soluble Sugar Assay
2.3. Antioxidant and Antioxidant Capacity Assays
2.3.1. Total Phenolic Content Assay
2.3.2. Total Flavonoids
2.3.3. Total Antioxidant Capacity Assay
2.4. Extraction of Desulfo-Glucosinolates and Ultra-Performance Liquid Chromatography
2.5. Library Preparation and RNA Sequencing
2.6. Transcript Quantification, Differential Expression Analysis, and Gene Annotation
2.7. Quantitative Real-Time PCR Expression Analysis in B. rapa
2.8. Statistical Analysis
2.9. Data Depositon
3. Results
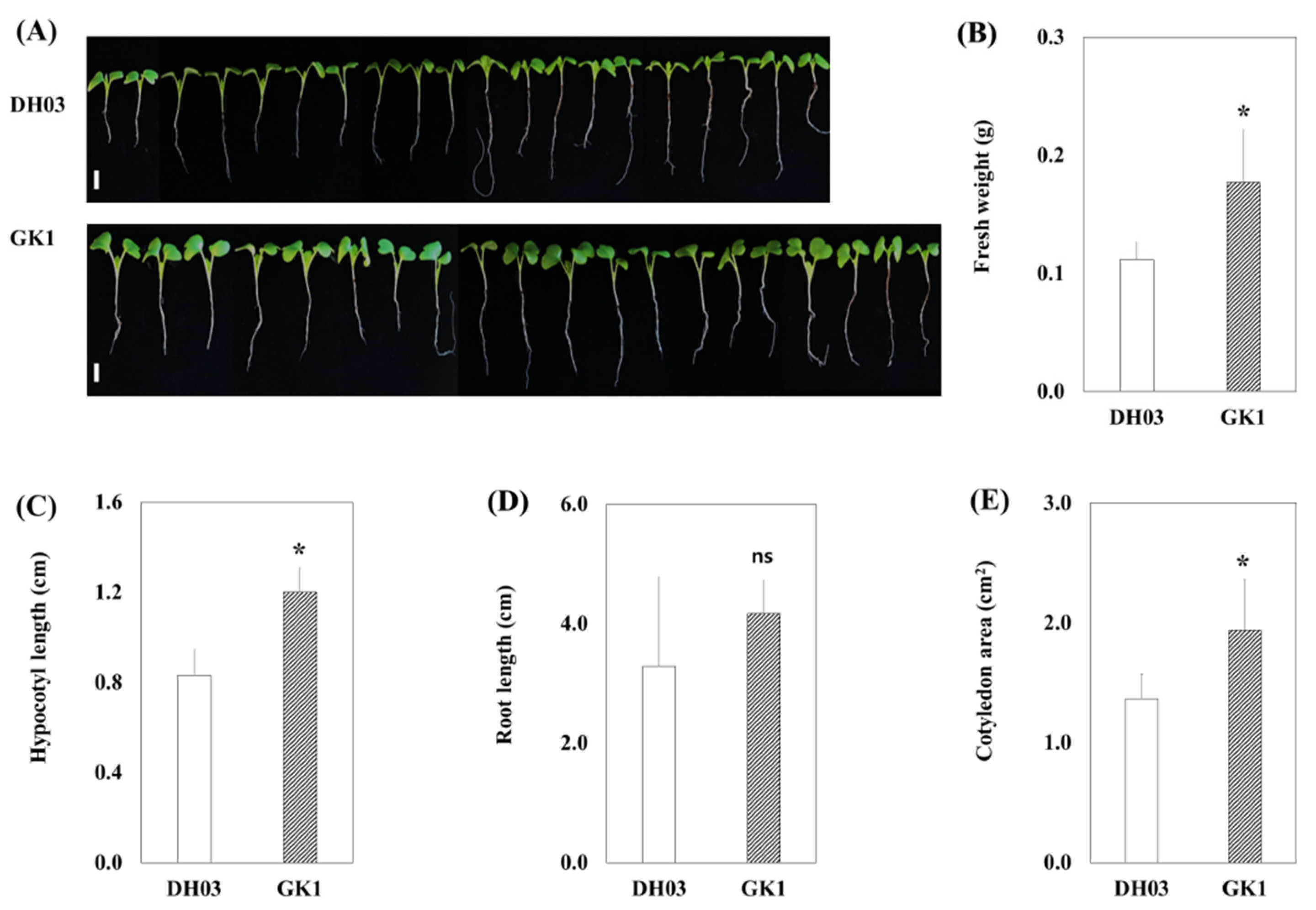
3.1. Promotion of Growth and Secondary Metabolites in GI Knockdown Chinese Cabbage
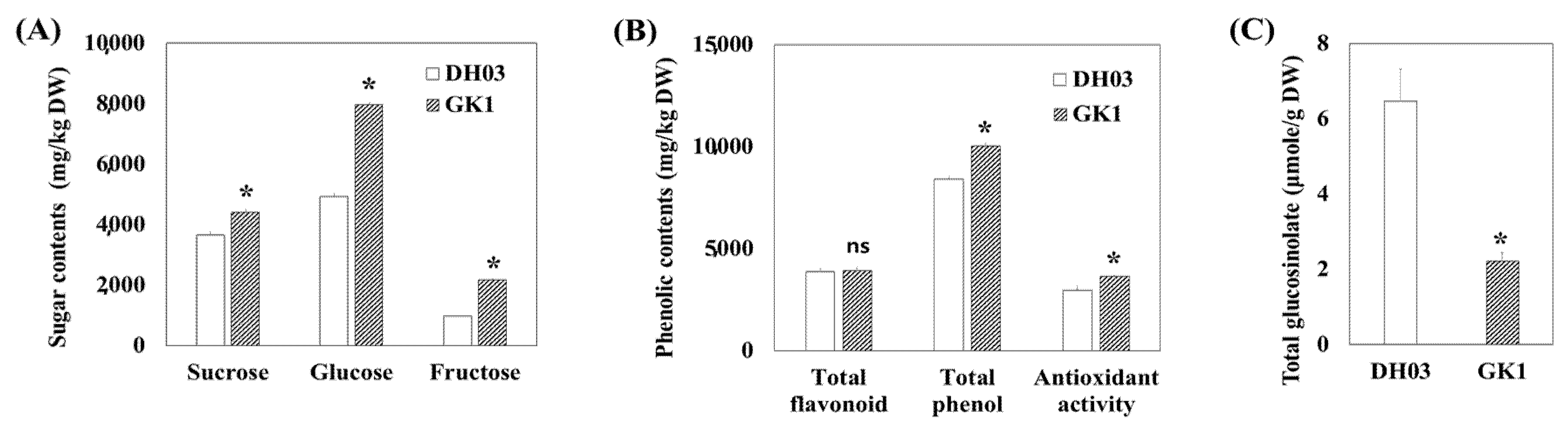
3.2. Primary and Secondary Metabolite Contents
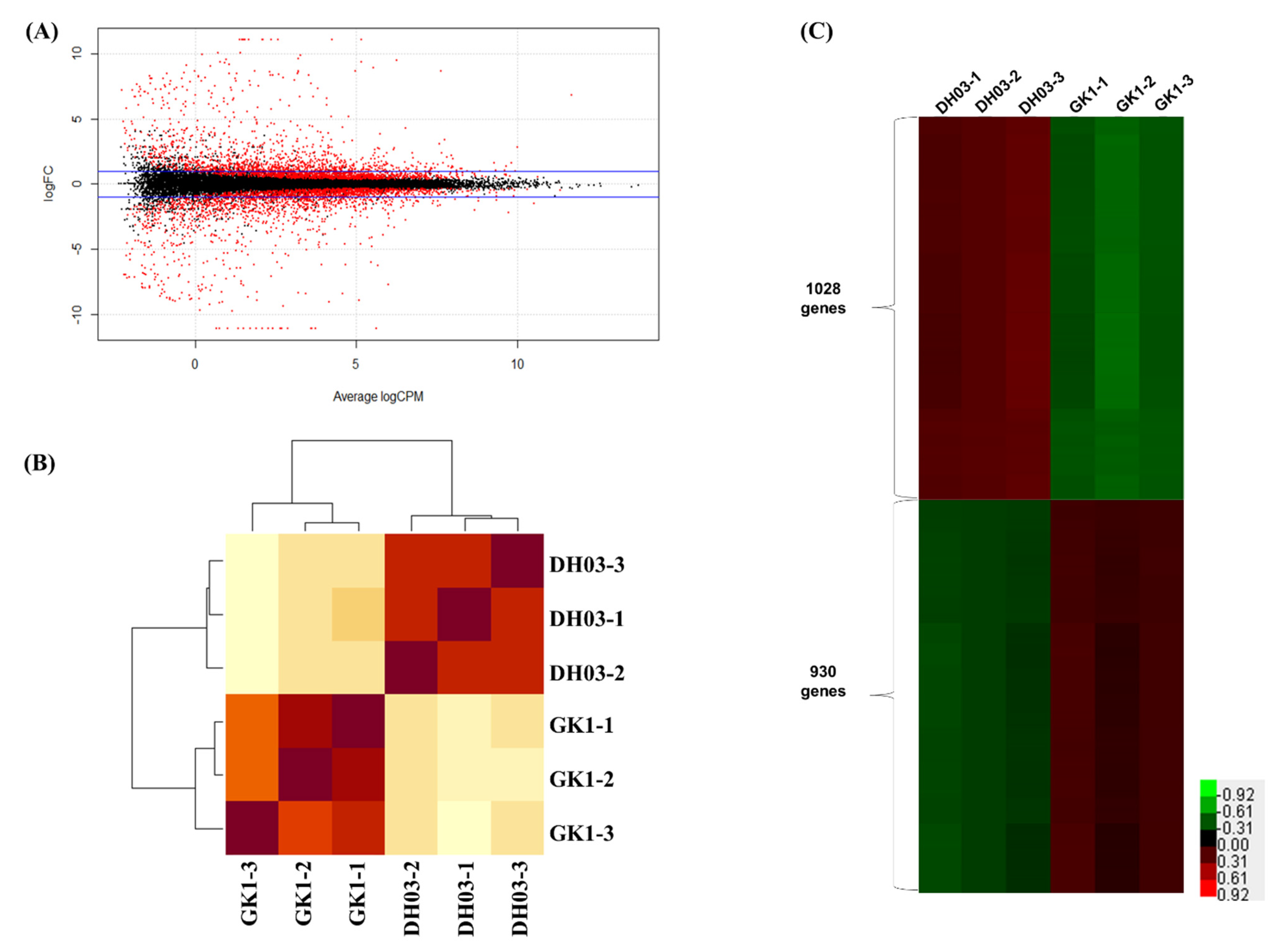
3.3. Transcriptomic Analysis
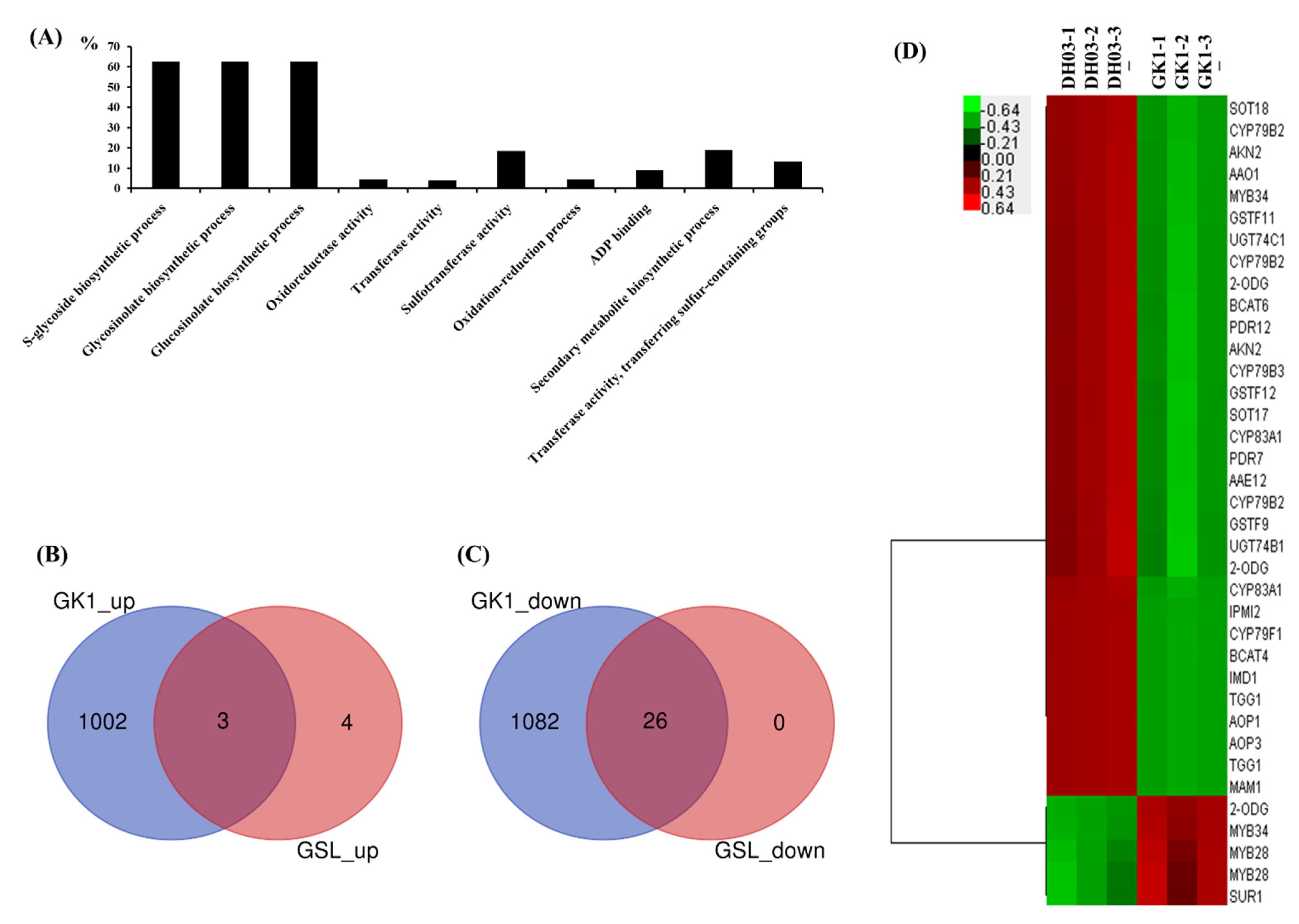
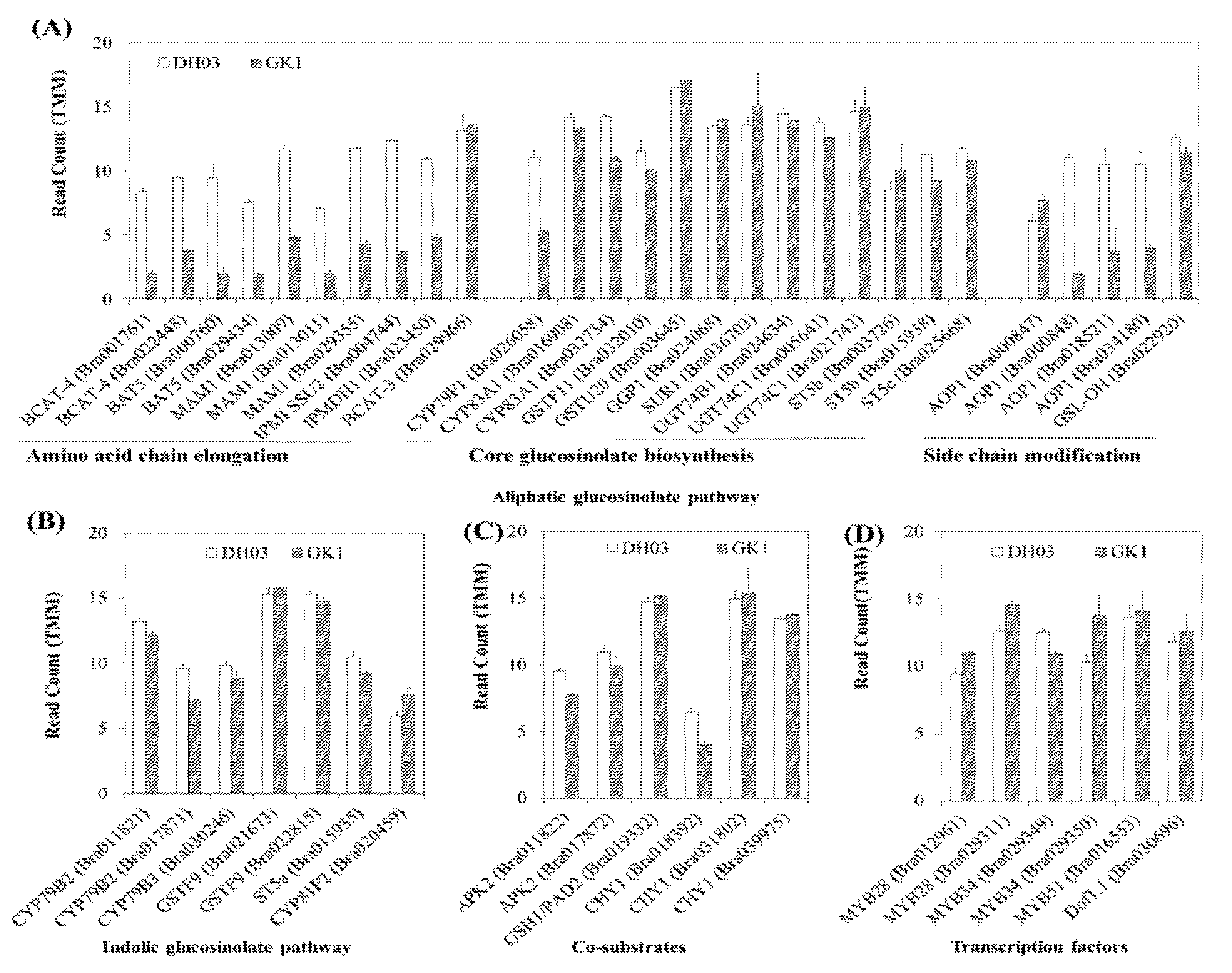
3.4. Expression of Genes Related to GSL
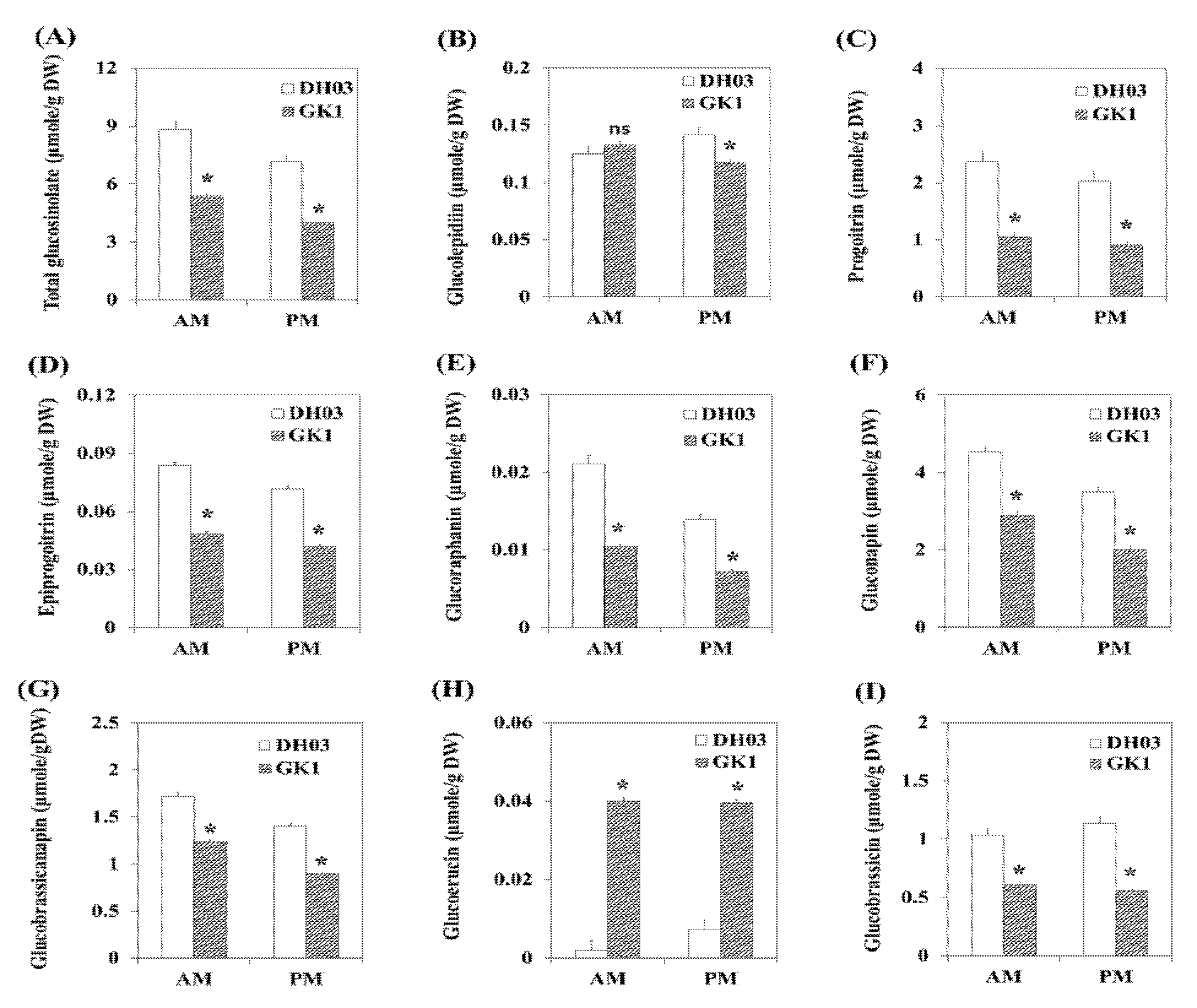
3.5. GSL Compounds in GK1
4. Discussion
4.1. Reduced Expression of GI Alters the Main Traits of Crop Plants
4.2. Reduced Expression of GI Could Affect the GSL Pathway
4.3. Clock Genes Could Be Modified to Improve Crop Productivity and Quality
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Farre, E.M.; Liu, T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 2013, 16, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soengas, P.; Cartea, M.E.; Velasco, P.; Francisco, M. Brassica glucosinolate rhythmicity in response to light-dark entrainment cycles is cultivar-dependent. Plant Sci. 2018, 275, 28–35. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Comes a time. Curr. Opin. Plant Biol. 2008, 11, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Farre, E.M.; Weise, S.E. The interactions between the circadian clock and primary metabolism. Curr. Opin. Plant Biol. 2012, 15, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef] [PubMed]
- Yazdanbakhsh, N.; Sulpice, R.; Graf, A.; Stitt, M.; Fisahn, J. Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ. 2011, 34, 877–894. [Google Scholar] [CrossRef]
- Webb, A.A.; Satake, A. Understanding circadian regulation of carbohydrate metabolism in Arabidopsis using mathematical models. Plant Cell Physiol. 2015, 56, 586–593. [Google Scholar]
- Haydon, M.J.; Roman, A.; Arshad, W. Nutrient homeostasis within the plant circadian network. Front. Plant Sci. 2015, 6, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sweet immunity in the plant circadian regulatory network. J. Exp. Bot. 2013, 64, 1439–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Shin, J.; Davis, S.J. Abiotic stress and the plant circadian clock. Plant Signal. Behav. 2011, 6, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanowski, A.; Yanovsky, M.J. Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 2015, 6, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, C.J.; Kay, S.A. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010, 44, 419–444. [Google Scholar] [PubMed] [Green Version]
- Kerwin, R.E.; Jimenez-Gomez, J.M.; Fulop, D.; Harmer, S.L.; Maloof, J.N.; Kliebenstein, D.J. Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 2011, 23, 471–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.A.; Jung, H.E.; Hong, J.K.; Hermand, V.; Robertson McClung, C.; Lee, Y.H.; Kim, J.Y.; Lee, S.I.; Jeong, M.J.; Kim, J.; et al. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Jayaprakasha, G.K.; Singh, J.; Uckoo, R.; Borrego, E.J.; Finlayson, S.; Kolomiets, M.; Patil, B.S.; Braam, J.; Zhu-Salzman, K. CIRCADIAN CLOCK-ASSOCIATED1 Controls Resistance to Aphids by Altering Indole Glucosinolate Production. Plant Physiol. 2019, 181, 1344–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.J.; Yeh, K.W.; Tsai, C.Y. Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 2001, 161, 635–644. [Google Scholar] [CrossRef]
- Mishra, P.; Panigrahi, K.C. GIGANTEA––An emerging story. Front. Plant Sci. 2015, 6, 8. [Google Scholar] [PubMed] [Green Version]
- Fowler, S.; Lee, K.; Onouchi, H.; Samach, A.; Richardson, K.; Morris, B.; Coupland, G.; Putterill, J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999, 18, 4679–4688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Tryon, E.L.; Kreps, J.A.; Harmer, S.L. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007, 143, 473–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [PubMed] [Green Version]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Q.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C.; et al. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, D.; Kim, W.Y.; Cha, J.Y.; Park, H.J.; Shin, G.; Park, J.; Lim, C.J.; Chun, H.J.; Li, N.; Kim, D.H.; et al. The GIGANTEA-ENHANCED EM LEVEL Complex Enhances Drought Tolerance via Regulation of Abscisic Acid Synthesis. Plant Physiol. 2020, 184, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Kim, H.S.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.S.; Choi, Y.I.; Xu, B.; Deng, X.; Yun, D.J.; et al. Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar. Plant Biotechnol. J. 2017, 15, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Padilla, G.; Cartea, M.E.; Velasco, P.; de Haro, A.; Ordas, A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 2007, 68, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Martinez-Ballesta, M.; Moreno, D.A.; Carvajal, M. The physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef] [Green Version]
- Mugford, S.G.; Yoshimoto, N.; Reichelt, M.; Wirtz, M.; Hill, L.; Mugford, S.T.; Nakazato, Y.; Noji, M.; Takahashi, H.; Kramell, R.; et al. Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 2009, 21, 910–927. [Google Scholar] [CrossRef] [Green Version]
- Huseby, S.; Koprivova, A.; Lee, B.R.; Saha, S.; Mithen, R.; Wold, A.B.; Bengtsson, G.B.; Kopriva, S. Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis. J. Exp. Bot. 2013, 64, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Lou, P.; Wu, J.; Cheng, F.; Cressman, L.G.; Wang, X.W.; McClung, C.R. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 2012, 24, 2415–2426. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.A.; Shim, D.; Kumari, S.; Jung, H.E.; Jung, K.H.; Jeong, H.; Kim, W.Y.; Lee, S.I.; Jeong, M.J. Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage. Genes 2019, 10, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Menichini, F.; Conforti, F.; Rigano, D.; Formisano, C.; Piozzi, F.; Senatore, F. Phytochemical composition, anti-inflammatory and antitumour activities of four Teucrium essential oils from Greece. Food Chem. 2009, 115, 679–686. [Google Scholar] [CrossRef]
- Shiva Ram Bhandari, J.-H.K. Seasonal variation in phytochemicals and antioxidant activities in different tissues of various Broccoli cultivars. Afr. J. Biotechnol. 2014, 13, 604–615. [Google Scholar] [CrossRef]
- Kumari, S.; Jo, J.S.; Choi, H.S.; Lee, J.G.; Lee, S.I.; Jeong, M.-J.; Kim, J.A. Molecular Characterization and Expression Analysis of MYB Transcription Factors Involved in the Glucosinolate Pathway in Chinese Cabbage (Brassica rapa ssp. pekinensis). Agronomy 2019, 9, 807. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, S.R.; Jo, J.S.; Lee, J.G. Comparison of Glucosinolate Profiles in Different Tissues of Nine Brassica Crops. Molecules 2015, 20, 15827–15841. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008, 9, 321–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gigolashvili, T.; Berger, B.; Mock, H.P.; Muller, C.; Weisshaar, B.; Flugge, U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 50, 886–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, M.Y.; Sugiyama, K.; Sawada, Y.; Tohge, T.; Obayashi, T.; Suzuki, A.; Araki, R.; Sakurai, N.; Suzuki, H.; Aoki, K.; et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 6478–6483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.B.; Li, X.; Kim, S.J.; Kim, H.H.; Lee, J.; Kim, H.; Park, S.U. MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 2013, 18, 8682–8695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Tang, J.; Hu, R.; Wu, P.; Hou, X.L.; Song, X.M.; Xiong, A.S. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics 2015, 16, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, J.Y.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Oliverio, K.A.; Crepy, M.; Martin-Tryon, E.L.; Milich, R.; Harmer, S.L.; Putterill, J.; Yanovsky, M.J.; Casal, J.J. GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 2007, 144, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, T.S.; Salome, P.A.; McClung, C.R.; Olszewski, N.E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16, 1550–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Kim, J.Y.; Lee, J.H.; Lee, B.D.; Paek, N.C.; Park, C.M. GIGANTEA Shapes the Photoperiodic Rhythms of Thermomorphogenic Growth in Arabidopsis. Mol. Plant 2020, 13, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Tseng, T.S.; Olszewski, N.E. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 2001, 126, 1174–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Chen, Q.; Botella, J.R.; Guo, S. Beyond Light: Insights into the Role of Constitutively Photomorphogenic1 in Plant Hormonal Signaling. Front. Plant Sci. 2019, 10, 557. [Google Scholar] [CrossRef] [PubMed]
- Halliday, K.J.; Martinez-Garcia, J.F.; Josse, E.M. Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. 2009, 1, a001586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Kumar, S.V. DET1 and COP1 Modulate the Coordination of Growth and Immunity in Response to Key Seasonal Signals in Arabidopsis. Cell Rep. 2018, 25, 29–37.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandoli, C.; Petri, C.; Egea-Cortines, M.; Weiss, J. The clock gene Gigantea 1 from Petunia hybrida coordinates vegetative growth and inflorescence architecture. Sci. Rep. 2020, 10, 275. [Google Scholar] [CrossRef]
- Cha, J.Y.; Lee, D.Y.; Ali, I.; Jeong, S.Y.; Shin, B.; Ji, H.; Kim, J.S.; Kim, M.G.; Kim, W.Y. Arabidopsis GIGANTEA negatively regulates chloroplast biogenesis and resistance to herbicide butafenacil. Plant Cell Rep. 2019, 38, 793–801. [Google Scholar] [CrossRef]
- Dalchau, N.; Baek, S.J.; Briggs, H.M.; Robertson, F.C.; Dodd, A.N.; Gardner, M.J.; Stancombe, M.A.; Haydon, M.J.; Stan, G.B.; Goncalves, J.M.; et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 2011, 108, 5104–5109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eimert, K.; Wang, S.M.; Lue, W.I.; Chen, J. Monogenic Recessive Mutations Causing Both Late Floral Initiation and Excess Starch Accumulation in Arabidopsis. Plant Cell 1995, 7, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, H.; Cai, C.; Wei, J.; Huang, J.; Chang, J.; Qian, H.; Zhang, X.; Zhao, Y.; Sun, B.; Wang, B.; et al. Glucose enhances indolic glucosinolate biosynthesis without reducing primary sulfur assimilation. Sci. Rep. 2016, 6, 31854. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Hansen, C.H.; Wittstock, U.; Halkier, B.A. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000, 275, 33712–33717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef] [Green Version]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, K.; Mithen, R.; Williamson, G. Selective increase of the potential anticarcinogen 4-methylsulphinylbutyl glucosinolate in Broccoli. Carcinogenesis 1998, 19, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Mithen, R.; Faulkner, K.; Magrath, R.; Rose, P.; Williamson, G.; Marquez, J. Development of isothiocyanate-enriched Broccoli, and its enhanced ability to induce phase 2 detoxification enzymes in mammalian cells. Theor. Appl. Genet. 2003, 106, 727–734. [Google Scholar] [CrossRef] [PubMed]

| Sample Name | Read-Pairs | Both Surviving | Overall Alignment Rate | Concordant Zero | Concordant Pair Alignment | Multiple Alignment |
|---|---|---|---|---|---|---|
| DH03_R1 | 39,266,003 | 36,514,950 (92.99%) | 91.71% | 12.79% | 84.82% | 2.39% |
| DH03_R2 | 41,520,758 | 38,946,152 (93.80%) | 91.87% | 12.46% | 85.04% | 2.49% |
| DH03_R3 | 43,990,750 | 41,288,526 (93.86%) | 91.92% | 12.54% | 84.99% | 2.47% |
| GK1_R1 | 39,987,718 | 37,533,690 (93.86%) | 91.95% | 12.70% | 84.69% | 2.61% |
| GK1_R2 | 31,435,216 | 29,695,080 (94.46%) | 92.16% | 12.32% | 85.34% | 2.34% |
| GK1_R3 | 38,946,087 | 36,703,007 (94.24%) | 92.03% | 12.55% | 84.90% | 2.55% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.S.; Kim, S.J.; Jo, J.S.; Lee, J.G.; Lee, S.I.; Kim, D.H.; Kim, J.A. The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage. Genes 2021, 12, 1664. https://doi.org/10.3390/genes12111664
Kim NS, Kim SJ, Jo JS, Lee JG, Lee SI, Kim DH, Kim JA. The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage. Genes. 2021; 12(11):1664. https://doi.org/10.3390/genes12111664
Chicago/Turabian StyleKim, Nan Sun, Su Jeong Kim, Jung Su Jo, Jun Gu Lee, Soo In Lee, Dong Hwan Kim, and Jin A. Kim. 2021. "The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage" Genes 12, no. 11: 1664. https://doi.org/10.3390/genes12111664
APA StyleKim, N. S., Kim, S. J., Jo, J. S., Lee, J. G., Lee, S. I., Kim, D. H., & Kim, J. A. (2021). The BrGI Circadian Clock Gene Is Involved in the Regulation of Glucosinolates in Chinese Cabbage. Genes, 12(11), 1664. https://doi.org/10.3390/genes12111664








