Single-Cell RNA-Seq Revealed the Gene Expression Pattern during the In Vitro Maturation of Donkey Oocytes
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. Chemicals
2.3. Collection and In Vitro Maturation (IVM) of Cumulus-Oocyte-Complexes (COCs)
2.4. Collection of Donkey Oocytes
2.5. Library Preparation and RNA Sequencing
2.6. Identification of Differentially Expressed Gene
2.7. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
2.8. qRT-PCR Analysis
2.9. Statistical Analysis
3. Results
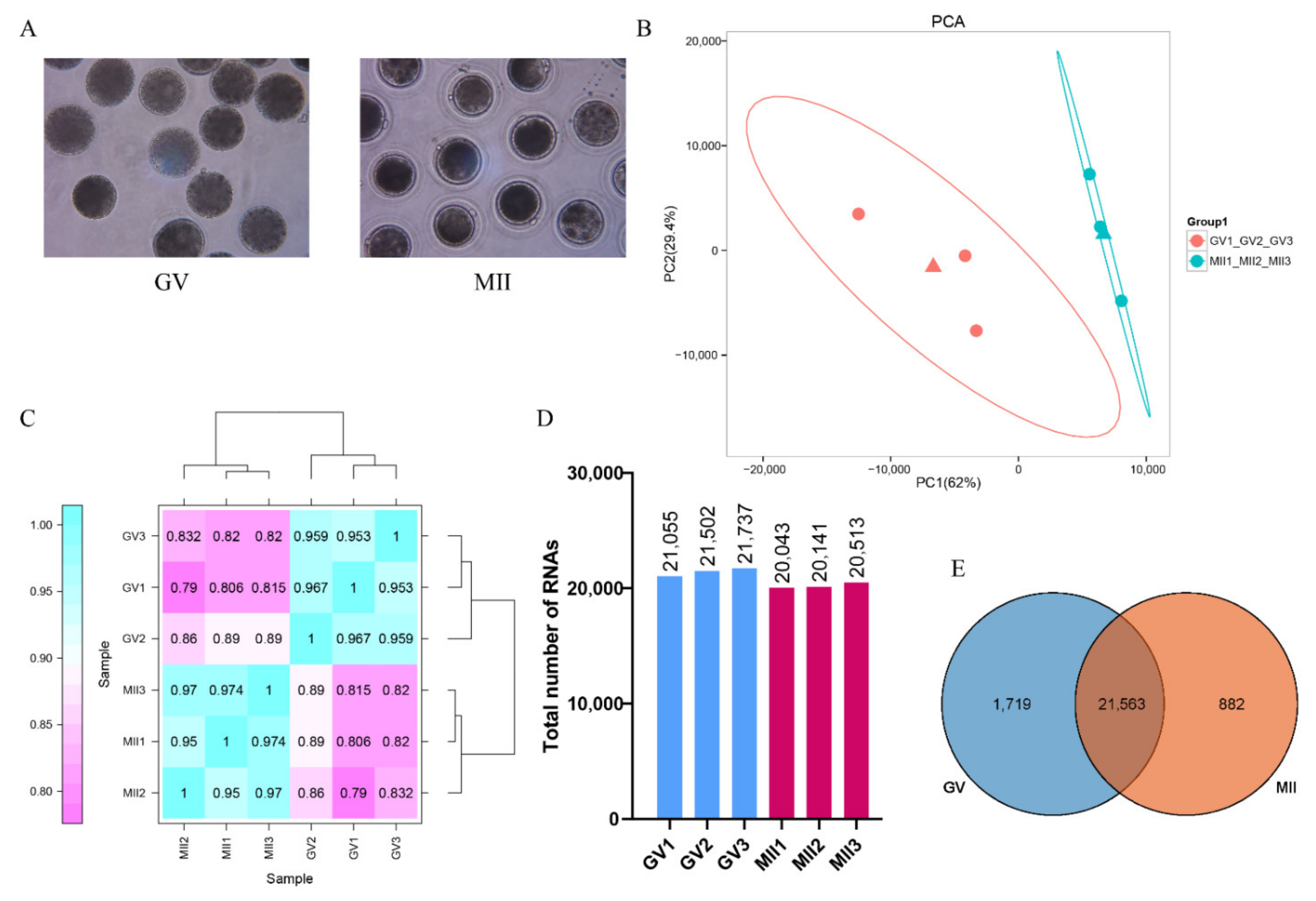
3.1. In Vitro Development of Donkey Oocytes
3.2. scRNA-seq of Donkey Oocytes
3.3. Abundant mRNAs in Oocyte Meiosis
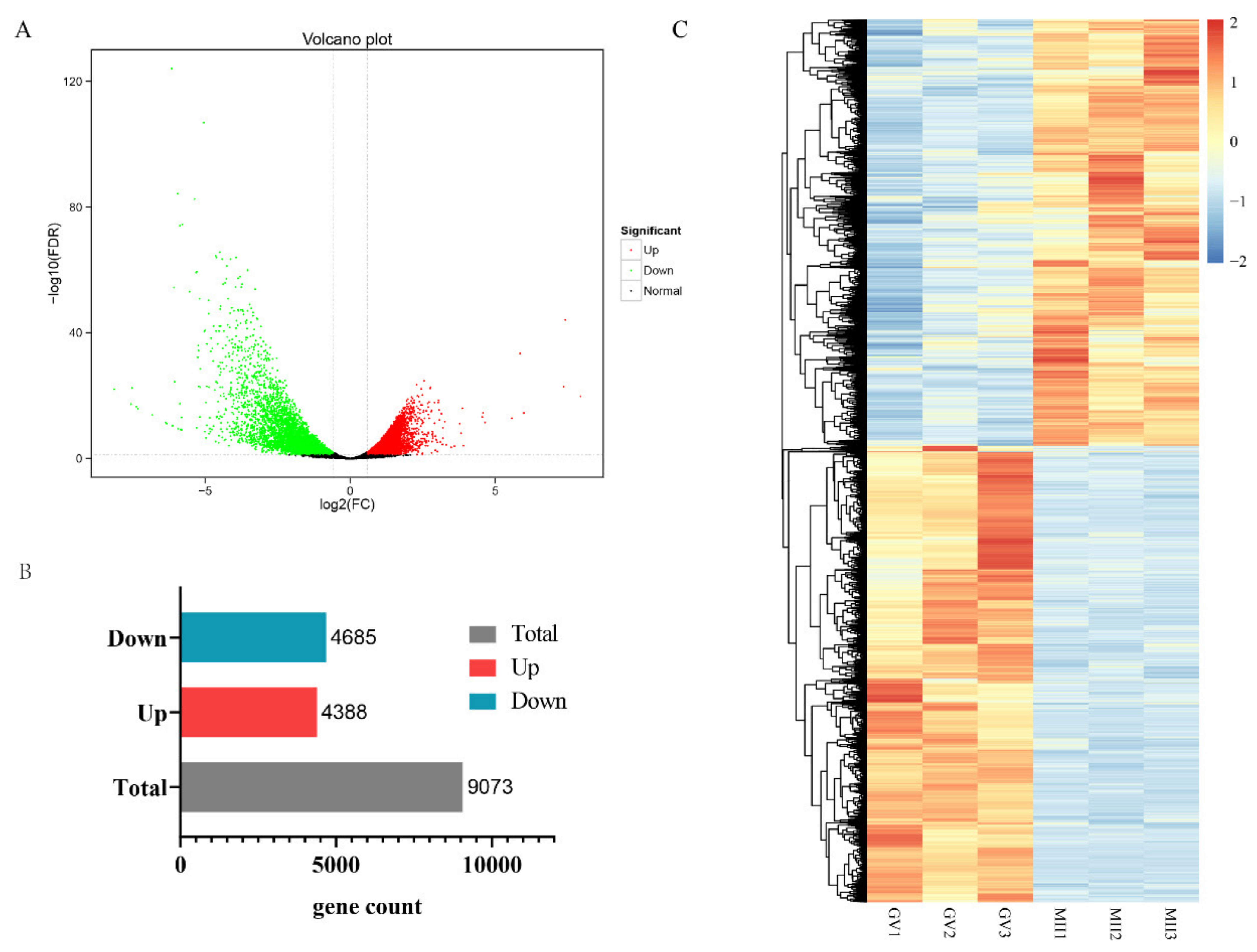
3.4. Differentially Expressed Genes (DEGs) in GV and MII Oocytes
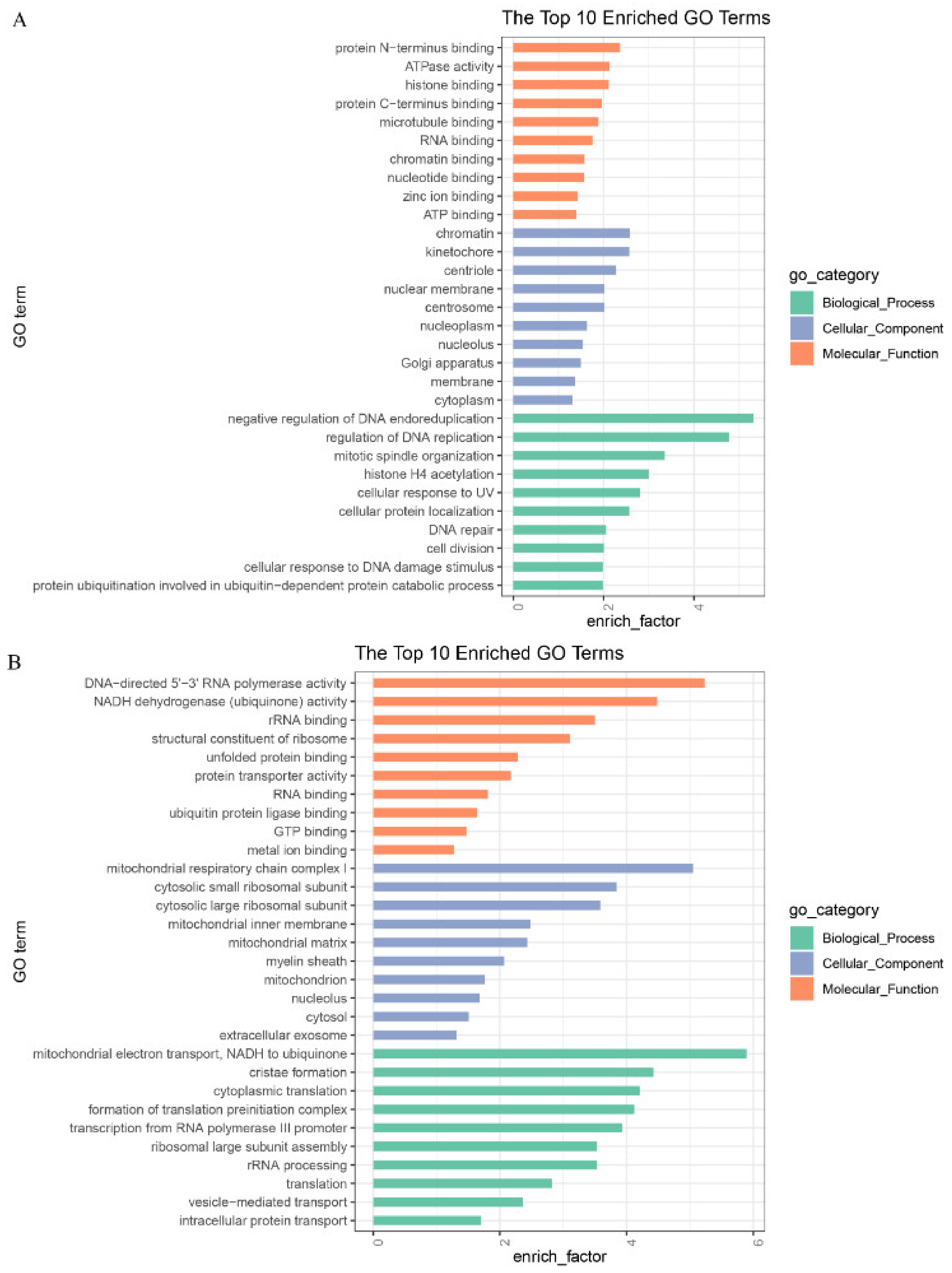
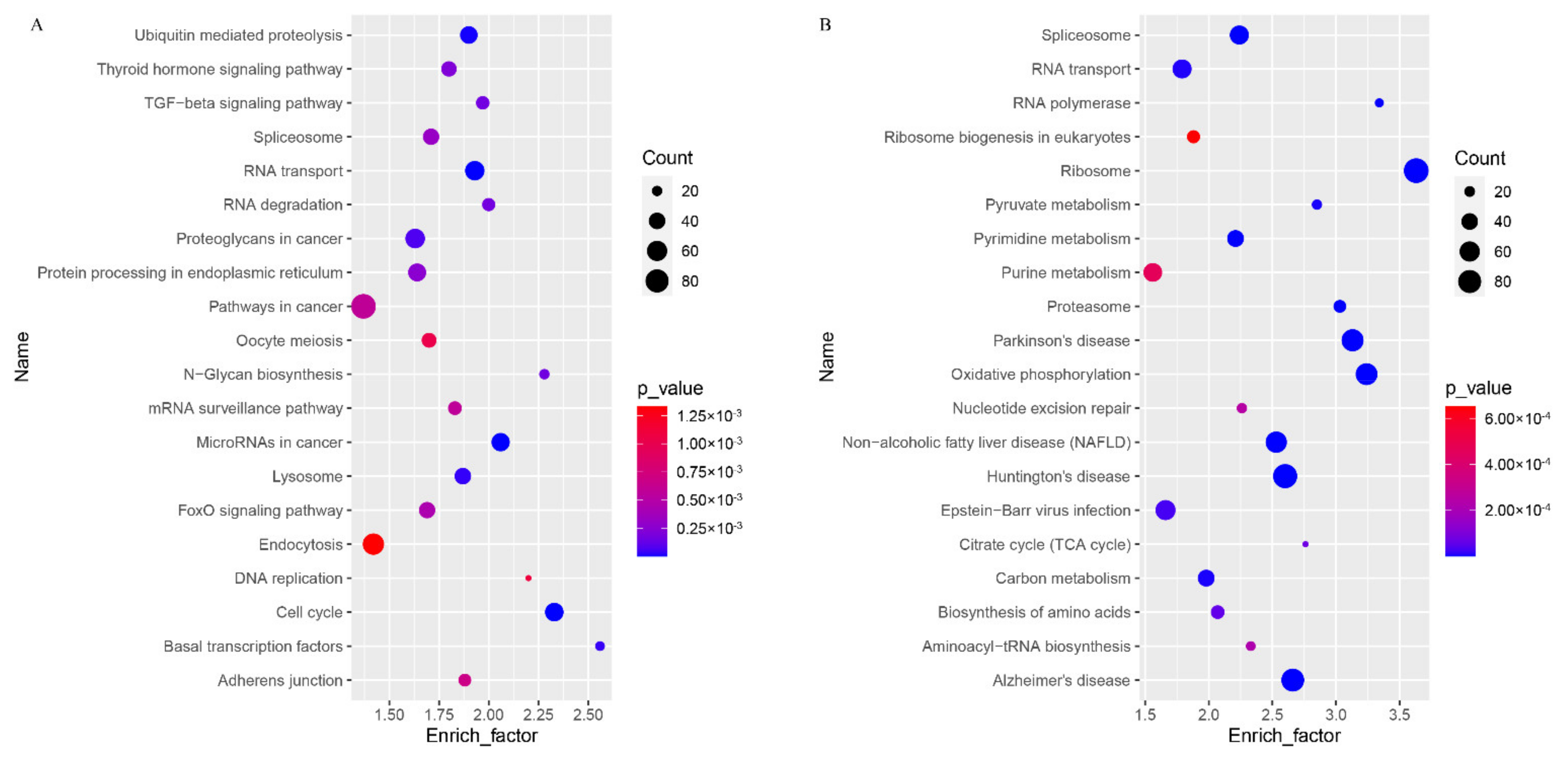
3.5. GO and KEGG Analyses of DEGs in GV and MII Oocytes
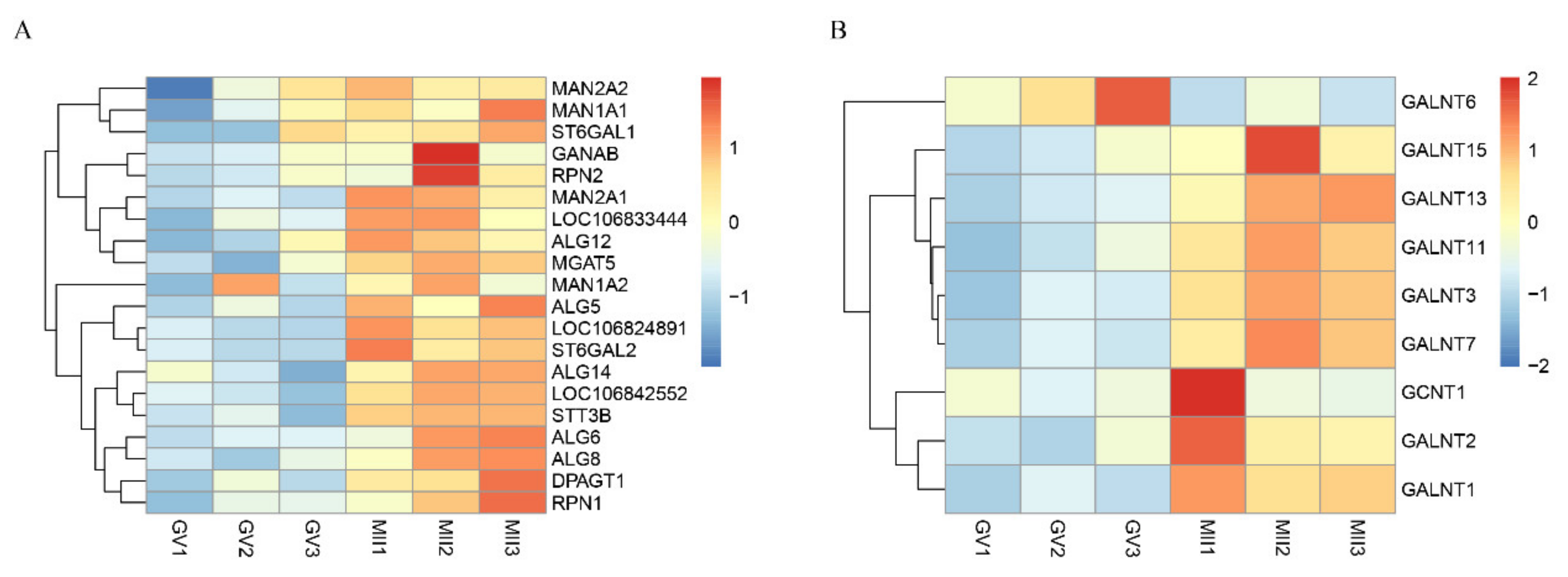
3.6. Glycosylation Vital for Oocyte In Vitro Maturation
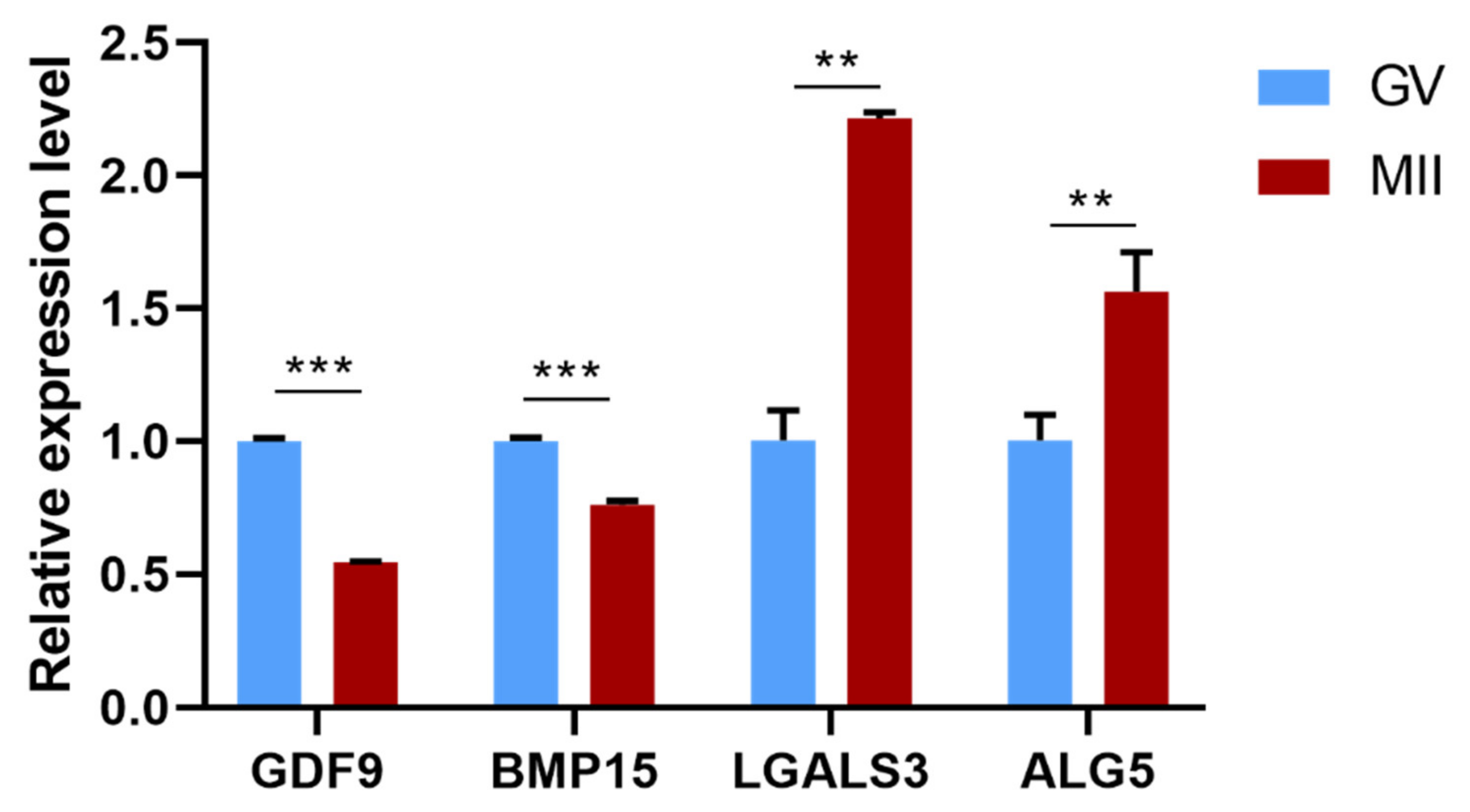
3.7. qPCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goudet, G.; Douet, C.; Kaabouba-Escurier, A.; Couty, I.; Nicolás, C.M.; Barrière, P.; Blard, T.; Reigner, F.; Deleuze, S.; Magistrini, M. Establishment of conditions for ovum pick up and IVM of jennies oocytes toward the setting up of efficient IVF and in vitro embryos culture procedures in donkey (Equus asinus). Theriogenology 2016, 86, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Abdoon, A.S.; Fathalla, S.I.; Shawky, S.M.; Kandil, O.M.; Kishta, A.A.; Masoud, S.W. In Vitro Maturation and Fertilization of Donkey Oocytes. J. Equine Veter. Sci. 2018, 65, 118–122. [Google Scholar] [CrossRef]
- Polidori, P.; Cavallucci, C.; Beghelli, D.; Vincenzetti, S. Physical and chemical characteristics of donkey meat from Martina Franca breed. Meat Sci. 2009, 82, 469–471. [Google Scholar] [CrossRef]
- Guo, H.; Pang, K.; Zhang, X.; Zhao, L.; Chen, S.; Dong, M.; Ren, F. Composition, Physiochemical Properties, Nitrogen Fraction Distribution, and Amino Acid Profile of Donkey Milk. J. Dairy Sci. 2007, 90, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.; Pfuderer, S. The Potential for New Donkey Farming Systems to Supply the Growing Demand for Hides. Animals 2020, 10, 718. [Google Scholar] [CrossRef]
- Appeltant, R.; Somfai, T.; Maes, M.; Van Soom, A.; Kikuchi, K. Porcine oocyte maturation in vitro: Role of cAMP and oocyte-secreted factors—A practical approach. J. Reprod. Dev. 2016, 62, 439–449. [Google Scholar] [CrossRef]
- Sun, Q.-Y.; Nagai, T. Molecular Mechanisms Underlying Pig Oocyte Maturation and Fertilization. J. Reprod. Dev. 2003, 49, 347–359. [Google Scholar] [CrossRef]
- Macaulay, I.; Voet, T. Single Cell Genomics: Advances and Future Perspectives. PLoS Genet. 2014, 10, e1004126. [Google Scholar] [CrossRef]
- Liu, X.-M.; Wang, Y.-K.; Liu, Y.-H.; Yu, X.-X.; Wang, P.-C.; Li, X.; Du, Z.-Q.; Yang, C.-X. Single-cell transcriptome sequencing reveals that cell division cycle 5-like protein is essential for porcine oocyte maturation. J. Biol. Chem. 2018, 293, 1767–1780. [Google Scholar] [CrossRef]
- Wang, N.; Li, C.-Y.; Zhu, H.-B.; Hao, H.-S.; Wang, H.-Y.; Yan, C.-L.; Zhao, S.-J.; Du, W.-H.; Wang, D.; Liu, Y.; et al. Effect of vitrification on the mRNA transcriptome of bovine oocytes. Reprod. Domest. Anim. 2017, 52, 531–541. [Google Scholar] [CrossRef]
- Huang, J.; Ma, Y.; Wei, S.; Pan, B.; Qi, Y.; Hou, Y.; Meng, Q.; Zhou, G.; Han, H. Dynamic changes in the global transcriptome of bovine germinal vesicle oocytes after vitrification followed by in vitro maturation. Reprod. Fertil. Dev. 2018, 30, 1298. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Jayavelu, N.D.; Battle, S.L.; Mar, J.C.; Schimmel, T.; Cohen, J.; Hawkins, R.D. Single-cell analysis of transcriptome and DNA methylome in human oocyte maturation. PLoS ONE 2020, 15, e0241698. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Gao, B.; Wang, G.; Li, H.; Ahmed, J.Z.; Zhang, D.; Ye, S.; Liu, S.; Li, M.; Shi, D.; et al. The key long non-coding RNA screening and validation between germinal vesicle and metaphase II of porcine oocyte in vitro maturation. Reprod. Domest. Anim. 2020, 55, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Niu, M.H.; Zhang, T.; Shen, W.; Cao, H.G. Genome-Wide Network of lncRNA-mRNA During Ovine Oocyte Development From Germinal Vesicle to Metaphase II in vitro. Front. Physiol. 2020, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.M.; Chitwood, J.L.; Ross, P.J. RNA-Seq profiling of single bovine oocyte transcript abundance and its modulation by cytoplasmic polyadenylation. Mol. Reprod. Dev. 2015, 82, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Jia, G.; Li, A.; Ma, H.; Huang, Z.; Zhu, S.; Hou, Y.; Fu, X. RNA-Seq transcriptome profiling of mouse oocytes after in vitro maturation and/or vitrification. Sci. Rep. 2017, 7, 13245. [Google Scholar] [CrossRef]
- Yang, C.-X.; Song, Z.-Q.; Pei, S.-R.; Yu, X.-X.; Miao, J.-K.; Liang, H.; Miao, Y.-L.; Du, Z.-Q. Single cell RNA-seq reveals molecular pathways altered by 7, 12-dimethylbenz[a]anthracene treatment on pig oocytes. Theriogenology 2020, 157, 449–457. [Google Scholar] [CrossRef]
- Hinrichs, K.; Schmidt, A.L.; Friedman, P.P.; Selgrath, J.P.; Martin, M.G. In Vitro Maturation of Horse Oocytes: Characterization of Chromatin Configuration Using Fluorescence Microscopy1. Biol. Reprod. 1993, 48, 363–370. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, C.; Han, S.J.; Daldello, E.M.; Cho, A.; Martins, J.P.S.; Xia, G.; Conti, M. Maternal mRNAs with distinct 3′ UTRs define the temporal pattern ofCcnb1synthesis during mouse oocyte meiotic maturation. Genes Dev. 2017, 31, 1302–1307. [Google Scholar] [CrossRef]
- Rong, Y.; Ji, S.-Y.; Zhu, Y.-Z.; Wu, Y.-W.; Shen, L.; Fan, H.-Y. ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res. 2019, 47, 11387–11402. [Google Scholar] [CrossRef]
- Song, Z.-Q.; Li, X.; Wang, Y.-K.; Du, Z.-Q.; Yang, C.-X. DMBA acts on cumulus cells to desynchronize nuclear and cytoplasmic maturation of pig oocytes. Sci. Rep. 2017, 7, 1687. [Google Scholar] [CrossRef]
- Su, Y.-Q.; Sugiura, K.; Woo, Y.; Wigglesworth, K.; Kamdar, S.; Affourtit, J.; Eppig, J.J. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 2007, 302, 104–117. [Google Scholar] [CrossRef]
- Filho, F.L.T.; Baracat, E.C.; Lee, T.H.; Suh, C.S.; Matsui, M.; Chang, R.J.; Shimasaki, S.; Erickson, G.F. Aberrant Expression of Growth Differentiation Factor-9 in Oocytes of Women with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2002, 87, 1337–1344. [Google Scholar] [CrossRef]
- Hussein, T.S.; Thompson, J.; Gilchrist, R. Oocyte-secreted factors enhance oocyte developmental competence. Dev. Biol. 2006, 296, 514–521. [Google Scholar] [CrossRef]
- Persani, L.; Rossetti, R.; Di Pasquale, E.; Cacciatore, C.; Fabre, S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum. Reprod. Updat. 2014, 20, 869–883. [Google Scholar] [CrossRef]
- Tong, B.; Wang, J.; Cheng, Z.; Liu, J.; Wu, Y.; Li, Y.; Bai, C.; Zhao, S.; Yu, H.; Li, G. Novel Variants in GDF9 Gene Affect Promoter Activity and Litter Size in Mongolia Sheep. Genes 2020, 11, 375. [Google Scholar] [CrossRef]
- Yang, J.-L.; Zhang, C.-P.; Li, L.; Huang, L.; Ji, S.-Y.; Lu, C.-L.; Fan, C.-H.; Cai, H.; Ren, Y.; Hu, Z.-Y.; et al. Testosterone Induces Redistribution of Forkhead Box-3a and Down-Regulation of Growth and Differentiation Factor 9 Messenger Ribonucleic Acid Expression at Early Stage of Mouse Folliculogenesis. Endocrinology 2010, 151, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li-Ling, J.; Xiong, D.; Wei, J.; Zhong, T.; Tan, H. Age-related decline in the expression of GDF9 and BMP15 genes in follicle fluid and granulosa cells derived from poor ovarian responders. J. Ovarian Res. 2021, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Qin, Z.; Liu, S.; Nong, S.; Ma, Q.; Chen, B.; Liu, M.; Pan, T.; Liao, D.J. Cloning of Porcine Pituitary Tumor Transforming Gene 1 and Its Expression in Porcine Oocytes and Embryos. PLoS ONE 2016, 11, e0153189. [Google Scholar] [CrossRef]
- Mei, S.; Chen, P.; Lee, C.-L.; Zhao, W.; Wang, Y.; Lam, K.K.W.; Ho, P.-C.; Yeung, W.S.B.; Fang, C.; Chiu, P.C.N. The role of galectin-3 in spermatozoa-zona pellucida binding and its association with fertilization in vitro. Mol. Hum. Reprod. 2019, 25, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.K.; Somashekar, L.; Selvaraju, S.; Jamuna, K.V.; Parthipan, S.; Binsila, B.K.; Prasad, R.V.; Ravindra, J.P. Immuno-histological mapping and functional association of seminal proteins in testis and excurrent ducts with sperm function in buffalo. Reprod. Domest. Anim. 2020, 55, 998–1010. [Google Scholar] [CrossRef]
- Hanaue, M.; Miwa, N.; Uebi, T.; Fukuda, Y.; Katagiri, Y.; Takamatsu, K. Characterization of S100A11, a suppressive factor of fertilization, in the mouse female reproductive tract. Mol. Reprod. Dev. 2011, 78, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Mamo, S.; Carter, F.; Lonergan, P.; Leal, C.L.; Al Naib, A.; Mcgettigan, P.; Mehta, J.P.; Evans, A.C.; Fair, T. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: Potential molecular markers of oocyte maturation. BMC Genom. 2011, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Webb, R.J.; Tinworth, L.; Thomas, G.M.; Zaccolo, M.; Carroll, J. Developmentally acquired PKA localisation in mouse oocytes and embryos. Dev. Biol. 2008, 317, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Turner, K.; Saunders, P.; Verhoeven, G.; De Gendt, K.; Atanassova, N.; Sharpe, R. Androgen Regulation of Stage-Dependent Cyclin D2 Expression in Sertoli Cells Suggests a Role in Modulating Androgen Action on Spermatogenesis1. Biol. Reprod. 2005, 72, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Skogestad, J.; Aronsen, J.M.; Tovsrud, N.; Wanichawan, P.; Hougen, K.; Stokke, M.K.; Carlson, C.R.; Sjaastad, I.; Sejersted, O.M.; Swift, F. Coupling of the Na+/K+-ATPase to Ankyrin B controls Na+/Ca2+ exchanger activity in cardiomyocytes. Cardiovasc. Res. 2020, 116, 78–90. [Google Scholar] [CrossRef]
- Tsai, M.F.; Phillips, C.B.; Ranaghan, M.; Tsai, C.W.; Wu, Y.; Willliams, C.; Miller, C. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. Elife 2016, 5, e15545. [Google Scholar] [CrossRef] [PubMed]
- Krisher, R.L. The effect of oocyte quality on development. J. Anim. Sci. 2004, 82 (Suppl. S13), E14–E23. [Google Scholar]
- Lewis, N.; Hinrichs, K.; Leese, H.J.; Argo, C.M.; Brison, D.R.; Sturmey, R. Energy metabolism of the equine cumulus oocyte complex during in vitro maturation. Sci. Rep. 2020, 10, 3493. [Google Scholar] [CrossRef]
- Steeves, T.; Gardner, D. Metabolism of glucose, pyruvate, and glutamine during the maturation of oocytes derived from pre-pubertal and adult cows. Mol. Reprod. Dev. 1999, 54, 92–101. [Google Scholar] [CrossRef]
- Li, L.; Zhu, S.; Shu, W.; Guo, Y.; Guan, Y.; Zeng, J.; Wang, H.; Han, L.; Zhang, J.; Liu, X.; et al. Characterization of Metabolic Patterns in Mouse Oocytes during Meiotic Maturation. Mol. Cell 2020, 80, 525–540.e9. [Google Scholar] [CrossRef] [PubMed]
- Markowska, A.I.; Jefferies, K.C.; Panjwani, N. Galectin-3 Protein Modulates Cell Surface Expression and Activation of Vascular Endothelial Growth Factor Receptor 2 in Human Endothelial Cells. J. Biol. Chem. 2011, 286, 29913–29921. [Google Scholar] [CrossRef] [PubMed]
- Priglinger, C.S.; Obermann, J.; Szober, C.M.; Merl-Pham, J.; Ohmayer, U.; Behler, J.; Gruhn, F.; Kreutzer, T.C.; Wertheimer, C.; Geerlof, A.; et al. Epithelial-to-Mesenchymal Transition of RPE Cells In Vitro Confers Increased beta1,6-N-Glycosylation and Increased Susceptibility to Galectin-3 Binding. PLoS ONE 2016, 11, e0146887. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; You, L.; Tian, Y.; Guo, J.; Fang, X.; Zhou, C.; Shi, L.; Su, Y. DPAGT1-Mediated ProteinN-Glycosylation Is Indispensable for Oocyte and Follicle Development in Mice. Adv. Sci. 2020, 7, 2000531. [Google Scholar] [CrossRef] [PubMed]
- Tanghe, S.; Van Soom, A.; Duchateau, L.; De Kruif, A. Inhibition of bovine sperm-oocyte fusion by the p-aminophenyl derivative of D-mannose. Mol. Reprod. Dev. 2004, 67, 224–232. [Google Scholar] [CrossRef] [PubMed]
| Repetitions | No. of Ovaries Per Experiment | No. of Oocytes Recovered | No. of Oocytes Per Ovaries |
|---|---|---|---|
| 1 | 48 | 361 | 7.52 |
| 2 | 45 | 291 | 6.47 |
| 3 | 48 | 365 | 7.60 |
| 4 | 55 | 445 | 8.09 |
| 5 | 33 | 230 | 6.96 |
| Total | 229 | 1692 | 7.33 ± 0.28 * |
| Oocytes Stage | No. of Oocytes (%) |
|---|---|
| GV | 1018 (63.68 ± 2.68 c) |
| MI | 150 (9.26 ± 1.18 a) |
| MII | 423 (27.08 ± 3.23 b) |
| GV Group (n = 3) | MII Group (n = 3) | ||||
|---|---|---|---|---|---|
| Gene Name | Gene Description | Average FPKM | Gene Name | Gene Description | Average FPKM |
| DPPA3 | Developmental Pluripotency-Associated Protein 3 | 20,656.8 | DPPA3 | Developmental Pluripotency-Associated Protein 3 | 20,223.6 |
| PTTG1 | Pituitary Tumor-Transforming 1 | 19,649.5 | PTTG1 | Pituitary Tumor-Transforming 1 | 19,585.9 |
| LOC106824246 | 10,616.3 | NewGene_15562 | 7462.3 | ||
| GDF9 | Growth Differentiation Factor 9 | 6493.5 | BTG4 | BTG Anti-Proliferation Factor 4 | 5170.7 |
| UBB | Ubiquitin B | 6060.0 | LOC106824246 | 5137.5 | |
| AGR2 | Anterior Gradient 2, Protein Disulphide Isomerase Family Member | 5435.3 | RASL11A | RAS Like Family 11 Member A | 4670.0 |
| NewGene_15562 | 4559.9 | KPNA7 | Karyopherin Subunit α 7 | 4645.6 | |
| ZP3 | Zona Pellucida Glycoprotein 3 | 4516.6 | CKS1B | CDC28 Protein Kinase Regulatory Subunit 1B | 4343.5 |
| CKS1B | CDC28 Protein Kinase Regulatory Subunit 1B | 4381.1 | NewGene_15553 | 3948.4 | |
| RNF34 | Ring Finger Protein 34 | 3478.1 | RNF34 | Ring Finger Protein 34 | 3662.4 |
| BTG4 | BTG Anti-Proliferation Factor 4 | 3358.6 | UBB | Ubiquitin B | 3030.3 |
| ZAR1L | Zygote Arrest 1 Like | 3323.5 | ZP3 | Zona Pellucida Glycoprotein 3 | 2997.1 |
| ARG2 | Arginase 2 | 3278.5 | WEE2 | WEE2 Oocyte Meiosis Inhibiting Kinase | 2327.4 |
| KPNA7 | Karyopherin Subunit α 7 | 3226.7 | LOC106829384 | 2294.0 | |
| LOC106840558 | 2823.5 | CNBP | CCHC-Type Zinc Finger Nucleic Acid Binding Protein | 2293.9 | |
| NewGene_26555 | 2684.6 | CCNB1 | Cyclin B1 | 2198.8 | |
| LOC106844365 | 2541.2 | CENPE | Centromere Protein E | 2163.2 | |
| BMP15 | Bone Morphogenetic Protein 15 | 2472.0 | GDF9 | Growth Differentiation Factor 9 | 2068.7 |
| CALM2 | Calmodulin 2 | 1989.0 | HNRNPA1 | Heterogeneous Nuclear Ribonucleoprotein A1 | 2042.1 |
| RFC3 | Replication Factor C Subunit 3 | 1935.0 | LOC106844365 | 1945.9 | |
| Gene Symbol | Gene Name | Fold Change | p-Value |
|---|---|---|---|
| LGALS3 | Galectin-3 | 7.94 | 1.42 × 10−20 |
| TFPI2 | Tissue Factor Pathway Inhibitor 2 | 7.42 | 7.93 × 10−45 |
| ANXA1 | Annexin A1 | 7.35 | 1.54 × 10−23 |
| S100A11 | S100 Calcium Binding Protein A11 | 5.99 | 2.95 × 10−15 |
| AKAP2 | A-Kinase Anchoring Protein 2 | 5.86 | 4.08 × 10−34 |
| ACTG2 | Actin γ 2, Smooth Muscle | 5.57 | 1.41 × 10−13 |
| INHBA | Inhibin Subunit β A | 4.57 | 3.00 × 10−15 |
| LOC106825888 | 4.56 | 5.77 × 10−14 | |
| VIM | Vimentin | 3.91 | 8.33 × 10−5 |
| CCND2 | Cyclin D2 | 3.87 | 8.85 × 10−17 |
| ATP5J2 | ATP Synthase Membrane Subunit F | −7.52 | 3.40 × 10−23 |
| LOC106837922 | −7.32 | 1.52 × 10−16 | |
| FKBP2 | FKBP Prolyl Isomerase 2 | −6.37 | 3.40 × 10−12 |
| SMDT1 | Single-Pass Membrane Protein With Aspartate Rich Tail 1 | −6.33 | 7.50 × 10−12 |
| TUBB4B | Tubulin β 4B Class IVb | −6.15 | 8.38 × 10−125 |
| LOC106847707 | −6.13 | 4.16 × 10−11 | |
| LOC106823315 | −6.08 | 3.44 × 10−55 | |
| TMEM11 | Transmembrane Protein 11 | −6.06 | 4.10 × 10−25 |
| NDUFA8 | NADH: Ubiquinone Oxidoreductase Subunit A8 | −6.06 | 4.10 × 10−25 |
| LOC106836642 | −5.95 | 4.85 × 10−85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Song, X.; Yin, S.; Yan, J.; Lv, P.; Shan, H.; Cui, K.; Liu, H.; Liu, Q. Single-Cell RNA-Seq Revealed the Gene Expression Pattern during the In Vitro Maturation of Donkey Oocytes. Genes 2021, 12, 1640. https://doi.org/10.3390/genes12101640
Li Z, Song X, Yin S, Yan J, Lv P, Shan H, Cui K, Liu H, Liu Q. Single-Cell RNA-Seq Revealed the Gene Expression Pattern during the In Vitro Maturation of Donkey Oocytes. Genes. 2021; 12(10):1640. https://doi.org/10.3390/genes12101640
Chicago/Turabian StyleLi, Zhipeng, Xinhui Song, Shan Yin, Jiageng Yan, Peiru Lv, Huiquan Shan, Kuiqing Cui, Hongbo Liu, and Qingyou Liu. 2021. "Single-Cell RNA-Seq Revealed the Gene Expression Pattern during the In Vitro Maturation of Donkey Oocytes" Genes 12, no. 10: 1640. https://doi.org/10.3390/genes12101640
APA StyleLi, Z., Song, X., Yin, S., Yan, J., Lv, P., Shan, H., Cui, K., Liu, H., & Liu, Q. (2021). Single-Cell RNA-Seq Revealed the Gene Expression Pattern during the In Vitro Maturation of Donkey Oocytes. Genes, 12(10), 1640. https://doi.org/10.3390/genes12101640








