Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions
Abstract
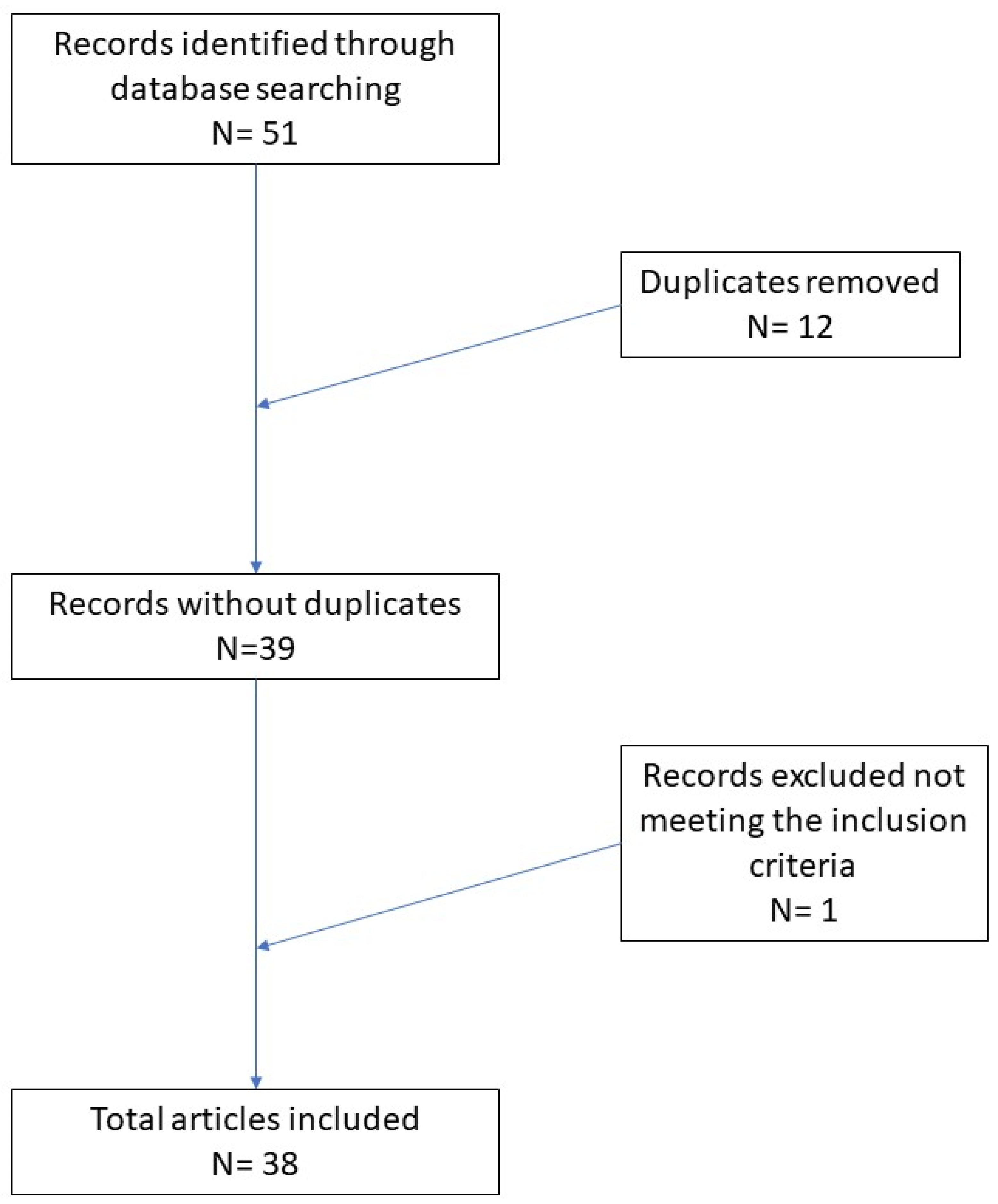
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Strategies for Minimization of Sequence Artifacts from FFPE DNA
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aly, S.; Sabri, D.M. Next generation sequencing (NGS): A golden tool in forensic toolkit. Arch. Forensic Med. Criminol. 2015, 4, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. J. Biomed. Biotechnol. 2012, 2012, 896451. [Google Scholar] [CrossRef]
- Lavacchi, D.; Roviello, G.; D’Angelo, A. Tumor-Agnostic Treatment for Cancer: When How is Better than Where. Clin. Drug Investig. 2020, 40, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oxford Centre for Evidence-Based Medicine 2011 Levels of Evidence. Available online: http://www.cebm.net/wp-content/uploads/2014/06/CEBM-Levels-of-Evidence-2.1.pdf (accessed on 31 August 2021).
- Loomis, T.A. Formaldehyde toxicity. Arch. Pathol. Lab. Med. 1979, 103, 321–324. [Google Scholar]
- Farooqui, M.Y. Formaldehyde. J. Appl Toxicol. 1983, 3, 264–265. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.A.; Heng, X.; Sundqvist, K.; Liu, Y.; Atzori, L.; Elfwing, A.; Arvidson, K.; Grafström, R.C. Toxicity of formaldehyde to human oral fibroblasts and epithelial cells: Influences of culture conditions and role of thiol status. J. Dent. Res. 1998, 77, 1896–1903. [Google Scholar] [CrossRef]
- Kwak, K.; Paek, D.; Park, J. Occupational exposure to formaldehyde and risk of lung cancer: A systematic review and meta-analysis. Am. J. Ind. Med. 2020, 63, 312–327. [Google Scholar] [CrossRef]
- Plénat, F.; Antunes, L.; Haller, T.; Piet-Ounnoughene, M.; Klein-Monhoven, N.; Champigneulle, J.; Chenal, P.; Bland, V.; Garcia-Pimenta, F.; Labouyrie, E. Formaldehyde fixation in the third millennium. Ann. de Pathol. 2001, 21, 29–47. [Google Scholar]
- Xuan, J.; Yu, Y.; Qing, T.; Guo, L.; Shi, L. Next-generation sequencing in the clinic: Promises and challenges. Cancer Lett. 2012, 340, 284–295. [Google Scholar] [CrossRef] [Green Version]
- Do, H.; Dobrovic, A. Sequence Artifacts in DNA from Formalin-Fixed Tissues: Causes and Strategies for Minimization. Clin. Chem. 2015, 61, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Schweiger, M.R.; Kerick, M.; Timmermann, B.; Albrecht, M.W.; Borodina, T.; Parkhomchuk, D.; Zatloukal, K.; Lehrach, H. Genome-Wide Massively Parallel Sequencing of Formaldehyde Fixed-Paraffin Embedded (FFPE) Tumor Tissues for Copy-Number- and Mutation-Analysis. PLoS ONE 2009, 4, e5548. [Google Scholar] [CrossRef] [Green Version]
- Feldman, M.Y. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog. Nucleic Acid Res. Mol. Biol. 1973, 13, 1–49. [Google Scholar] [PubMed]
- Fraenkel-Conrat, H.; Olcott, H.S. The Reaction of Formaldehyde with Proteins. V. Cross-linking between Amino and Primary Amide or Guanidyl Groups. J. Am. Chem. Soc. 1948, 70, 2673–2684. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; von Hippel, P.H. Formaldehyde as a probe of DNA structure. II. Reaction with endocyclic imino groups of DNA bases. Biochemistry 1975, 14, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Ludyga, N.; Grünwald, B.; Azimzadeh, O.; Englert, S.; Höfler, H.; Tapio, S.; Aubele, M. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch. 2012, 460, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Pontén, F.; Moberg, C.; Söderkvist, P.; Uhlen, M.; Pontén, J.; Sitbon, G.; Lundeberg, J. A High Frequency of Sequence Alterations Is Due to Formalin Fixation of Archival Specimens. Am. J. Pathol. 1999, 155, 1467–1471. [Google Scholar] [CrossRef] [Green Version]
- Quach, N.; Goodman, M.F.; Shibata, D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 2004, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.I.G.; Floor, K.; Rijmen, F.; Grünberg, K.; A Rodriguez, J.; Giaccone, G. EGFR and K-ras Mutation Analysis in Non-Small Cell Lung Cancer: Comparison of Paraffin Embedded versus Frozen Specimens. Cell Oncol. 2007, 29, 257–264. [Google Scholar] [CrossRef]
- Murray, S.; Dahabreh, I.J.; Linardou, H.; Manoloukos, M.; Bafaloukos, D.; Kosmidis, P. Somatic Mutations of the Tyrosine Kinase Domain of Epidermal Growth Factor Receptor and Tyrosine Kinase Inhibitor Response to TKIs in Non-small Cell Lung Cancer: An Analytical Database. J. Thorac. Oncol. 2008, 3, 832–839. [Google Scholar] [CrossRef]
- Tsao, M.; Sakurada, A.; Cutz, J.-C.; Zhu, C.; Kamel-Reid, S.; Squire, J.; Lorimer, I.; Zhang, T.; Liu, N.; Daneshmand, M.; et al. Erlotinib in Lung Cancer—Molecular and Clinical Predictors of Outcome. N. Engl. J. Med. 2005, 353, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Didelot, A.; Kotsopoulos, S.K.; Lupo, A.; Pekin, D.; Li, X.; Atochin, I.; Srinivasan, P.; Zhong, Q.; Olson, J.; Link, D.R.; et al. Multiplex Picoliter-Droplet Digital PCR for Quantitative Assessment of DNA Integrity in Clinical Samples. Clin. Chem. 2013, 59, 815–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Ohsumi, S.; Makino, K. Mechanistic studies on depurination and apurinic site chain breakage in oligodeoxyribonucleotides. Nucleic Acids Res. 1994, 22, 4997–5003. [Google Scholar] [CrossRef]
- Zsikla, V.; Baumann, M.; Cathomas, G. Effect of buffered formalin on amplification of DNA from paraffin wax embedded small biopsies using real-time PCR. J. Clin. Pathol. 2004, 57, 654–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ofner, R.; Ritter, C.; Ugurel, S.; Cerroni, L.; Stiller, M.; Bogenrieder, T.; Solca, F.; Schrama, D.; Becker, J.C. Non-reproducible sequence artifacts in FFPE tissue: An experience report. J. Cancer Res. Clin Oncol. 2017, 143, 1199–1207. [Google Scholar] [CrossRef]
- Högnäs, G.; Kivinummi, K.; Kallio, H.M.L.; Hieta, R.; Ruusuvuori, P.; Koskenalho, A.; Kesseli, J.; Tammela, T.L.J.; Riikonen, J.; Ilvesaro, J.; et al. Feasibility of Prostate PAXgene Fixation for Molecular Research and Diagnostic Surgical Pathology: Comparison of Matched Fresh Frozen, FFPE, and PFPE Tissues. Am. J. Surg. Pathol. 2018, 42, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Báez, W.; García-Latorre, E.A.; Maldonado-Martínez, H.A.; Coronado-Martínez, I.; Flores-García, L.; Taja-Chayeb, L. Impact of fixation artifacts and threshold selection on high resolution melting analysis for KRAS mutation screening. Mol. Cell. Probes 2017, 35, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.; Chen, L.; Houghton, J.; Kemppainen, J.; Marko, A.C.; Zeigler, R.; Latham, G.J. Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Med. 2013, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Jackson, V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell 1978, 15, 945–954. [Google Scholar] [CrossRef]
- Shi, S.-R.; Cote, R.J.; Wu, L.; Liu, C.; Datar, R.; Shi, Y.; Liu, D.; Lim, H.; Taylor, C.R. DNA Extraction from Archival Formalin-fixed, Paraffin-embedded Tissue Sections Based on the Antigen Retrieval Principle: Heating Under the Influence of pH. J. Histochem. Cytochem. 2002, 50, 1005–1011. [Google Scholar] [CrossRef] [Green Version]
- Campos, P.F.; Gilbert, T.M.P. DNA Extraction from Formalin-Fixed Material. Methods Mol. Biol. 2011, 840, 81–85. [Google Scholar] [CrossRef]
- Wu, L.; Patten, N.; Yamashiro, C.T.; Chui, B. Extraction and Amplification of DNA From Formalin-Fixed, Paraffin-Embedded Tissues. Appl. Immunohistochem. Mol. Morphol. 2002, 10, 269–274. [Google Scholar] [CrossRef]
- Do, H.; Dobrovic, A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil-DNA glycosylase. Oncotarget 2012, 3, 546–558. [Google Scholar] [CrossRef] [Green Version]
- Do, H.; Wong, S.Q.; Li, J.; Dobrovic, A. Reducing Sequence Artifacts in Amplicon-Based Massively Parallel Sequencing of Formalin-Fixed Paraffin-Embedded DNA by Enzymatic Depletion of Uracil-Containing Templates. Clin. Chem. 2013, 59, 1376–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonough, S.J.; Bhagwate, A.; Sun, Z.; Wang, C.; Zschunke, M.; Gorman, J.A.; Kopp, K.J.; Cunningham, J.M. Use of FFPE-derived DNA in next generation sequencing: DNA extraction methods. PLoS ONE 2019, 14, e0211400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagwate, A.V.; Liu, Y.; Winham, S.J.; McDonough, S.J.; Stallings-Mann, M.L.; Heinzen, E.P.; Davila, J.I.; Vierkant, R.A.; Hoskin, T.L.; Frost, M.; et al. Bioinformatics and DNA-extraction strategies to reliably detect genetic variants from FFPE breast tissue samples. BMC Genom. 2019, 20, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kresse, S.H.; Namløs, H.M.; Lorenz, S.; Berner, J.-M.; Myklebost, O.; Bjerkehagen, B.; Meza-Zepeda, L.A. Evaluation of commercial DNA and RNA extraction methods for high-throughput sequencing of FFPE samples. PLoS ONE 2018, 13, e0197456. [Google Scholar] [CrossRef]
- Mullegama, S.V.; Alberti, M.O.; Au, C.; Li, Y.; Toy, T.; Tomasian, V.; Xian, R.R. Nucleic Acid Extraction from Human Biological Samples. Methods Mol. Biol. 2018, 359–383. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cazzato, G.; Caporusso, C.; Arezzo, F.; Cimmino, A.; Colagrande, A.; Loizzi, V.; Cormio, G.; Lettini, T.; Maiorano, E.; Scarcella, V.S.; et al. Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions. Genes 2021, 12, 1472. https://doi.org/10.3390/genes12101472
Cazzato G, Caporusso C, Arezzo F, Cimmino A, Colagrande A, Loizzi V, Cormio G, Lettini T, Maiorano E, Scarcella VS, et al. Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions. Genes. 2021; 12(10):1472. https://doi.org/10.3390/genes12101472
Chicago/Turabian StyleCazzato, Gerardo, Concetta Caporusso, Francesca Arezzo, Antonietta Cimmino, Anna Colagrande, Vera Loizzi, Gennaro Cormio, Teresa Lettini, Eugenio Maiorano, Vincenza Sara Scarcella, and et al. 2021. "Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions" Genes 12, no. 10: 1472. https://doi.org/10.3390/genes12101472
APA StyleCazzato, G., Caporusso, C., Arezzo, F., Cimmino, A., Colagrande, A., Loizzi, V., Cormio, G., Lettini, T., Maiorano, E., Scarcella, V. S., Tarantino, P., Marrone, M., Stellacci, A., Parente, P., Romita, P., De Marco, A., Venerito, V., Foti, C., Ingravallo, G., ... Resta, L. (2021). Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions. Genes, 12(10), 1472. https://doi.org/10.3390/genes12101472












