DEAD-Box RNA Helicases and Genome Stability
Abstract
1. Introduction
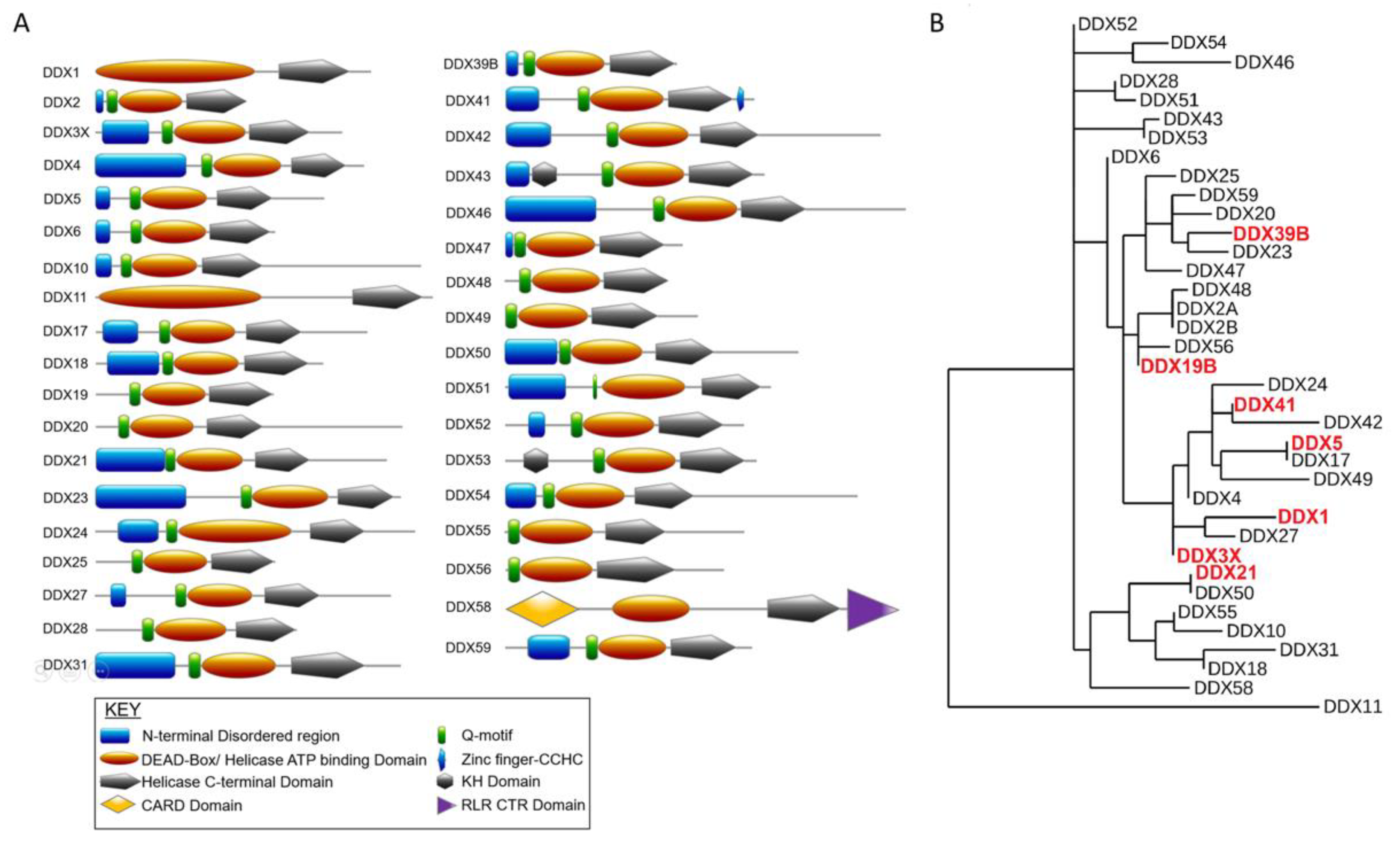
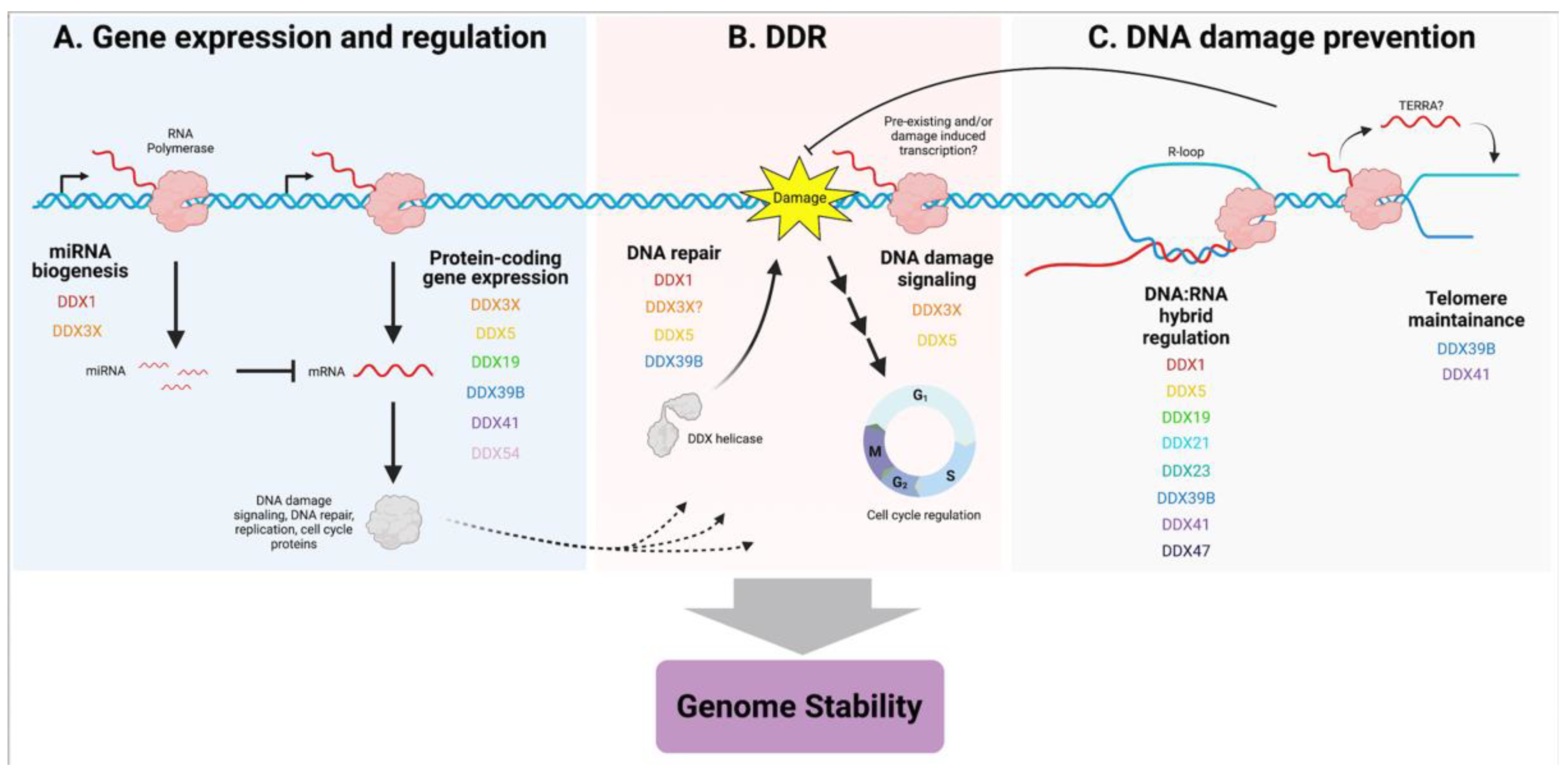
2. DEAD-Box RNA Helicase Studies
2.1. DDX1
2.2. DDX3X
2.3. DDX5
2.4. DDX19
2.5. DDX21
2.6. DDX39B
2.7. DDX41
3. Implications for Cancer
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AID | activation-induced deaminase |
| AML | acute myelogenous leukemia |
| ATM | ataxia-telangiectasia mutated |
| ATR | ataxia telangiectasia and Rad3 related |
| BLM | Bloom syndrome protein |
| CtIP | C terminal-binding protein—interacting protein |
| DDR | DNA damage response |
| DDX | DEAD-box helicase |
| DRIP | DNA:RNA hybrid immunoprecipitation |
| G4 | G-quadruplex |
| Ig | Immunoglobulin |
| HDR | homology directed repair |
| PARP1 | poly (ADP-ribose) polymerase 1 |
| rRNA | ribosomal RNA |
| RPA | replication protein A |
| TERRA | telomeric repeat-containing RNA |
References
- Rocak, S.; Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef]
- Bleichert, F.; Baserga, S.J. The Long Unwinding Road of RNA Helicases. Mol. Cell 2007, 27, 339–352. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Fuller-Pace, F.V. DEAD box RNA helicase functions in cancer. RNA Biol. 2013, 10, 121–132. [Google Scholar] [CrossRef]
- Paine, I.; Posey, J.; Grochowski, C.M.; Jhangiani, S.N.; Rosenheck, S.; Kleyner, R.; Marmorale, T.; Yoon, M.; Wang, K.; Robison, R.; et al. Paralog Studies Augment Gene Discovery: DDX and DHX Genes. Am. J. Hum. Genet. 2019, 105, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Lambert, S.; Carreira, A.; Amor-Guéret, M.; Vagner, S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem. Sci. 2014, 39, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Hieter, P.; Stirling, P.C. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014, 30, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Vagner, S. DNA-Damage Response RNA-Binding Proteins (DDRBPs): Perspectives from a New Class of Proteins and Their RNA Targets. J. Mol. Biol. 2017, 429, 3139–3145. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Hulo, N.; Bairoch, A.; Bulliard, V.; Cerutti, L.; Cuche, B.A.; de Castro, E.; Lachaize, C.; Langendijk-Genevaux, P.; Sigrist, C.J.A. The 20 years of PROSITE. Nucleic Acids Res. 2007, 36, D245–D249. [Google Scholar] [CrossRef]
- Li, L.; Monckton, E.A.; Godbout, R. A Role for DEAD Box 1 at DNA Double-Strand Breaks. Mol. Cell. Biol. 2008, 28, 6413–6425. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Germain, D.R.; Poon, H.-Y.; Hildebrandt, M.R.; Monckton, E.A.; McDonald, D.; Hendzel, M.; Godbout, R. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Mol. Cell. Biol. 2016, 36, 2794–2810. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Poon, H.-Y.; Hildebrandt, M.; Monckton, E.A.; Germain, D.R.; Fahlman, R.P.; Godbout, R. Role for RIF1-interacting partner DDX1 in BLM recruitment to DNA double-strand breaks. DNA Repair 2017, 55, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chedin, F.; Hsieh, C.-L.; Wilson, T.E.; Lieber, M. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, C.R.; Dhir, S.; Dhir, A.; Moghaddam, A.E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 2018, 70, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Liu, Y.; Wan, G.; Choi, H.J.; Zhao, L.; Ivan, C.; He, X.; Sood, A.K.; Zhang, X.; Lu, X. The RNA-Binding Protein DDX1 Promotes Primary MicroRNA Maturation and Inhibits Ovarian Tumor Progression. Cell Rep. 2014, 8, 1447–1460. [Google Scholar] [CrossRef]
- Wang, Y.; Taniguchi, T. MicroRNAs and DNA damage response. Cell Cycle 2012, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Bucci, J.; Chang, L.; Malouf, D.; Graham, P.; Li, Y. Targeting MicroRNAs in Prostate Cancer Radiotherapy. Theranostics 2017, 7, 3243–3259. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 1–20. [Google Scholar] [CrossRef]
- Wang, T.; Birsoy, K.; Hughes, N.W.; Krupczak, K.M.; Post, Y.; Wei, J.J.; Lander, E.S.; Sabatini, D.M. Identification and characterization of essential genes in the human genome. Science 2015, 350, 1096–1101. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chan, C.-H.; Chen, C.-M.; Tsai, Y.-S.; Tsai, T.-Y.; Lee, Y.-H.W.; You, L.-R. Targeted inactivation of murineDdx3x: Essential roles of Ddx3xin placentation and embryogenesis. Hum. Mol. Genet. 2016, 25, 2905–2922. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bol, G.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX 3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015, 7, 648–669. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Putnam, A.A.; Jankowsky, E. Biochemical Differences and Similarities between the DEAD-Box Helicase Orthologs DDX3X and Ded1p. J. Mol. Biol. 2017, 429, 3730–3742. [Google Scholar] [CrossRef]
- Cristini, A.; Groh, M.; Kristiansen, M.S.; Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018, 23, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Riva, V.; Garbelli, A.; Casiraghi, F.; Arena, F.; Trivisani, C.I.; Gagliardi, A.; Bini, L.; Schroeder, M.; Maffia, A.; Sabbioneda, S.; et al. Novel alternative ribonucleotide excision repair pathways in human cells by DDX3X and specialized DNA polymerases. Nucleic Acids Res. 2020, 48, 11551–11565. [Google Scholar] [CrossRef]
- Chan, C.-H.; Chen, C.-M.; Lee, Y.-H.W.; You, L.-R. DNA Damage, Liver Injury, and Tumorigenesis: Consequences of DDX3X Loss. Mol. Cancer Res. 2019, 17, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Y.; Zhao, Y.; He, Y. DDX3X promotes the biogenesis of a subset of miRNAs and the potential roles they played in cancer development. Sci. Rep. 2016, 6, 32739. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhou, T.; Jonasch, E.; Jope, R.S. DDX3 regulates DNA damage-induced apoptosis and p53 stabilization. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1833, 1489–1497. [Google Scholar] [CrossRef]
- Chen, W.-J.; Wang, W.-T.; Tsai, T.-Y.; Li, H.-K.; Lee, Y.-H.W. DDX3 localizes to the centrosome and prevents multipolar mitosis by epigenetically and translationally modulating p53 expression. Sci. Rep. 2017, 7, 9411. [Google Scholar] [CrossRef] [PubMed]
- Cargill, M.J.; Morales, A.; Ravishankar, S.; Warren, E.H. RNA helicase, DDX3X, is actively recruited to sites of DNA damage in live cells. DNA Repair 2021, 103, 103137. [Google Scholar] [CrossRef] [PubMed]
- Kai, M. Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 2016, 17, 310. [Google Scholar] [CrossRef] [PubMed]
- Giraud, G.; Terrone, S.; Bourgeois, S.T.A.C.F. Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep. 2018, 51, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, A.; Luo, W.; Krasnitz, A.; Hicks, J.; Powers, R.S.; Stillman, B. DDX5 Regulates DNA Replication and Is Required for Cell Proliferation in a Subset of Breast Cancer Cells. Cancer Discov. 2012, 2, 812–825. [Google Scholar] [CrossRef] [PubMed]
- Bates, G.J.; Nicol, S.M.; Wilson, B.J.; Jacobs, A.-M.F.; Bourdon, J.-C.; Wardrop, J.; Gregory, D.; Lane, D.; Perkins, N.D.; Fuller-Pace, F.V. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005, 24, 543–553. [Google Scholar] [CrossRef]
- Nicol, S.M.; Bray, S.E.; Black, H.D.; Lorimore, S.A.; Wright, E.G.; Lane, D.; Meek, D.W.; Coates, P.; Fuller-Pace, F.V. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 2013, 32, 3461–3469. [Google Scholar] [CrossRef]
- Yu, Z.; Mersaoui, S.Y.; Guitton-Sert, L.; Coulombe, Y.; Song, J.; Masson, J.-Y.; Richard, S. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer 2020, 2, 2. [Google Scholar] [CrossRef]
- Mersaoui, S.Y.; Yu, Z.; Coulombe, Y.; Karam, M.; Busatto, F.F.; Masson, J.; Richard, S. Arginine methylation of the DDX 5 helicase RGG / RG motif by PRMT 5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019, 38, e100986. [Google Scholar] [CrossRef]
- Villarreal, O.D.; Mersaoui, S.Y.; Yu, Z.; Masson, J.-Y.; Richard, S. Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution. Life Sci. Alliance 2020, 3, e202000762. [Google Scholar] [CrossRef]
- Abbasi, S.; Schild-Poulter, C. Mapping the Ku Interactome Using Proximity-Dependent Biotin Identification in Human Cells. J. Proteome Res. 2018, 18, 1064–1077. [Google Scholar] [CrossRef]
- Sessa, G.; Gómez-González, B.; Silva, S.; Pérez-Calero, C.; Beaurepere, R.; Barroso, S.; Martineau, S.; Martin, C.; Ehlén, Å.; Martínez, J.S.; et al. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repair. EMBO J. 2021, 40, e106018. [Google Scholar] [CrossRef]
- Adamson, B.; Smogorzewska, A.; Sigoillot, F.D.; King, R.; Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012, 14, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Fortuna, T.R.; Kour, S.; Anderson, E.N.; Ward, C.; Rajasundaram, D.; Donnelly, C.J.; Hermann, A.; Wyne, H.; Shewmaker, F.; Pandey, U.B. DDX17 is involved in DNA damage repair and modifies FUS toxicity in an RGG-domain dependent manner. Acta Neuropathol. 2021, 142, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Hodroj, D.; Recolin, B.; Serhal, K.; Martinez, S.; Tsanov, N.; Merhi, R.A.; Maiorano, D. An ATR -dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017, 36, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Abe, M.; Itaya, A.; Tomida, J.; Ishiai, M.; Takaori-Kondo, A.; Taoka, M.; Isobe, T.; Takata, M. FANCD 2 protects genome stability by recruiting RNA processing enzymes to resolve R-loops during mild replication stress. FEBS J. 2018, 286, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Yamanaka, Y.; Shigemoto, M.; Kitadani, Y.; Kobayashi, Y.; Kambe, T.; Nagao, M.; Kobayashi, I.; Okumura, K.; Masuda, S. Depletion of mRNA export regulator DBP5/DDX19, GLE1 or IPPK that is a key enzyme for the production of IP6, resulting in differentially altered cytoplasmic mRNA expression and specific cell defect. PLoS ONE 2018, 13, e0197165. [Google Scholar] [CrossRef]
- Zhang, Y.; Baysac, K.C.; Yee, L.-F.; Saporita, A.J.; Weber, J.D. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014, 16, 449. [Google Scholar] [CrossRef]
- Tanaka, A.; Wang, J.Y.; Shia, J.; Zhou, Y.; Ogawa, M.; Hendrickson, R.C.; Klimstra, D.S.; Roehrl, M.H. DEAD-box RNA helicase protein DDX21 as a prognosis marker for early stage colorectal cancer with microsatellite instability. Sci. Rep. 2020, 10, 22085. [Google Scholar] [CrossRef]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Lindström, M.S.; Jurada, D.; Bursac, S.; Oršolić, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef]
- Aguilera, A.; Klein, H.L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell. Biol. 1990, 10, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-Y.; Merker, R.J.; Klein, H.L. High-Copy-Number Expression of Sub2p, a Member of the RNA Helicase Superfamily, Suppresses hpr1 -Mediated Genomic Instability. Mol. Cell. Biol. 2001, 21, 5459–5470. [Google Scholar] [CrossRef] [PubMed]
- Jimeno, S.; Rondón, A.G.; Luna, R.; Aguilera, A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002, 21, 3526–3535. [Google Scholar] [CrossRef]
- Huertas, P.; Aguilera, A. Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Saguez, C.; Gonzales, F.A.; Schmid, M.; Bøggild, A.; Latrick, C.M.; Malagon, F.; Putnam, A.; Sanderson, L.; Jankowsky, E.; Brodersen, D.E.; et al. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. RNA 2013, 19, 1363–1371. [Google Scholar] [CrossRef]
- Domínguez-Sánchez, M.S.; Barroso, S.; Gómez-González, B.; Luna, R.; Aguilera, A. Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREX. PLoS Genet. 2011, 7, e1002386. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Calero, C.; Bayona-Feliu, A.; Xue, X.; Barroso, S.I.; Muñoz, S.; González-Basallote, V.M.; Sung, P.; Aguilera, A. UAP56/DDX39B is a major cotranscriptional RNA–DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020, 34, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.H.; Chung, I.K. Requirement of DDX39 DEAD box RNA helicase for genome integrity and telomere protection. Aging Cell 2011, 10, 557–571. [Google Scholar] [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef]
- Gaillard, H.; Wellinger, R.E.; Aguilera, A. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res. 2007, 35, 3893–3906. [Google Scholar] [CrossRef]
- Xu, Z.; Li, X.; Li, H.; Nie, C.; Liu, W.; Li, S.; Liu, Z.; Wang, W.; Wang, J. Suppression of DDX39B sensitizes ovarian cancer cells to DNA-damaging chemotherapeutic agents via destabilizing BRCA1 mRNA. Oncogene 2020, 39, 7051–7062. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-G.; Kim, S.S.-Y.; Kui, L.; Voon, D.; Mauduit, M.; Bist, P.; Bi, X.; Pereira, N.A.; Liu, C.; Sukumaran, B.; et al. Bruton’s Tyrosine Kinase Phosphorylates DDX41 and Activates Its Binding of dsDNA and STING to Initiate Type 1 Interferon Response. Cell Rep. 2015, 10, 1055–1065. [Google Scholar] [CrossRef]
- Weinreb, J.T.; Gupta, V.; Sharvit, E.; Weil, R.; Bowman, T.V. Ddx41 inhibition of DNA damage signaling permits erythroid progenitor expansion in zebrafish. Haematologia 2021. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, J.T.; Ghazale, N.; Pradhan, K.; Gupta, V.; Potts, K.S.; Tricomi, B.; Daniels, N.J.; Padgett, R.A.; De Oliveira, S.; Verma, A.; et al. Excessive R-loops trigger an inflammatory cascade leading to increased HSPC production. Dev. Cell 2021, 56, 627–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.X.; Grunseich, C.; Fox, J.; Burdick, J.; Zhu, Z.; Ravazian, N.; Hafner, M.; Cheung, V.G. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018, 28, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Polprasert, C.; Schulze, I.; Sekeres, M.; Makishima, H.; Przychodzen, B.; Hosono, N.; Singh, J.; Padgett, R.; Gu, X.; Phillips, J.G.; et al. Inherited and Somatic Defects in DDX41 in Myeloid Neoplasms. Cancer Cell 2015, 27, 658–670. [Google Scholar] [CrossRef]
- Mosler, T.; Conte, F.; Mikicic, I.; Kreim, N.; Möckel, M.; Flach, J.; Luke, B.; Beli, P. R-Loop Proximity Proteomics Identifies a Role of DDX41 in Transcription-Associated Genomic Instability; Research Square Platform LLC: Durham, NC, USA, 2021. [Google Scholar] [CrossRef]
- Cardoso, S.R.; Ryan, G.; Walne, A.J.; Ellison, A.; Lowe, R.; Tummala, H.; Rio-Machin, A.; Collopy, L.; Al Seraihi, A.; Wallis, Y.; et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia 2016, 30, 2083–2086. [Google Scholar] [CrossRef]
- Seo, M.; Park, S.; Gil Nam, H.; Lee, S.-J.V. RNA helicase SACY-1 is required for longevity caused by various genetic perturbations in Caenorhabditis elegans. Cell Cycle 2016, 15, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.; Radine, C.; Reese, A.; Budach, W.; Sohn, D.; Jänicke, R.U. The DEAD-box RNA helicase DDX41 is a novel repressor of p21WAF1/CIP1 mRNA translation. J. Biol. Chem. 2017, 292, 8331–8341. [Google Scholar] [CrossRef]
- Cai, W.; Chen, Z.X.; Rane, G.; Singh, S.S.; Choo, Z.; Wang, C.; Yuan, Y.; Tan, T.Z.; Arfuso, F.; Yap, C.T.; et al. Wanted DEAD/H or Alive: Helicases Winding Up in Cancers. J. Natl. Cancer Inst. 2017, 109, 278. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Sridhara, S.C.; Carvalho, S.; Grosso, A.R.; Páez, L.M.G.; Carmo-Fonseca, M.; de Almeida, S.F. Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation. Cell Rep. 2017, 18, 334–343. [Google Scholar] [CrossRef]
- Milek, M.; Imami, K.; Mukherjee, N.; De Bortoli, F.; Zinnall, U.; Hazapis, O.; Trahan, C.; Oeffinger, M.; Heyd, F.; Ohler, U.; et al. DDX54 regulates transcriptome dynamics during DNA damage response. Genome Res. 2017, 27, 1344–1359. [Google Scholar] [CrossRef] [PubMed]
- McKay, B.C. Post-Transcriptional Regulation of DNA Damage-Responsive Gene Expression. Antioxid. Redox Signal. 2014, 20, 640–654. [Google Scholar] [CrossRef]
- Wu, D.-W.; Lee, M.-C.; Wang, J.; Chen, C.-Y.; Cheng, Y.-W.; Lee, H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/Slug/E-cadherin pathway and poor patient outcome in non-small-cell lung cancer. Oncogene 2013, 33, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Rossow, K.L.; Grande, J.P.; Janknecht, R. Involvement of RNA Helicases p68 and p72 in Colon Cancer. Cancer Res. 2007, 67, 7572–7578. [Google Scholar] [CrossRef]
- Wortham, N.C.; Ahamed, E.; Nicol, S.M.; Thomas, R.S.; Periyasamy, M.; Jiang, J.; Ochocka, A.M.; Shousha, S.; Huson, L.; E Bray, S.; et al. The DEAD-box protein p72 regulates ERα-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERα-positive breast cancer. Oncogene 2009, 28, 4053–4064. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, Z.; Liu, L.; Sun, J.; Tang, H.; Zhang, B.; Zou, Y.; Li, H. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/β-catenin pathway. Cell Death Dis. 2018, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, P.; Wang, C.; Zhang, D.; Zeng, L.; Liu, X.; Lin, J. DEAD-box RNA Helicase 39 Promotes Invasiveness and Chemoresistance of ER-positive Breast Cancer. J. Cancer 2020, 11, 1846–1858. [Google Scholar] [CrossRef]
- Lans, H.; Hoeijmakers, J.H.J.; Vermeulen, W.; Marteijn, J.A. The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 2019, 20, 766–784. [Google Scholar] [CrossRef]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Xia, T.; Konno, H.; Konno, K.; Ruiz, P.; Barber, G.N. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 2014, 5, 5166. [Google Scholar] [CrossRef] [PubMed]
- Chabanon, R.M.; Rouanne, M.; Lord, C.J.; Soria, J.-C.; Pasero, P.; Postel-Vinay, S. Targeting the DNA damage response in immuno-oncology: Developments and opportunities. Nat. Rev. Cancer 2021, 1–17. [Google Scholar] [CrossRef]
- Kouyama, Y.; Masuda, T.; Fujii, A.; Ogawa, Y.; Sato, K.; Tobo, T.; Wakiyama, H.; Yoshikawa, Y.; Noda, M.; Tsuruda, Y.; et al. Oncogenic splicing abnormalities induced by DEAD -Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 2019, 110, 3132–3144. [Google Scholar] [CrossRef] [PubMed]
- National Institute of Health. August 2013–November 2019. A Multi-Center, Dose Finding, Open Label, Phase 1 Study of RX-5902 in Subjects with Advanced or Metastatic Solid Tumors. Identifier NCT02003092. Available online: https://clinicaltrials.gov/ct2/show/record/NCT02003092 (accessed on 20 July 2021).
- Tentler, J.J.; Lang, J.; Capasso, A.; Kim, D.J.; Benaim, E.; Lee, Y.B.; Eisen, A.; Bagby, S.M.; Hartman, S.J.; Yacob, B.W.; et al. RX-5902, a novel β-catenin modulator, potentiates the efficacy of immune checkpoint inhibitors in preclinical models of triple-negative breast Cancer. BMC Cancer 2020, 20, 1063. [Google Scholar] [CrossRef]
- van Voss, M.H.; Brilliant, J.D.; Vesuna, F.; Bol, G.M.; Van Der Wall, E.; Van Diest, P.J.; Raman, V. Combination treatment using DDX3 and PARP inhibitors induces synthetic lethality in BRCA1-proficient breast cancer. Med Oncol. 2017, 34, 33. [Google Scholar] [CrossRef]
- Isabelle, M.; Moreel, X.; Gagné, J.-P.; Rouleau, M.; Ethier, C.; Gagné, P.; Hendzel, M.J.; Poirier, G.G. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Boeing, S.; Williamson, L.; Encheva, V.; Gori, I.; Saunders, R.E.; Instrell, R.; Aygün, O.; Rodriguez-Martinez, M.; Weems, J.C.; Kelly, G.; et al. Multiomic Analysis of the UV-Induced DNA Damage Response. Cell Rep. 2016, 15, 1597–1610. [Google Scholar] [CrossRef]
- Zhang, X.; Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef]
- Hussain, T.; Saha, D.; Purohit, G.; Kar, A.; Mukherjee, A.K.; Sharma, S.; Sengupta, S.; Dhapola, P.; Maji, B.; Vedagopuram, S.; et al. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci. Rep. 2017, 7, 11541. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, O.; Zatsepin, T. RNA Helicases as Shadow Modulators of Cell Cycle Progression. Int. J. Mol. Sci. 2021, 22, 2984. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Huang, J.T.J.; Hiom, K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018, 9, 4346. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Hiom, K. DHX9-dependent recruitment of BRCA1 to RNA promotes DNA end resection in homologous recombination. Nat. Commun. 2021, 12, 4126. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cargill, M.; Venkataraman, R.; Lee, S. DEAD-Box RNA Helicases and Genome Stability. Genes 2021, 12, 1471. https://doi.org/10.3390/genes12101471
Cargill M, Venkataraman R, Lee S. DEAD-Box RNA Helicases and Genome Stability. Genes. 2021; 12(10):1471. https://doi.org/10.3390/genes12101471
Chicago/Turabian StyleCargill, Michael, Rasika Venkataraman, and Stanley Lee. 2021. "DEAD-Box RNA Helicases and Genome Stability" Genes 12, no. 10: 1471. https://doi.org/10.3390/genes12101471
APA StyleCargill, M., Venkataraman, R., & Lee, S. (2021). DEAD-Box RNA Helicases and Genome Stability. Genes, 12(10), 1471. https://doi.org/10.3390/genes12101471





