Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds
Abstract
1. Introduction
2. The Fungal KDAC Repertoire: More Than Just the Number of Enzymes
3. The Role of Lysine Deacetylases in Fungal Pathogenicity and as Drug Targets for Novel Antifungal Agents
3.1. KDACs as Virulence Determinants of Human Fungal Pathogens
3.2. Putative KDAC Targets Involved in Virulence and Antifungal Resistance
- Is the virulence phenotype a result of changed gene expression patterns caused by altered chromatin regulation (i.e., hyperacetylation of histone tails at specific genomic loci like those coding for virulence factors or other proteins critical for fungal fitness)?
- Are (hyper)acetylated transcription factors of such critical genes responsible for the altered virulence behavior?
- Is a single or are multiple (hyper)acetylated protein(s) contributing to attenuated virulence?
- Is it a multifactorial process (i.e., a combination of the aforementioned points)?
3.3. KDACs as Modulators of Fungal Drug Resistance
3.4. KDAC Inhibitors for the Treatment of Invasive Fungal Infections
3.5. Targeting Protein–Protein Interaction within KDAC Complexes: Another Possibility to Combat Fungal Infections?
4. Lysine Deacetylases as Regulators of Small Fungal Natural Products
4.1. Impact of KDACs in the Regulation of SM Production: First Evidence
4.2. Versatile Functions of KDACs in the Expression of SMs: First Functional Hypothesis
4.3. Teamwork: KDACs Are Rarely Acting Alone
4.4. Pan KDAC Inhibitors for Mining New SMs: Quick and Easy but Not Always Valid
4.5. Deletion of KDACs or Their Complex Partners for Mining New SMs: Reliable but Elaborate and Not Always Feasible
4.6. An Alternative Approach: Depletion of (Essential) KDACs via the Promoter Rundown Technique
4.7. Microbial Interactions and the Induction of SMs: Are KDACs Involved in a Biochemical Warfare for Resources?
5. Concluding Remarks and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grimm, L.H.; Kelly, S.; Krull, R.; Hempel, D.C. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 2005, 69, 375–384. [Google Scholar] [CrossRef]
- Brakhage, A.A. Systemic fungal infections caused by Aspergillus species: Epidemiology, infection process and virulence determinants. Curr. Drug Targets 2005, 6, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Bahn, Y.-S.; Nielsen, K.; Lin, X.; Fraser, J.A.; Heitman, J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 2005, 3, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Alternative Candida albicans lifestyles: Growth on surfaces. Annu. Rev. Microbiol. 2005, 59, 113–133. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.N.; Fisher, M.C.; Bates, K.A. Diagnosing Emerging Fungal Threats: A One Health Perspective. Front. Genet. 2018, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.A. Superficial fungal infections. Lancet 2004, 364, 1173–1182. [Google Scholar] [CrossRef]
- Cornely, O.A. Aspergillus to Zygomycetes: Causes, risk factors, prevention, and treatment of invasive fungal infections. Infection 2008, 36, 296–313. [Google Scholar] [CrossRef]
- Tudesq, J.-J.; Peyrony, O.; Lemiale, V.; Azoulay, E. Invasive Pulmonary Aspergillosis in Nonimmunocompromised Hosts. Semin. Respir. Crit. Care Med. 2019, 40, 540–547. [Google Scholar] [CrossRef]
- Denning, D.W. Invasive aspergillosis. Clin. Infect. Dis. 1998, 26, 781–803. [Google Scholar] [CrossRef]
- Lamoth, F.; Juvvadi, P.R.; Steinbach, W.J. Histone deacetylase inhibition as an alternative strategy against invasive aspergillosis. Front. Microbiol. 2015, 6, 96. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Aigner, M.; Lass-Flörl, C. Treatment of drug-resistant Aspergillus infection. Expert Opin. Pharmacother. 2015, 16, 2267–2270. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Meis, J.F. Emergence of azole resistant Aspergillus fumigatus and One Health: Time to implement environmental stewardship. Environ. Microbiol. 2018, 20, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.J.; Pasqualotto, A.C.; Anderson, M.J.; Leatherbarrow, H.; Albarrag, A.M.; Harrison, E.; Gregson, L.; Bowyer, P.; Denning, D.W. Major variations in Aspergillus fumigatus arising within aspergillomas in chronic pulmonary aspergillosis. Mycoses 2013, 56, 434–441. [Google Scholar] [CrossRef]
- Meis, J.F.; Chowdhary, A.; Rhodes, J.L.; Fisher, M.C.; Verweij, P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2016, 371, 20150460. [Google Scholar] [CrossRef]
- Campoy, S.; Adrio, J.L. Antifungals. Biochem. Pharmacol. 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Tribus, M.; Bauer, I.; Galehr, J.; Rieser, G.; Trojer, P.; Brosch, G.; Loidl, P.; Haas, H.; Graessle, S. A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell 2010, 21, 345–353. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Chen, X.; Wang, J.; Zhao, Y.; Li, Y.; He, B. Zinc-dependent Deacetylase (HDAC) Inhibitors with Different Zinc Binding Groups. Curr. Top. Med. Chem. 2019, 19, 223–241. [Google Scholar] [CrossRef]
- Pitt, J.I. Toxigenic fungi and mycotoxins. Br. Med. Bull. 2000, 56, 184–192. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Brosch, G.; Loidl, P.; Graessle, S. Histone modifications and chromatin dynamics: A focus on filamentous fungi. FEMS Microbiol. Rev. 2008, 32, 409–439. [Google Scholar] [CrossRef]
- Gacek, A.; Strauss, J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012, 95, 1389–1404. [Google Scholar] [CrossRef] [PubMed]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, T.; Jamin, E.L.; Debrauwer, L.; Puel, O.; Oswald, I.P. From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 2018, 35, 147–173. [Google Scholar] [CrossRef]
- Collemare, J.; Seidl, M.F. Chromatin-dependent regulation of secondary metabolite biosynthesis in fungi: Is the picture complete? FEMS Microbiol. Rev. 2019, 43, 591–607. [Google Scholar] [CrossRef]
- Poças-Fonseca, M.J.; Cabral, C.G.; Manfrão-Netto, J.H.C. Epigenetic manipulation of filamentous fungi for biotechnological applications: A systematic review. Biotechnol. Lett. 2020, 42, 885–904. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef]
- Roze, L.V.; Arthur, A.E.; Hong, S.-Y.; Chanda, A.; Linz, J.E. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol. 2007, 66, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Leipe, D.D.; Landsman, D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 1997, 25, 3693–3697. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Wu, L.; Sun, R.; Keller, N.P.; Yang, K.; Ye, L.; He, S.; Zhang, F.; Wang, S. The HosA Histone Deacetylase Regulates Aflatoxin Biosynthesis Through Direct Regulation of Aflatoxin Cluster Genes. Mol. Plant-Microbe Interact. MPMI 2019, 32, 1210–1228. [Google Scholar] [CrossRef]
- Xie, J.; Jenull, S.; Tscherner, M.; Kuchler, K. The Paralogous Histone Deacetylases Rpd3 and Rpd31 Play Opposing Roles in Regulating the White-Opaque Switch in the Fungal Pathogen Candida albicans. mBio 2016, 7, e01807-16. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, M.; Jamnischek, A.; Weinzierl, G.; Ladendorf, O.; Huber, S.; Kahmann, R.; Kämper, J. The histone deacetylase Hda1 from Ustilago maydis is essential for teliospore development. Mol. Microbiol. 2002, 46, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- González-Prieto, J.M.; Domínguez, A.; Rosas-Quijano, R.; Cervantes-Chávez, J.A.; Ruiz-Herrera, J. Isolation and molecular analysis of Umhda2 a gene encoding a histone deacetylase from Ustilago maydis. DNA Seq. 2004, 15, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Sui, M.; Li, M.; Wang, J.; Meng, Y.; Sun, T.; Liang, Q.; Suo, C.; Gao, X.; et al. Fungal acetylome comparative analysis identifies an essential role of acetylation in human fungal pathogen virulence. Commun. Biol. 2019, 2, 154. [Google Scholar] [CrossRef] [PubMed]
- Carmen, A.A.; Rundlett, S.E.; Grunstein, M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 1996, 271, 15837–15844. [Google Scholar] [CrossRef] [PubMed]
- Rundlett, S.E.; Carmen, A.A.; Kobayashi, R.; Bavykin, S.; Turner, B.M.; Grunstein, M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 1996, 93, 14503–14508. [Google Scholar] [CrossRef] [PubMed]
- Lechner, T.; Carrozza, M.J.; Yu, Y.; Grant, P.A.; Eberharter, A.; Vannier, D.; Brosch, G.; Stillman, D.J.; Shore, D.; Workman, J.L. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3[middle dot]Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 2000, 275, 40961–40966. [Google Scholar] [CrossRef] [PubMed]
- Pijnappel, W.W.; Schaft, D.; Roguev, A.; Shevchenko, A.; Tekotte, H.; Wilm, M.; Rigaut, G.; Séraphin, B.; Aasland, R.; Stewart, A.F. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001, 15, 2991–3004. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Li, B.; Florens, L.; Suganuma, T.; Swanson, S.K.; Lee, K.K.; Shia, W.-J.; Anderson, S.; Yates, J.; Washburn, M.P.; et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005, 123, 581–592. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Florens, L.; Swanson, S.K.; Shia, W.-J.; Anderson, S.; Yates, J.; Washburn, M.P.; Workman, J.L. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 2005, 1731, 77–87. [Google Scholar] [CrossRef]
- Keogh, M.-C.; Kurdistani, S.K.; Morris, S.A.; Ahn, S.H.; Podolny, V.; Collins, S.R.; Schuldiner, M.; Chin, K.; Punna, T.; Thompson, N.J.; et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 2005, 123, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Rodríguez, M.; Heitman, J. Cyclophilin A is localized to the nucleus and controls meiosis in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Maskos, K.; Huber, R. Structural and functional studies of the yeast class II Hda1 histone deacetylase complex. J. Mol. Biol. 2009, 391, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.-I.; Xiao, G.; Noma, K.-I.; Malikzay, A.; Bjerling, P.; Ekwall, K.; Kobayashi, R.; Grewal, S.I.S. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003, 22, 2776–2787. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, E.; Yamada, T.; Cam, H.P.; Fitzgerald, P.C.; Kobayashi, R.; Grewal, S.I.S. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007, 14, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Cam, H.P.; Sugiyama, R.; Noma, K.-I.; Zofall, M.; Kobayashi, R.; Grewal, S.I.S. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 2007, 128, 491–504. [Google Scholar] [CrossRef]
- Zilio, N.; Codlin, S.; Vashisht, A.A.; Bitton, D.A.; Head, S.R.; Wohlschlegel, J.A.; Bähler, J.; Boddy, M.N. A novel histone deacetylase complex in the control of transcription and genome stability. Mol. Cell. Biol. 2014, 34, 3500–3514. [Google Scholar] [CrossRef] [PubMed]
- Grewal, C.; Hickmott, J.; Rentas, S.; Karagiannis, J. A conserved histone deacetylase with a role in the regulation of cytokinesis in Schizosaccharomyces pombe. Cell Div. 2012, 7, 1–14. [Google Scholar] [CrossRef]
- Baidyaroy, D.; Brosch, G.; Ahn, J.H.; Graessle, S.; Wegener, S.; Tonukari, N.J.; Caballero, O.; Loidl, P.; Walton, J.D. A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell 2001, 13, 1609–1624. [Google Scholar] [CrossRef]
- Wang, A.; Kurdistani, S.K.; Grunstein, M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 2002, 298, 1412–1414. [Google Scholar] [CrossRef]
- Kim, T.; Buratowski, S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5’ transcribed regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef]
- Ryu, H.-Y.; Zhao, D.; Li, J.; Su, D.; Hochstrasser, M. Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 2020, 48, 12151–12168. [Google Scholar] [CrossRef]
- Baker, L.A.; Ueberheide, B.M.; Dewell, S.; Chait, B.T.; Zheng, D.; Allis, C.D. The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol. Cell. Biol. 2013, 33, 3735–3748. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.E.; Chandru, A.; Cowley, S.M. Co-repressor, co-activator and general transcription factor: The many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 2018, 475, 3921–3932. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.M.; Tomar, R.S.; Dempsey, A.E.; Reese, J.C. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 2007, 27, 3199–3210. [Google Scholar] [CrossRef]
- Takahata, S.; Yu, Y.; Stillman, D.J. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009, 28, 3378–3389. [Google Scholar] [CrossRef] [PubMed]
- Bosio, M.C.; Fermi, B.; Spagnoli, G.; Levati, E.; Rubbi, L.; Ferrari, R.; Pellegrini, M.; Dieci, G. Abf1 and other general regulatory factors control ribosome biogenesis gene expression in budding yeast. Nucleic Acids Res. 2017, 45, 4493–4506. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.B.; Choi, A.; Kim, J.H.; Jun, Y.; Woo, H.; Ha, S.D.; Yoon, C.Y.; Hwang, J.-T.; Steinmetz, L.; Buratowski, S.; et al. Rpd3L HDAC links H3K4me3 to transcriptional repression memory. Nucleic Acids Res. 2018, 46, 8261–8274. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.A.; Struhl, K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 2005, 20, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.S.; Strålfors, A.; Catania, S.; Castillo, A.G.; Svensson, J.P.; Pidoux, A.L.; Ekwall, K.; Allshire, R.C. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 2012, 8, e1002985. [Google Scholar] [CrossRef]
- Hnisz, D.; Bardet, A.F.; Nobile, C.J.; Petryshyn, A.; Glaser, W.; Schöck, U.; Stark, A.; Kuchler, K. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012, 8, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Hartooni, N.; Mitchell, K.F.; Hnisz, D.; Andes, D.R.; Kuchler, K.; Johnson, A.D. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 2014, 5, e01201-14. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-L.; Liu, W.; Iliuk, A.; Ribot, C.; Vallet, J.; Tao, A.; Wang, Y.; Lebrun, M.-H.; Xu, J.-R. The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell 2010, 22, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Liang, S.; Zhang, P.; Kang, R.; Zhang, M.; Wang, M.; Chen, L.; Yuan, H.; Ding, S.; et al. FpDep1, a component of Rpd3L histone deacetylase complex, is important for vegetative development, ROS accumulation, and pathogenesis in Fusarium pseudograminearum. Fungal Genet. Biol. 2020, 135, 103299. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Majer, O.; Frohner, I.E.; Komnenovic, V.; Kuchler, K. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 2010, 6, e1000889. [Google Scholar] [CrossRef]
- Shevchenko, A.; Roguev, A.; Schaft, D.; Buchanan, L.; Habermann, B.; Sakalar, C.; Thomas, H.; Krogan, N.J.; Shevchenko, A.; Stewart, A.F. Chromatin Central: Towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008, 9, R167. [Google Scholar] [CrossRef]
- Bauer, I.; Gross, S.; Merschak, P.; Kremser, L.; Karahoda, B.; Bayram, Ö.S.; Abt, B.; Binder, U.; Gsaller, F.; Lindner, H.; et al. RcLS2F—A Novel Fungal Class 1 KDAC Co-repressor Complex in Aspergillus nidulans. Front. Microbiol. 2020, 11, 43. [Google Scholar] [CrossRef]
- Almeida, R.S.; Loss, O.; Colabardini, A.C.; Brown, N.A.; Bignell, E.; Savoldi, M.; Pantano, S.; Goldman, M.H.S.; Arst, H.N., Jr.; Goldman, G.H. Genetic bypass of Aspergillus nidulans crzA function in calcium homeostasis. G3 (Bethesda) 2013, 3, 1129–1141. [Google Scholar] [CrossRef]
- Juvvadi, P.R.; Lee, S.C.; Heitman, J.; Steinbach, W.J. Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence 2017, 8, 186–197. [Google Scholar] [CrossRef]
- Lee, J.-E.; Oh, J.-H.; Ku, M.; Kim, J.; Lee, J.-S.; Kang, S.-O. Ssn6 has dual roles in Candida albicans filament development through the interaction with Rpd31. FEBS Lett. 2015, 589, 513–520. [Google Scholar] [CrossRef]
- Wu, J.; Carmen, A.A.; Kobayashi, R.; Suka, N.; Grunstein, M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc. Natl. Acad. Sci. USA 2001, 98, 4391–4396. [Google Scholar] [CrossRef] [PubMed]
- Trojer, P.; Brandtner, E.M.; Brosch, G.; Loidl, P.; Galehr, J.; Linzmaier, R.; Haas, H.; Mair, K.; Tribus, M.; Graessle, S. Histone deacetylases in fungi: Novel members, new facts. Nucleic Acids Res. 2003, 31, 3971–3981. [Google Scholar] [CrossRef] [PubMed]
- Amorim-Vaz, S.; Tran, V.D.T.; Pradervand, S.; Pagni, M.; Coste, A.T.; Sanglard, D. RNA Enrichment Method for Quantitative Transcriptional Analysis of Pathogens In Vivo Applied to the Fungus Candida albicans. mBio 2015, 6, e00942-15. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Solis, N.V.; Ehrlich, R.L.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol. 2015, 13, e1002076. [Google Scholar] [CrossRef] [PubMed]
- Studt, L.; Schmidt, F.J.; Jahn, L.; Sieber, C.M.K.; Connolly, L.R.; Niehaus, E.-M.; Freitag, M.; Humpf, H.-U.; Tudzynski, B. Two histone deacetylases, FfHda1 and FfHda2, are important for Fusarium fujikuroi secondary metabolism and virulence. Appl. Environ. Microbiol. 2013, 79, 7719–7734. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Liu, W.; Wang, G.; Kang, Z.; Kistler, H.C.; Xu, J.-R. The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum. Mol. Plant-Microbe Interact. MPMI 2011, 24, 487–496. [Google Scholar] [CrossRef]
- Jiang, H.; Xia, A.; Ye, M.; Ren, J.; Li, D.; Liu, H.; Wang, Q.; Lu, P.; Wu, C.; Xu, J.-R.; et al. Opposing functions of Fng1 and the Rpd3 HDAC complex in H4 acetylation in Fusarium graminearum. PLoS Genet. 2020, 16, e1009185. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Xu, Y.; Chen, J.; Luo, Y.; Lv, Y.; Su, J.; Kershaw, M.J.; Li, W.; Wang, J.; Yin, J.; et al. MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 2018, 14, 1543–1561. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.-J.; Jeon, J. A histone deacetylase, MoHOS2 regulates asexual development and virulence in the rice blast fungus. J. Microbiol. 2019, 57, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Lee, S.H.; Oh, Y.T.; Jeon, J. A Histone Deacetylase, MoHDA1 Regulates Asexual Development and Virulence in the Rice Blast Fungus. Plant Pathol. J. 2020, 36, 314–322. [Google Scholar] [CrossRef]
- Izawa, M.; Takekawa, O.; Arie, T.; Teraoka, T.; Yoshida, M.; Kimura, M.; Kamakura, T. Inhibition of histone deacetylase causes reduction of appressorium formation in the rice blast fungus Magnaporthe oryzae. J. Gen. Appl. Microbiol. 2009, 55, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Cao, X.; Qu, Z.; Zhang, S.; Naqvi, N.I.; Deng, Y.Z. The Histone Deacetylases MoRpd3 and MoHst4 Regulate Growth, Conidiation, and Pathogenicity in the Rice Blast Fungus Magnaporthe oryzae. mSphere 2021, 6, e0011821. [Google Scholar] [CrossRef] [PubMed]
- Elías-Villalobos, A.; Fernández-Álvarez, A.; Moreno-Sánchez, I.; Helmlinger, D.; Ibeas, J.I. The Hos2 Histone Deacetylase Controls Ustilago maydis Virulence through Direct Regulation of Mating-Type Genes. PLoS Pathog. 2015, 11, e1005134. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Z.-K.; Shao, W.; Ying, S.-H.; Feng, M.-G. Essential role of Rpd3-dependent lysine modification in the growth, development and virulence of Beauveria bassiana. Environ. Microbiol. 2018, 20, 1590–1606. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Tong, S.-M.; Shao, W.; Ying, S.-H.; Feng, M.-G. Pleiotropic effects of the histone deacetylase Hos2 linked to H4-K16 deacetylation, H3-K56 acetylation, and H2A-S129 phosphorylation in Beauveria bassiana. Cell. Microbiol. 2018, 13, e12839. [Google Scholar] [CrossRef] [PubMed]
- Bauer, I.; Varadarajan, D.; Pidroni, A.; Gross, S.; Vergeiner, S.; Faber, B.; Hermann, M.; Tribus, M.; Brosch, G.; Graessle, S. A Class 1 Histone Deacetylase with Potential as an Antifungal Target. mBio 2016, 7, e00831-16. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, Z.; Zhang, Z.; Liang, W. BcRPD3-Mediated Histone Deacetylation Is Involved in Growth and Pathogenicity of Botrytis cinerea. Front. Microbiol. 2020, 11, 1832. [Google Scholar] [CrossRef]
- Lamoth, F.; Juvvadi, P.R.; Gehrke, C.; Asfaw, Y.G.; Steinbach, W.J. Transcriptional activation of heat shock protein 90 mediated via a proximal promoter region as trigger of caspofungin resistance in Aspergillus fumigatus. J. Infect. Dis. 2014, 209, 473–481. [Google Scholar] [CrossRef]
- Bauer, I.; Misslinger, M.; Shadkchan, Y.; Dietl, A.-M.; Petzer, V.; Orasch, T.; Abt, B.; Graessle, S.; Osherov, N.; Haas, H. The Lysine Deacetylase RpdA Is Essential for Virulence in Aspergillus fumigatus. Front. Microbiol. 2019, 10, 2773. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Samardzic, E.; Knoll, M. Serology anno 2021-fungal infections: From invasive to chronic. Clinical microbiology and infection. Clin. Microbiol. Infect. Dis. 2021, 27, 1230–1241. [Google Scholar] [CrossRef]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Liu, O.W.; Chun, C.D.; Chow, E.D.; Chen, C.; Madhani, H.D.; Noble, S.M. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 2008, 135, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Saenz, H.L.; Dehio, C. Signature-tagged mutagenesis: Technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 2005, 8, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Brandão, F.; Esher, S.K.; Ost, K.S.; Pianalto, K.; Nichols, C.B.; Fernandes, L.; Bocca, A.L.; Poças-Fonseca, M.J.; Alspaugh, J.A. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci. Rep. 2018, 8, 5209–5217. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Oh, J.-H.; Shwab, E.K.; Dagenais, T.R.T.; Andes, D.; Keller, N.P. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. FG B 2009, 46, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yang, Q.; Yin, C.; Liu, L.; Liang, W. Systematic analysis of the lysine acetylome in Fusarium graminearum. BMC Genom. 2016, 17, 1019. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef] [PubMed]
- James, A.M.; Smith, C.L.; Smith, A.C.; Robinson, A.J.; Hoogewijs, K.; Murphy, M.P. The Causes and Consequences of Nonenzymatic Protein Acylation. Trends Biochem. Sci. 2018, 43, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.J., Jr.; Kissinger, J.C.; et al. FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef]
- Köhler, J.R.; Hube, B.; Puccia, R.; Casadevall, A.; Perfect, J.R. Fungi that Infect Humans. Microbiol. Spectr. 2017, 5, 1–29. [Google Scholar] [CrossRef]
- Gacek-Matthews, A.; Noble, L.M.; Gruber, C.; Berger, H.; Sulyok, M.; Marcos, A.T.; Strauss, J.; Andrianopoulos, A. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol. 2015, 96, 839–860. [Google Scholar] [CrossRef] [PubMed]
- Gacek-Matthews, A.; Berger, H.; Sasaki, T.; Wittstein, K.; Gruber, C.; Lewis, Z.A.; Strauss, J. KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus nidulans. PLoS Genet. 2016, 12, e1006222. [Google Scholar] [CrossRef] [PubMed]
- Pidroni, A.; Faber, B.; Brosch, G.; Bauer, I.; Graessle, S. A Class 1 Histone Deacetylase as Major Regulator of Secondary Metabolite Production in Aspergillus nidulans. Front. Microbiol. 2018, 9, 2212. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef]
- Schilling, B.; Basisty, N.; Christensen, D.G.; Sorensen, D.; Orr, J.S.; Wolfe, A.J.; Rao, C.V. Global Lysine Acetylation in Escherichia coli Results from Growth Conditions That Favor Acetate Fermentation. J. Bacteriol. 2019, 201, e00768-18. [Google Scholar] [CrossRef]
- Zhou, X.; Qian, G.; Yi, X.; Li, X.; Liu, W. Systematic Analysis of the Lysine Acetylome in Candida albicans. J. Proteome Res. 2016, 15, 2525–2536. [Google Scholar] [CrossRef]
- Xu, X.; Liu, T.; Yang, J.; Chen, L.; Liu, B.; Wang, L.; Jin, Q. The First Whole-Cell Proteome- and Lysine-Acetylome-Based Comparison between Trichophyton rubrum Conidial and Mycelial Stages. J. Proteome Res. 2018, 17, 1436–1451. [Google Scholar] [CrossRef]
- Lv, B.; Yang, Q.; Li, D.; Liang, W.; Song, L. Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea. Sci. Rep. 2016, 6, 29313–29319. [Google Scholar] [CrossRef]
- Shapiro, R.S.; Uppuluri, P.; Zaas, A.K.; Collins, C.; Senn, H.; Perfect, J.R.; Heitman, J.; Cowen, L.E. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. CB 2009, 19, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Juvvadi, P.R.; Soderblom, E.J.; Moseley, M.A.; Asfaw, Y.G.; Steinbach, W.J. Identification of a key lysine residue in heat shock protein 90 required for azole and echinocandin resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2014, 58, 1889–1896. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, X.; Guo, Z.S.; Marcu, M.G.; Neckers, L.; Nguyen, D.M.; Chen, G.A.; Schrump, D.S. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J. Natl. Cancer Inst. 2002, 94, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Nimmanapalli, R.; Bali, P.; O’Bryan, E.; Fuino, L.; Guo, F.; Wu, J.; Houghton, P.; Bhalla, K. Arsenic trioxide inhibits translation of mRNA of bcr-abl, resulting in attenuation of Bcr-Abl levels and apoptosis of human leukemia cells. Cancer Res. 2003, 63, 7950–7958. [Google Scholar] [PubMed]
- Scroggins, B.T.; Robzyk, K.; Wang, D.; Marcu, M.G.; Tsutsumi, S.; Beebe, K.; Cotter, R.J.; Felts, S.; Toft, D.; Karnitz, L.; et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell 2007, 25, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.; Leach, M.D.; Cowen, L.E. Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2012, 2, 878–888. [Google Scholar] [CrossRef]
- Li, X.; Robbins, N.; O’Meara, T.R.; Cowen, L.E. Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol. 2017, 103, 635–656. [Google Scholar] [CrossRef]
- Beardsley, J.; Halliday, C.L.; Chen, S.C.-A.; Sorrell, T.C. Responding to the emergence of antifungal drug resistance: Perspectives from the bench and the bedside. Future Microbiol. 2018, 13, 1175–1191. [Google Scholar] [CrossRef]
- Nywening, A.V.; Rybak, J.M.; Rogers, P.D.; Fortwendel, J.R. Mechanisms of triazole resistance in Aspergillus fumigatus. Environ. Microbiol. 2020, 22, 4934–4952. [Google Scholar] [CrossRef]
- Lee, Y.; Puumala, E.; Robbins, N.; Cowen, L.E. Antifungal Drug Resistance: Molecular Mechanisms in Candida albicans and beyond. Chem. Rev. 2021, 121, 3390–3411. [Google Scholar] [CrossRef]
- Smith, W.L.; Edlind, T.D. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: Correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 2002, 46, 3532–3539. [Google Scholar] [CrossRef]
- Mai, A.; Rotili, D.; Massa, S.; Brosch, G.; Simonetti, G.; Passariello, C.; Palamara, A.T. Discovery of uracil-based histone deacetylase inhibitors able to reduce acquired antifungal resistance and trailing growth in Candida albicans. Bioorganic Med. Chem. Lett. 2007, 17, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Lindquist, S. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science 2005, 309, 2185–2189. [Google Scholar] [CrossRef] [PubMed]
- Harvey, Z.H.; Chakravarty, A.K.; Futia, R.A.; Jarosz, D.F. A Prion Epigenetic Switch Establishes an Active Chromatin State. Cell 2020, 180, 928–940.e14. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.; Kobayashi, M.; Nagashima, K.; Wakisaka, Y.; Koizumi, K. A new antifungal antibiotic, trichostatin. J. Antibiot. 1976, 29, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Kijima, M.; Akita, M.; Beppu, T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990, 265, 17174–17179. [Google Scholar] [CrossRef]
- Tang, J.; Yan, H.; Zhuang, S. Histone deacetylases as targets for treatment of multiple diseases. Clin. Sci. 2013, 124, 651–662. [Google Scholar] [CrossRef]
- McClure, J.J.; Li, X.; Chou, C.J. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv. Cancer Res. 2018, 138, 183–211. [Google Scholar]
- Chun, P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch. Pharmacal. Res. 2015, 38, 933–949. [Google Scholar] [CrossRef]
- Chun, P. Therapeutic effects of histone deacetylase inhibitors on heart disease. Arch. Pharmacal. Res. 2020, 43, 1276–1296. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam. Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Roger, T.; Lugrin, J.; Le Roy, D.; Goy, G.; Mombelli, M.; Koessler, T.; Ding, X.C.; Chanson, A.-L.; Reymond, M.K.; Miconnet, I.; et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 2011, 117, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Yamashita, T.; Mariko, Y.; Nosaka, Y.; Tsuchiya, K.; Ando, T.; Suzuki, T.; Tsuruo, T.; Nakanishi, O. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. USA 1999, 96, 4592–4597. [Google Scholar] [CrossRef] [PubMed]
- Brandão, F.A.; Derengowski, L.S.; Albuquerque, P.; Nicola, A.M.; Silva-Pereira, I.; Poças-Fonseca, M.J. Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence 2015, 6, 618–630. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Georgopapadakou, N.; Martell, L.A.; Besterman, J.M.; Diekema, D.J. Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J. Clin. Microbiol. 2009, 47, 3797–3804. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Rhomberg, P.R.; Messer, S.A.; Castanheira, M. In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn. Microbiol. Infect. Dis. 2015, 81, 259–263. [Google Scholar] [CrossRef]
- Han, G.; Liu, N.; Li, C.; Tu, J.; Li, Z.; Sheng, C. Discovery of Novel Fungal Lanosterol 14α-Demethylase (CYP51)/Histone Deacetylase Dual Inhibitors to Treat Azole-Resistant Candidiasis. J. Med. Chem. 2020, 63, 5341–5359. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tu, J.; Han, G.; Liu, N.; Sheng, C. Novel Carboline Fungal Histone Deacetylase (HDAC) Inhibitors for Combinational Treatment of Azole-Resistant Candidiasis. J. Med. Chem. 2021, 64, 1116–1126. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, W.; Fang, G. Targeting protein-protein interaction by small molecules. Annu. Rev. Pharm. Toxicol. 2014, 54, 435–456. [Google Scholar] [CrossRef]
- Chung, C.-W.; Coste, H.; White, J.H.; Mirguet, O.; Wilde, J.; Gosmini, R.L.; Delves, C.; Magny, S.M.; Woodward, R.; Hughes, S.A.; et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem. 2011, 54, 3827–3838. [Google Scholar] [CrossRef]
- Mossessova, E.; Corpina, R.A.; Goldberg, J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 2003, 12, 1403–1411. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.-T.; Wang, W.-T.; Zhang, X.-R.; Suo, F.; Ren, J.-Y.; Bi, Y.; Xue, Y.-X.; Hu, W.; Dong, M.-Q.; et al. Systematic analysis reveals the prevalence and principles of bypassable gene essentiality. Nat. Commun. 2019, 10, 1002. [Google Scholar] [CrossRef]
- Yaegashi, J.; Oakley, B.R.; Wang, C.C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotechnol. 2014, 41, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, M. The fungi: 1, 2, 3… 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P.; Kurumizaka, H. The nucleosome: A powerful regulator of transcription. Prog. Nucleic Acid Res. Mol. Biol. 1998, 61, 379–422. [Google Scholar] [PubMed]
- Graessle, S.; Dangl, M.; Haas, H.; Mair, K.; Trojer, P.; Brandtner, E.M.; Walton, J.D.; Loidl, P.; Brosch, G. Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta 2000, 1492, 120–126. [Google Scholar] [CrossRef]
- Borkovich, K.A.; Alex, L.A.; Yarden, O.; Freitag, M.; Turner, G.E.; Read, N.D.; Seiler, S.; Bell-Pedersen, D.; Paietta, J.; Plesofsky, N.; et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. MMBR 2004, 68, 1–108. [Google Scholar] [CrossRef]
- Baidyaroy, D.; Brosch, G.; Graessle, S.; Trojer, P.; Walton, J.D. Characterization of inhibitor-resistant histone deacetylase activity in plant-pathogenic fungi. Eukaryot. Cell 2002, 1, 538–547. [Google Scholar] [CrossRef][Green Version]
- Tribus, M.; Galehr, J.; Trojer, P.; Brosch, G.; Loidl, P.; Marx, F.; Haas, H.; Graessle, S. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell 2005, 4, 1736–1745. [Google Scholar] [CrossRef]
- Niehaus, E.-M.; Studt, L.; von Bargen, K.W.; Kummer, W.; Humpf, H.-U.; Reuter, G.; Tudzynski, B. Sound of silence: The beauvericin cluster in Fusarium fujikuroi is controlled by cluster-specific and global regulators mediated by H3K27 modification. Environ. Microbiol. 2016, 18, 4282–4302. [Google Scholar] [CrossRef]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Carmen, A.A.; Griffin, P.R.; Calaycay, J.R.; Rundlett, S.E.; Suka, Y.; Grunstein, M. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 12356–12361. [Google Scholar] [CrossRef] [PubMed]
- Bauer, I.; Pidroni, A.; Bayram, Ö.; Brosch, G.; Graessle, S. Single-Step Enrichment of a TAP-Tagged Histone Deacetylase of the Filamentous Fungus Aspergillus nidulans for Enzymatic Activity Assay. J. Vis. Exp. JoVE 2019, 147, e59527. [Google Scholar] [CrossRef]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Zutz, C.; Gacek, A.; Sulyok, M.; Wagner, M.; Strauss, J.; Rychli, K. Small chemical chromatin effectors alter secondary metabolite production in Aspergillus clavatus. Toxins 2013, 5, 1723–1741. [Google Scholar] [CrossRef] [PubMed]
- Albright, J.C.; Henke, M.T.; Soukup, A.A.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. Large-Scale Metabolomics Reveals a Complex Response of Aspergillus nidulans to Epigenetic Perturbation. ACS Chem. Biol. 2015, 10, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Henke, M.T.; Soukup, A.A.; Goering, A.W.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. New Aspercryptins, Lipopeptide Natural Products, Revealed by HDAC Inhibition in Aspergillus nidulans. ACS Chem. Biol. 2016, 11, 2117–2123. [Google Scholar] [CrossRef]
- Guzmán-Chávez, F.; Zwahlen, R.D.; Bovenberg, R.A.L.; Driessen, A.J.M. Engineering of the Filamentous Fungus Penicillium chrysogenum as Cell Factory for Natural Products. Front. Microbiol. 2018, 9, 2768. [Google Scholar] [CrossRef]
- Aldholmi, M.; Wilkinson, B.; Ganesan, A. Epigenetic modulation of secondary metabolite profiles in Aspergillus calidoustus and Aspergillus westerdijkiae through histone deacetylase (HDAC) inhibition by vorinostat. J. Antibiot. 2020, 73, 410–413. [Google Scholar] [CrossRef]
- Wu, J.-S.; Yao, G.-S.; Shi, X.-H.; Rehman, S.U.; Xu, Y.; Fu, X.-M.; Zhang, X.-L.; Liu, Y.; Wang, C.-Y. Epigenetic Agents Trigger the Production of Bioactive Nucleoside Derivatives and Bisabolane Sesquiterpenes from the Marine-Derived Fungus Aspergillus versicolor. Front. Microbiol. 2020, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Shao, C.-L.; Liu, Y.; Zhao, D.-L.; Cao, F.; Fu, X.-M.; Yu, J.-Y.; Wu, J.-S.; Zhang, Z.-K.; Wang, C.-Y. Terpenoids from the Coral-Derived Fungus Trichoderma harzianum (XS-20090075) Induced by Chemical Epigenetic Manipulation. Front. Microbiol. 2020, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Izawa, M.; Nakajima, Y.; Jin, Q.; Hirose, T.; Nakamura, T.; Koshino, H.; Kanamaru, K.; Ohsato, S.; Kamakura, T.; et al. Increased metabolite production by deletion of an HDA1-type histone deacetylase in the phytopathogenic fungi, Magnaporthe oryzae (Pyricularia oryzae) and Fusarium asiaticum. Lett. Appl. Microbiol. 2017, 65, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, L.; Wang, B.; Pan, L. The Histone Deacetylases HosA and HdaA Affect the Phenotype and Transcriptomic and Metabolic Profiles of Aspergillus niger. Toxins 2019, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yue, Y.; Ren, S.; Yang, M.; Zhang, Y.; Cao, X.; Wang, Y.; Zhang, J.; Ge, F.; Wang, S. Lysine acetylation contributes to development, aflatoxin biosynthesis and pathogenicity in Aspergillus flavus. Environ. Microbiol. 2019, 21, 4792–4807. [Google Scholar] [CrossRef]
- Zadra, I.; Abt, B.; Parson, W.; Haas, H. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 2000, 66, 4810–4816. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.-W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef]
- Falkenberg, K.J.; Johnstone, R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014, 13, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Fallon, J.; Kelly, J.; Kavanagh, K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol. Biol. 2012, 845, 469–485. [Google Scholar] [PubMed]
- Binder, U.; Maurer, E.; Lass-Flörl, C. Galleria mellonella: An invertebrate model to study pathogenicity in correctly defined fungal species. Fungal Biol. 2016, 120, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Maurer, E.; Browne, N.; Surlis, C.; Jukic, E.; Moser, P.; Kavanagh, K.; Lass-Flörl, C.; Binder, U. Galleria mellonella as a host model to study Aspergillus terreus virulence and amphotericin B resistance. Virulence 2015, 6, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Orasch, T.; Dietl, A.-M.; Shadkchan, Y.; Binder, U.; Bauer, I.; Lass-Flörl, C.; Osherov, N.; Haas, H. The leucine biosynthetic pathway is crucial for adaptation to iron starvation and virulence in Aspergillus fumigatus. Virulence 2019, 10, 925–934. [Google Scholar] [CrossRef] [PubMed]
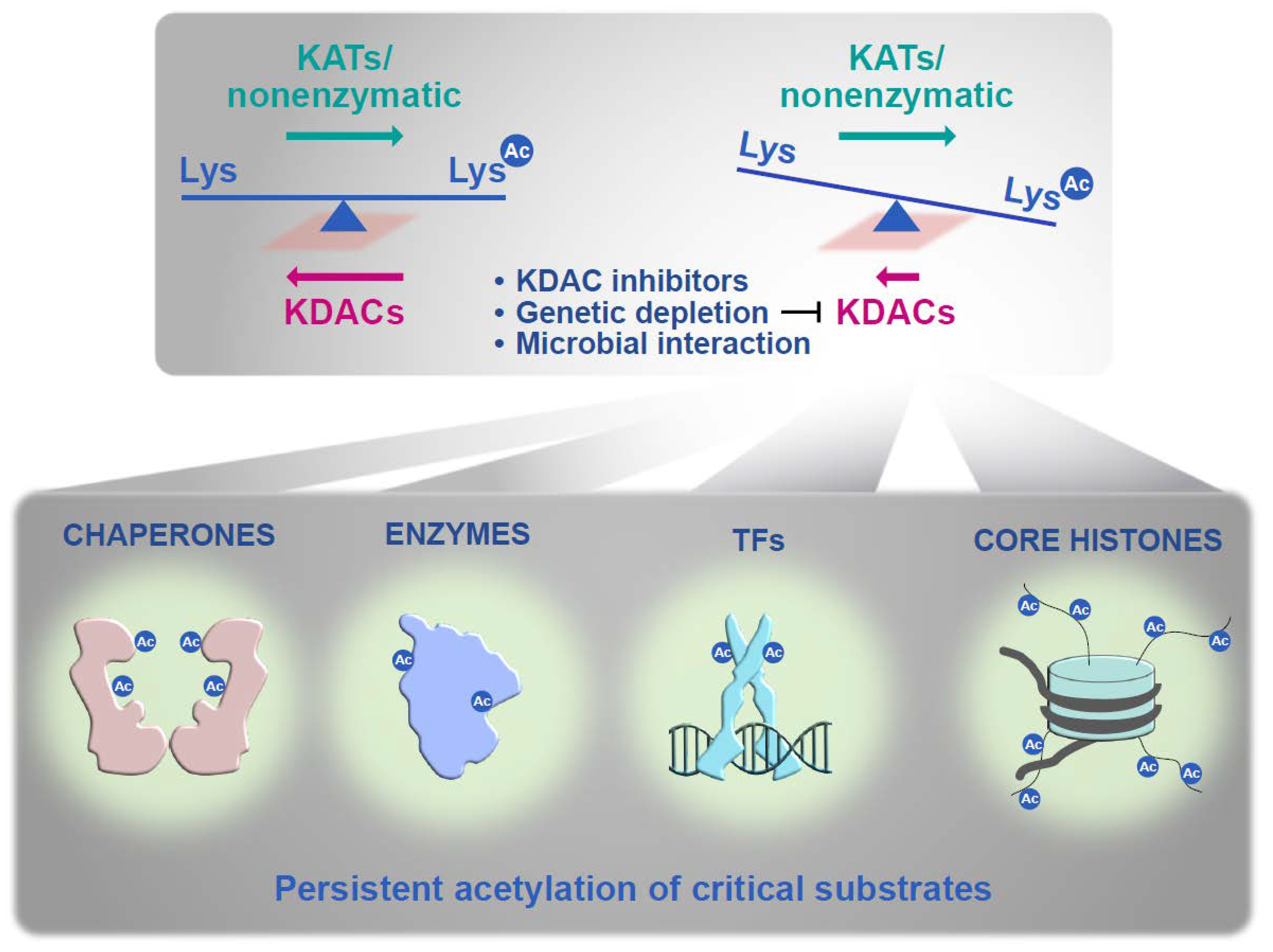
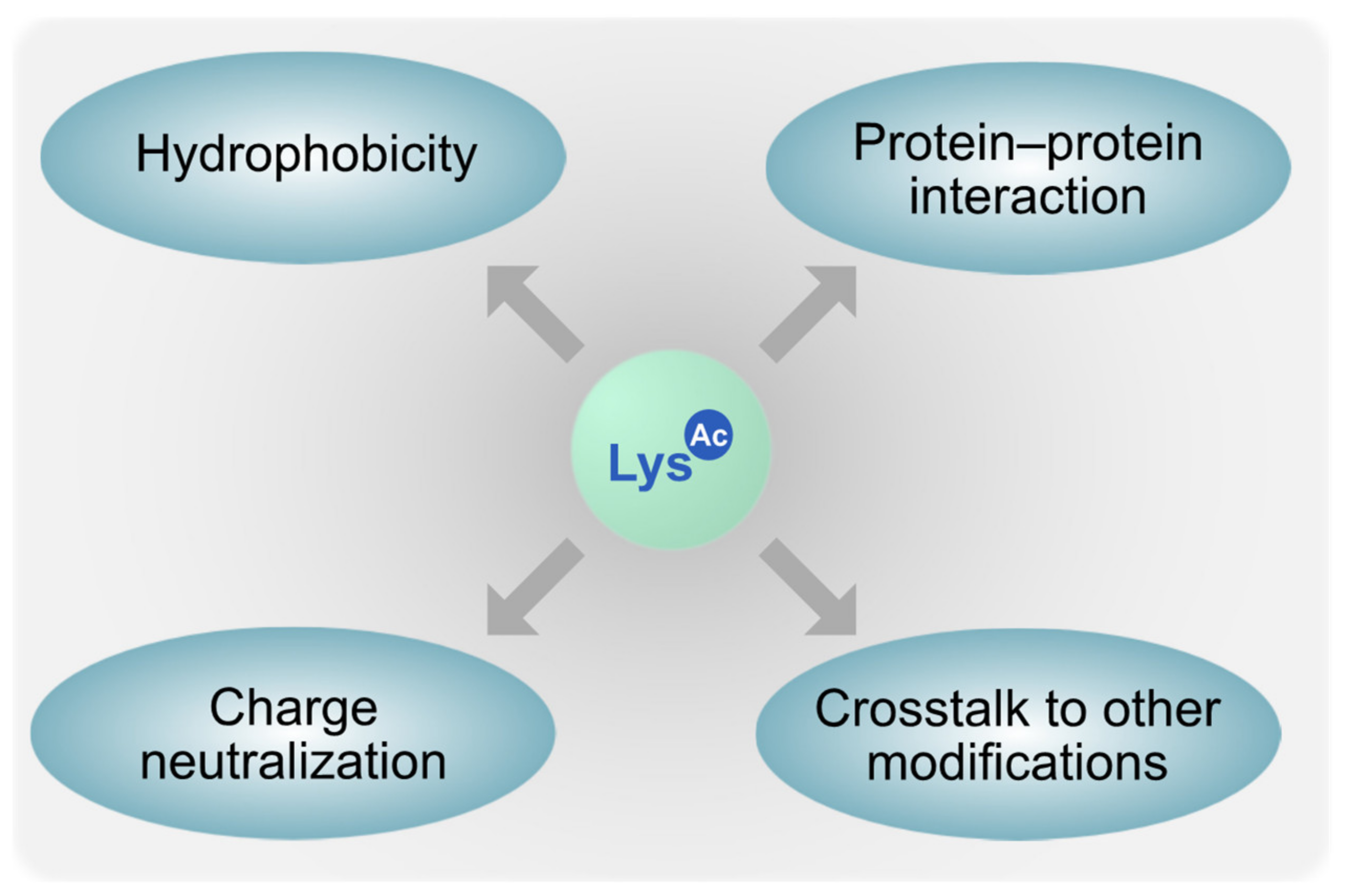
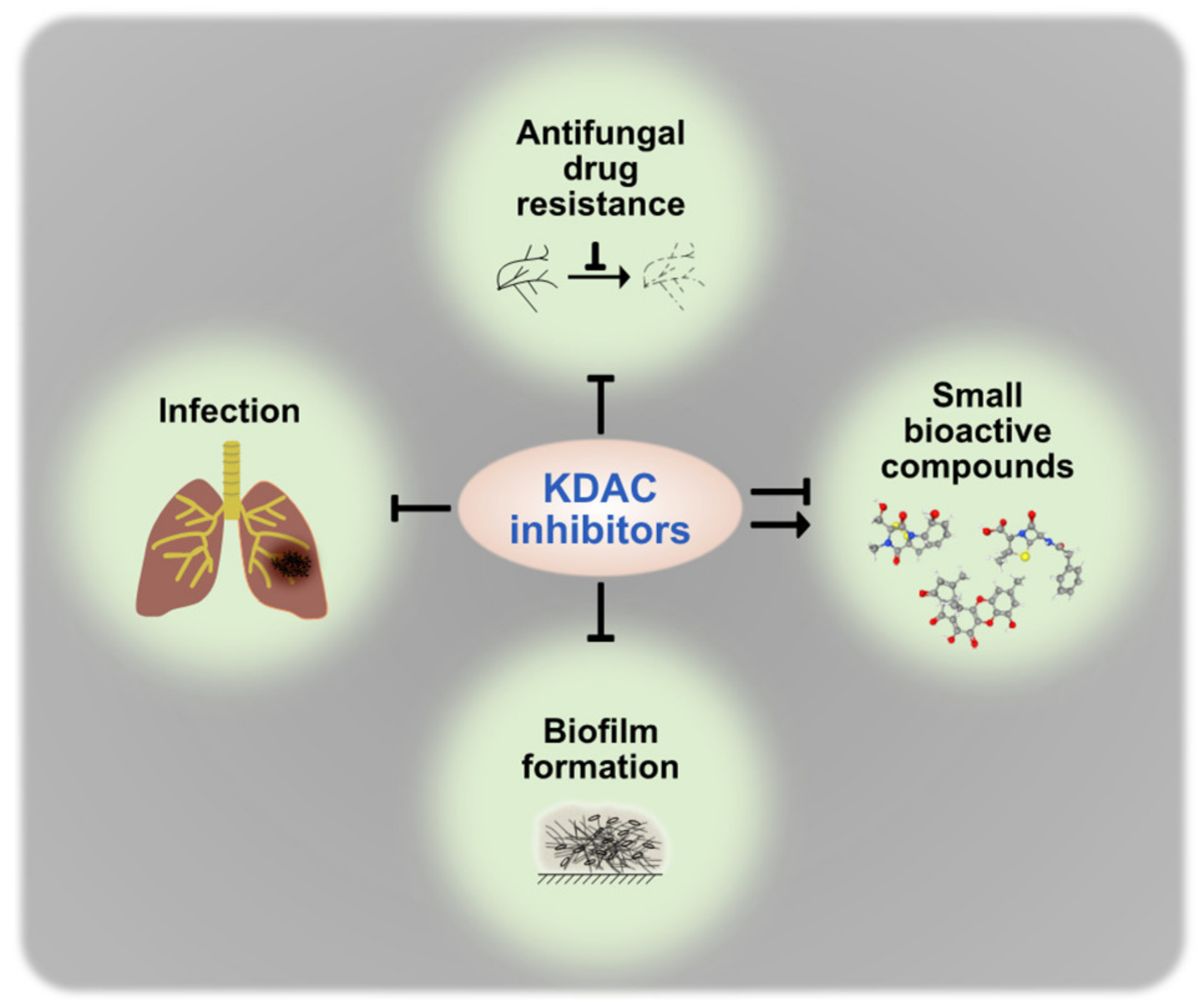
| Pathogen | Rpd3 Homolog | Hos2 Homolog | Hda1 Homolog |
|---|---|---|---|
| Aspergillus fumigatus | RpdA (Afu2g03390)/ KD 1/av 2/mouse [89] | HosA (Afu2g03810) n.d. 3 | HdaA (Afu5g01980)/ KO/fv/mouse [95] |
| Candida albicans | Rpd31 (C3_07000W_A) KO/av/mouse [70] | Hos2 (C3_00780W_A) n.d. | Hda1 (CR_02050C_A) n.d. |
| Cryptococcus neoformans | Rpd302 (CNAG_05096) KO/att/mouse [34] Rpd303 (CNAG_05276) KO/att/mouse [34] Rpd304 (CNAG_05690) KO/att/mouse [92] | Hos2 (CNAG_05563) KO/att/mouse [34] | Hda1 (CNAG_01563) KO/av/mouse [94] KO/att/mouse [34] |
| Beauveria bassiana | Rpd3 (EJP69682.1) KO/att/Galleria [84] | Hos2 (XP_008603650.1) KO/att/Galleria [85] | Hda1/Clr3 (EJP66596.1) n.d. |
| Botrytis cinerea | Rpd3 (Bcin05g02590) OE/att/tomato [64] | Hos2 (Bcin01g03610) n.d. | Hda1 (Bcin15g02130) n.d. |
| Cochliobolus carbonum | Hdc2 (AAK35180.1) n.d. | Hdc1 (AAL56814.1) KO/att/maize [49] | AAP95014.1 n.d. |
| Fusarium graminearum | Rpd3/Hda3 (FGRAMPH1_01G01959) KO/av/wheat [77] | Hdf1/Hda2 (FGRAMPH1_01G03337) KO/att/wheat, corn [76] | Hdf2/Hda1 (FGRAMPH1_01G15009) n.d. |
| Ustilago maydis | Hda1 (U6MAG_02065) KO/fv/maize [32] KO/fv/maize [83] Hda2 (UMAG_11308) KO/fv/maize [83] | Hos2 (UMAG_11828) KO/att/maize [83] | Hda1/Clr3 (UMAG_02102) KO/att/maize [83] |
| Magnaporthe oryzae | Rpd3 (MGG_05857) OE/av/rice, barley [82] | Hos2 (MGG_01633) KO/av/rice, barley [63] KO/att/rice [79] | Hda1 (MGG_01076) KO/att/rice [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, I.; Graessle, S. Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds. Genes 2021, 12, 1470. https://doi.org/10.3390/genes12101470
Bauer I, Graessle S. Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds. Genes. 2021; 12(10):1470. https://doi.org/10.3390/genes12101470
Chicago/Turabian StyleBauer, Ingo, and Stefan Graessle. 2021. "Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds" Genes 12, no. 10: 1470. https://doi.org/10.3390/genes12101470
APA StyleBauer, I., & Graessle, S. (2021). Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds. Genes, 12(10), 1470. https://doi.org/10.3390/genes12101470






