Merging Genomics and Transcriptomics for Predicting Fusarium Head Blight Resistance in Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Statistical Analysis of the Phenotypic Data
2.3. Genotypic Data and Transcriptome Profiling
2.4. Single-Trait Omics-Based Prediction
2.5. Trait-Assisted and Single-Step Prediction Models
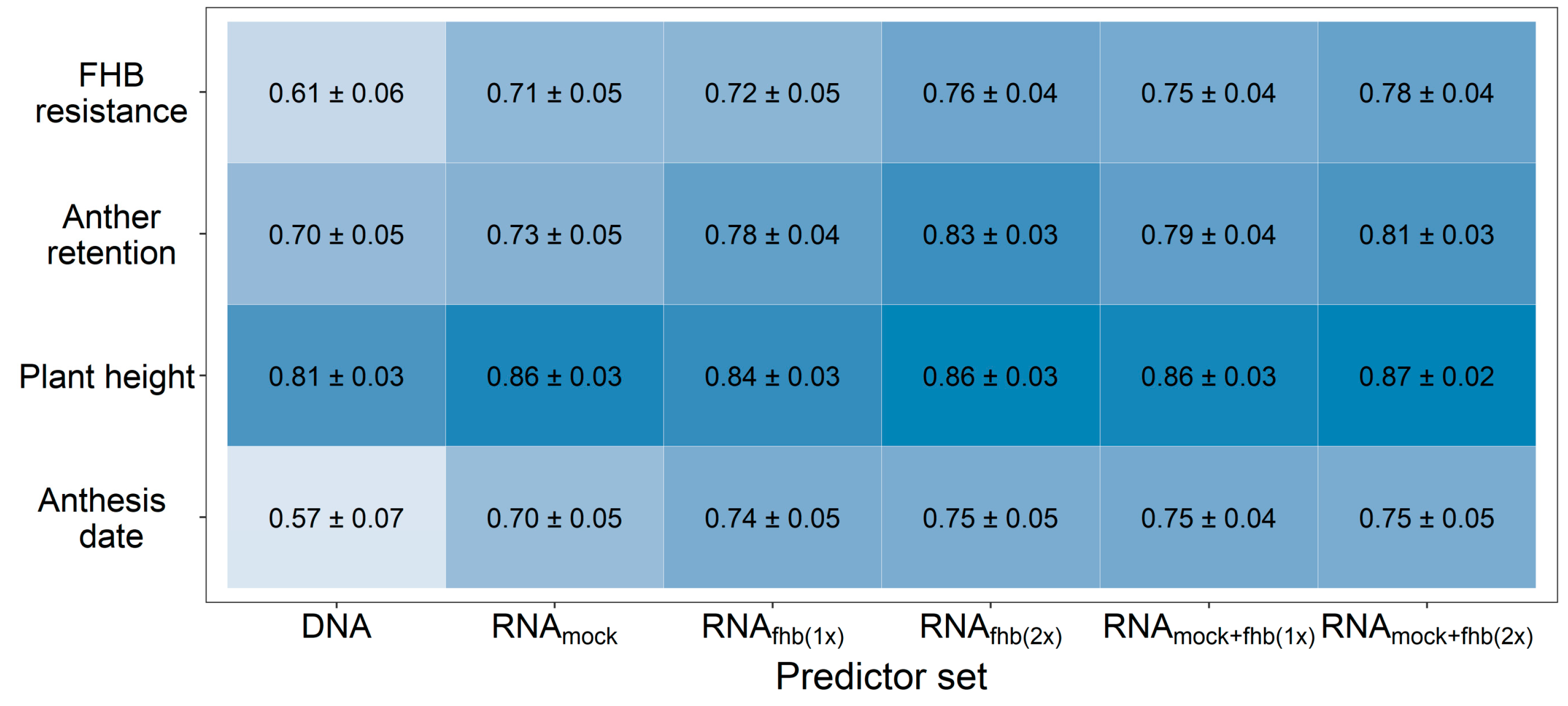
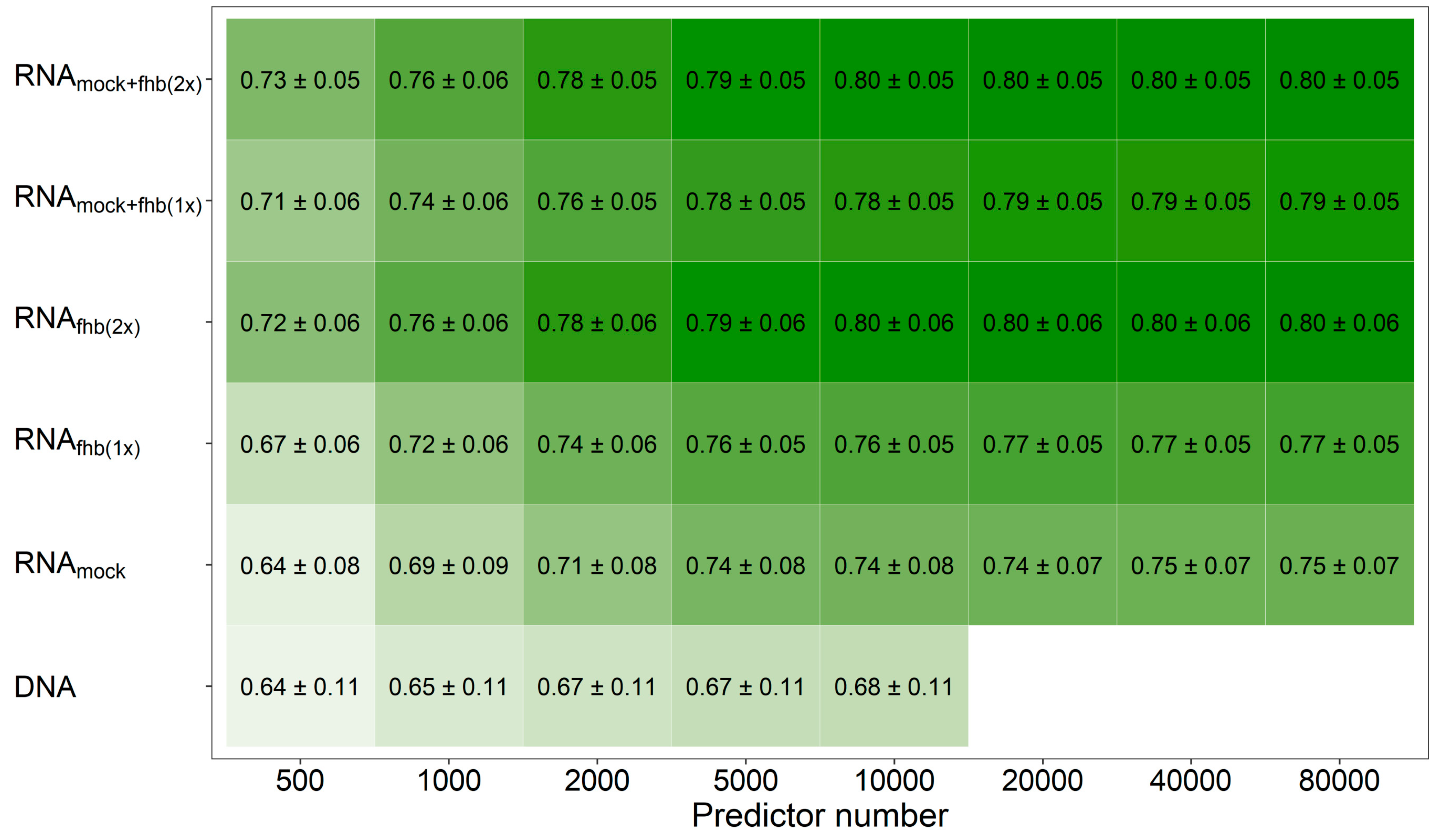
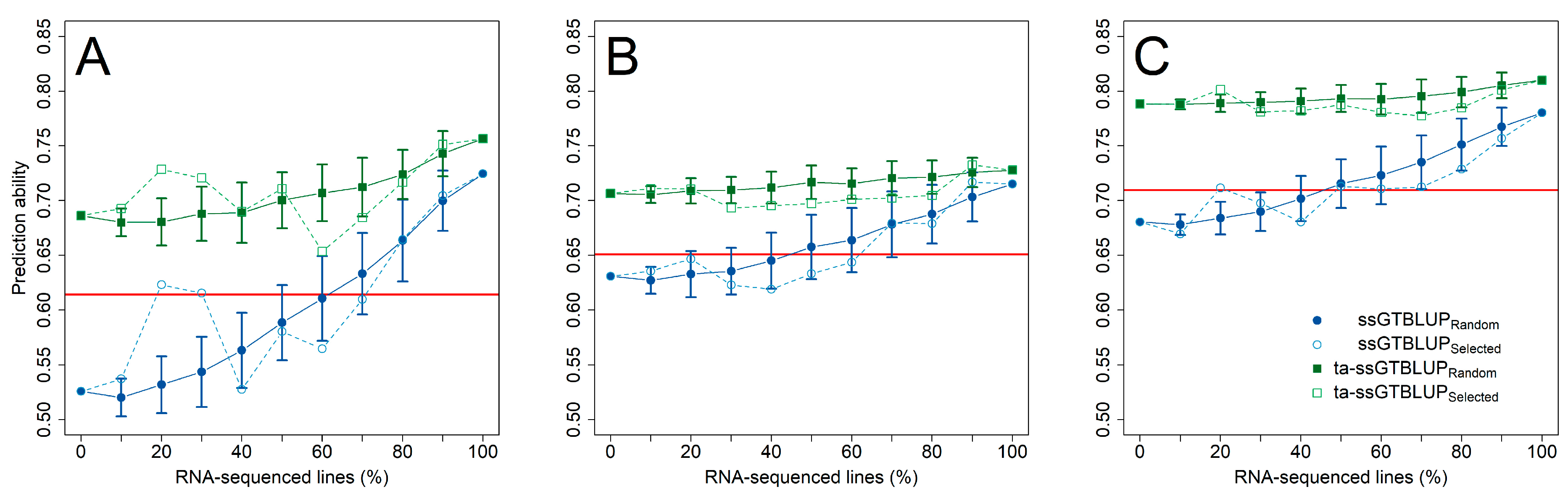
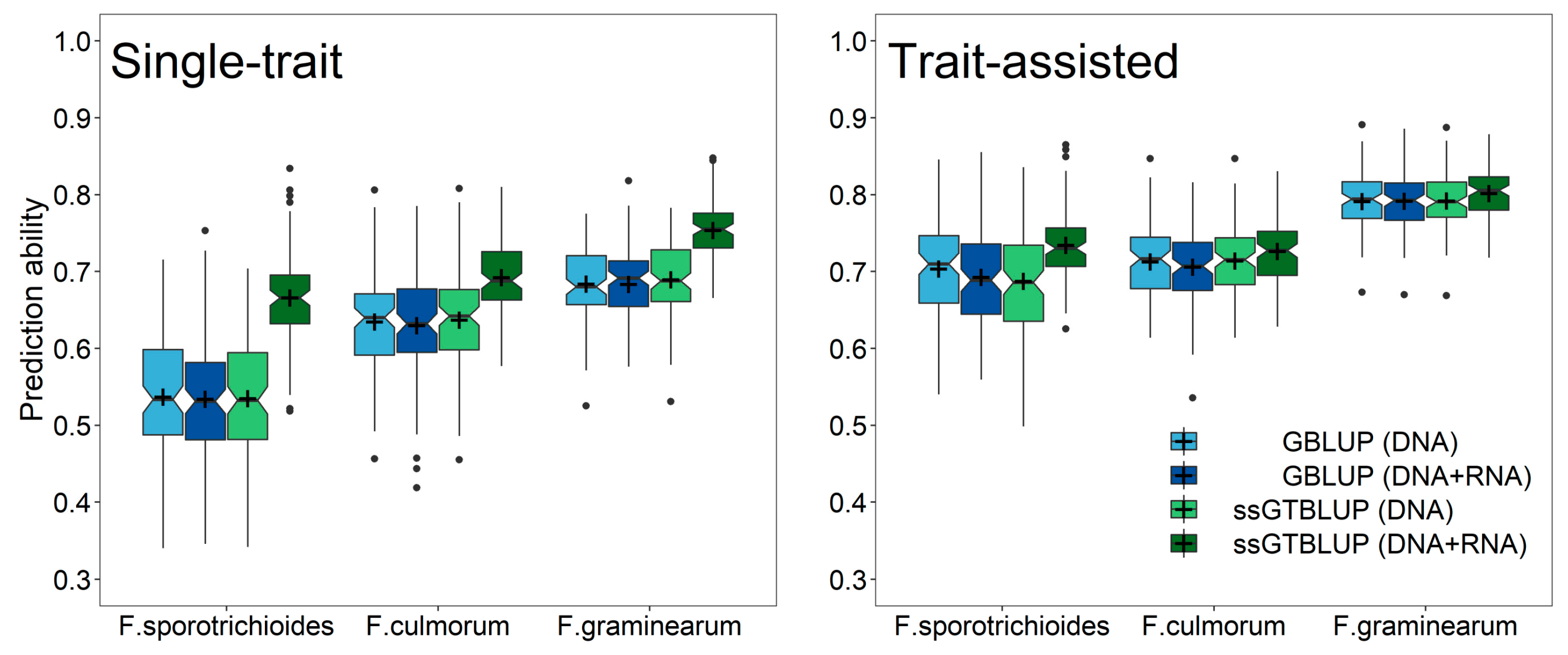
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robertsen, C.; Hjortshøj, R.; Janss, L. Genomic Selection in Cereal Breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef]
- Poland, J.; Rutkoski, J. Advances and Challenges in Genomic Selection for Disease Resistance. Annu. Rev. Phytopathol. 2016, 54, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Haile, J.K.; Diaye, A.N.; Sari, E.; Walkowiak, S. Potential of Genomic Selection and Integrating “Omics” Data for Disease Evaluation in Wheat. Crop. Breed. Genet. Genom. 2020, 2, e200016. [Google Scholar] [CrossRef]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1541–1568. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.P.; Lipka, A.E.; Brown, P.J.; Krill, A.M.; Thurber, C.; Brown-Guedira, G.; Dong, Y.; Foresman, B.J.; Kolb, F.L. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Lorenz, A.J.; Smith, K.P.; Jannink, J.L. Potential and optimization of genomic selection for Fusarium head blight resistance in six-row barley. Crop. Sci. 2012, 52, 1609–1621. [Google Scholar] [CrossRef]
- Galiano-Carneiro, A.L.; Boeven, P.H.G.; Maurer, H.P.; Würschum, T.; Miedaner, T. Genome-wide association study for an efficient selection of Fusarium head blight resistance in winter triticale. Euphytica 2019, 215, 4. [Google Scholar] [CrossRef]
- Rutkoski, J.; Benson, J.; Jia, Y.; Brown-Guedira, G.; Jannink, J.-L.; Sorrells, M. Evaluation of Genomic Prediction Methods for Fusarium Head Blight Resistance in Wheat. Plant. Genome 2012, 5, 51–61. [Google Scholar] [CrossRef]
- Borrenpohl, D.; Huang, M.; Olson, E.; Sneller, C. The value of early-stage phenotyping for wheat breeding in the age of genomic selection. Theor. Appl. Genet. 2020, 133, 2499–2520. [Google Scholar] [CrossRef]
- Schulthess, A.W.; Zhao, Y.; Longin, C.F.H.; Reif, J.C. Advantages and limitations of multiple-trait genomic prediction for Fusarium head blight severity in hybrid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2018, 131, 685–701. [Google Scholar] [CrossRef]
- Larkin, D.L.; Holder, A.L.; Mason, R.E.; Moon, D.E.; Brown-Guedira, G.; Price, P.P.; Harrison, S.A.; Dong, Y. Genome-wide analysis and prediction of Fusarium head blight resistance in soft red winter wheat. Crop. Sci. 2020, 60, 2882–2900. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Buerstmayr, H. The Semidwarfing Alleles Rht-D1b and Rht-B1b Show Marked Differences in Their Associations with Anther-Retention in Wheat Heads and with Fusarium Head Blight Susceptibility. Phytopathology 2016, 106, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; He, X.; Dreisigacker, S.; He, Z.; Singh, P.K. Anther extrusion and its association with Fusarium head blight in CIMMYT wheat germplasm. Agronomy 2020, 10, 47. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M. Transcriptomics of cereal–Fusarium graminearum interactions: What we have learned so far. Mol. Plant. Pathol. 2018, 19, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Riedelsheimer, C.; Czedik-Eysenberg, A.; Grieder, C.; Lisec, J.; Technow, F.; Sulpice, R.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat. Genet. 2012, 44, 217–220. [Google Scholar] [CrossRef]
- Schrag, T.A.; Westhues, M.; Schipprack, W.; Seifert, F.; Thiemann, A.; Scholten, S.; Melchinger, A.E. Beyond genomic prediction: Combining different types of omics data can improve prediction of hybrid performance in maize. Genetics 2018, 208, 1373–1385. [Google Scholar] [CrossRef]
- Gemmer, M.R.; Richter, C.; Jiang, Y.; Schmutzer, T.; Raorane, M.L.; Junker, B.; Pillen, K.; Maurer, A. Can metabolic prediction be an alternative to genomic prediction in barley? PLoS ONE 2020, 15, e0234052. [Google Scholar] [CrossRef]
- Guo, Z.; Magwire, M.M.; Basten, C.J.; Xu, Z.; Wang, D. Evaluation of the utility of gene expression and metabolic information for genomic prediction in maize. Theor. Appl. Genet. 2016, 129, 2413–2427. [Google Scholar] [CrossRef]
- Frisch, M.; Thiemann, A.; Fu, J.; Schrag, T.A.; Scholten, S.; Melchinger, A.E. Transcriptome-based distance measures for grouping of germplasm and prediction of hybrid performance in maize. Theor. Appl. Genet. 2010, 120, 441–450. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, Y.; Wang, X.; Ma, Y.; Li, P.; Yang, Z.; Zhang, X.; Xu, C.; Xu, S. Incorporation of parental phenotypic data into multi-omic models improves prediction of yield-related traits in hybrid rice. Plant. Biotechnol. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wei, J.; Li, R.; Qu, H.; Chater, J.M.; Ma, R.; Li, Y.; Xie, W.; Jia, Z. Identification of optimal prediction models using multi-omic data for selecting hybrid rice. Heredity 2019, 123, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Westhues, M.; Schrag, T.A.; Heuer, C.; Thaller, G.; Utz, H.F.; Schipprack, W.; Thiemann, A.; Seifert, F.; Ehret, A.; Schlereth, A.; et al. Omics-based hybrid prediction in maize. Theor. Appl. Genet. 2017, 130, 1927–1939. [Google Scholar] [CrossRef] [PubMed]
- Schrauf, M.F.; Martini, J.W.R.; Simianer, H.; de los Campos, G.; Cantet, R.; Freudenthal, J.; Korte, A.; Munilla, S. Phantom Epistasis in Genomic Selection: On the Predictive Ability of Epistatic Models. G3 2020, 10, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Weisweiler, M.; De Montaigu, A.; Ries, D.; Pfeifer, M.; Stich, B. Transcriptomic and presence/absence variation in the barley genome assessed from multi-tissue mRNA sequencing and their power to predict phenotypic traits. BMC Genom. 2019, 20, 787. [Google Scholar] [CrossRef] [PubMed]
- Westhues, M.; Heuer, C.; Thaller, G.; Fernando, R.; Melchinger, A.E. Efficient genetic value prediction using incomplete omics data. Theor. Appl. Genet. 2019, 132, 1211–1222. [Google Scholar] [CrossRef]
- Lourenco, D.; Legarra, A.; Tsuruta, S.; Masuda, Y.; Aguilar, I.; Misztal, I. Single-Step Genomic Evaluations from Theory to Practice: Using SNP Chips and Sequence Data in BLUPF90. Genes 2020, 11, 790. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Steiner, B.; Lemmens, M.; Ruckenbauer, P. Resistance to Fusarium Head Blight in Winter Wheat: Heritability and Trait Associations. Crop. Sci. 2000, 40, 1012–1018. [Google Scholar] [CrossRef]
- Runcie, D.; Cheng, H. Pitfalls and remedies for cross validation with multi-trait genomic prediction methods. G3 2019, 9, 3727–3741. [Google Scholar] [CrossRef]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the R Package sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020.
- Rimbert, H.; Darrier, B.; Navarro, J.; Kitt, J.; Choulet, F.; Leveugle, M.; Duarte, J.; Rivière, N.; Eversole, K.; Le Gouis, J.; et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 2018, 13, e0186329. [Google Scholar] [CrossRef] [PubMed]
- Stekhoven, D.J.; Bühlmann, P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Samad-Zamini, M.; Schweiger, W.; Nussbaumer, T.; Mayer, K.F.X.; Buerstmayr, H. Time-course expression QTL-atlas of the global transcriptional response of wheat to Fusarium graminearum. Plant. Biotechnol. J. 2017, 15, 1453–1464. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Endelman, J.B.; Jannink, J.-L. Shrinkage estimation of the realized relationship matrix. G3 2012, 2, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, L.; Rouseeuw, P. Finding Groups in Data: An Introduction to Cluster Analysis; Kaufman, L., Rousseeuw, P.J., Eds.; Wiley Series in Probability and Statistics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1990; ISBN 9780470316801. [Google Scholar]
- Hennig, C. Package ‘fpc’. Flexible Procedures for Clustering; University of Bologna: Bologna, Italy, 2020. [Google Scholar]
- Bustos-Korts, D.; Malosetti, M.; Chapman, S.; Biddulph, B.; van Eeuwijk, F. Improvement of Predictive Ability by Uniform Coverage of the Target Genetic Space. G3 2016, 6, 3733–3747. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Christensen, O.; Lund, M.S. Genomic relationship matrix when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef]
- Amadeu, R.R.; Cellon, C.; Olmstead, J.W.; Garcia, A.A.F.; Resende, M.F.R.; Muñoz, P.R. AGHmatrix: R Package to Construct Relationship Matrices for Autotetraploid and Diploid Species: A Blueberry Example. Plant. Genome 2016, 9, 1–10. [Google Scholar] [CrossRef]
- Hänsel, H. Yield potential of barley corrected for disease infection by regression residuals. Plant. Breed. 2001, 120, 223–226. [Google Scholar] [CrossRef]
- Kugler, K.G.; Siegwart, G.; Nussbaumer, T.; Ametz, C.; Spannagl, M.; Steiner, B.; Lemmens, M.; Mayer, K.F.X.; Buerstmayr, H.; Schweiger, W. Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genom. 2013, 14, 728. [Google Scholar] [CrossRef]
- Zenke-Philippi, C.; Thiemann, A.; Seifert, F.; Schrag, T.; Melchinger, A.E.; Scholten, S.; Frisch, M. Prediction of hybrid performance in maize with a ridge regression model employed to DNA markers and mRNA transcription profiles. BMC Genom. 2016, 17, 262. [Google Scholar] [CrossRef]
- Azodi, C.B.; Pardo, J.; VanBuren, R.; de Los Campos, G.; Shiu, S.H. Transcriptome-Based Prediction of Complex Traits in Maize. Plant. Cell 2020, 32, 139–151. [Google Scholar] [CrossRef]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant. Sci. 2017, 8, 592. [Google Scholar] [CrossRef]
- Wagner, C.; Buerstmayr, M.; Omony, J.; Nosenko, T.; Steiner, B.; Nussbaumer, T.; Samad-Zamini, M.; Mayer, K.F.X.; Buerstmayr, H. Fusarium head blight resistance in European winter wheat: Insights from genome-wide transcriptome analysis. manuscript in preparation.
- Lamarre, S.; Frasse, P.; Zouine, M.; Labourdette, D.; Sainderichin, E.; Hu, G.; Le Berre-Anton, V.; Bouzayen, M.; Maza, E. Optimization of an RNA-seq differential gene expression analysis depending on biological replicate number and library size. Front. Plant. Sci. 2018, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Uzbas, F.; Opperer, F.; Sönmezer, C.; Shaposhnikov, D.; Sass, S.; Krendl, C.; Angerer, P.; Theis, F.J.; Mueller, N.S.; Drukker, M. BART-Seq: Cost-effective massively parallelized targeted sequencing for genomics, transcriptomics, and single-cell analysis. Genome Biol. 2019, 20, 155. [Google Scholar] [CrossRef] [PubMed]
- Alpern, D.; Gardeux, V.; Russeil, J.; Mangeat, B.; Meireles-Filho, A.C.A.; Breysse, R.; Hacker, D.; Deplancke, B. BRB-seq: Ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef]
| Trial | |||
|---|---|---|---|
| Year | FS | FC | FG |
| 2011 | 26 (2) | 26 (4) | |
| 2012 | 58 (2) | 58 (2) | |
| 2013 | 96 (2) | 96 (2) | |
| 2014 | 96 (2) | ||
| 2015 | 96 (2) | 96 (2) | |
| Set | Trait | Min | Mean | Max | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All trials | FHB | 51.89 | 114.62 | 282.47 | 2338.49 | 0.00 | 629.45 | 10226.20 | 0.18 | 0.94 |
| AR | 0.00 | 52.96 | 93.13 | 734.03 | 68.81 | 22.42 | 156.29 | 0.75 | 0.97 | |
| PH | 65.71 | 78.23 | 104.68 | 72.36 | 3.74 | 0.00 | 10.84 | 0.83 | 0.98 | |
| AD | 20.20 | 24.67 | 30.00 | 5.62 | 0.85 | 0.00 | 1.36 | 0.72 | 0.97 | |
| Isolates † | FHBFS | 0.00 | 39.40 | 177.92 | 1308.28 | 335.18 | 831.78 | 0.53 | 0.91 | |
| FHBFC | 17.04 | 227.36 | 846.55 | 35605.89 | 3776.73 | 6610.13 | 0.77 | 0.95 | ||
| FHBFG | 69.34 | 201.10 | 452.40 | 7572.85 | 1877.78 | 5436.90 | 0.51 | 0.87 | ||
| Ind. Sel. ‡ | ARwoFS | 0.00 | 50.07 | 91.55 | 744.94 | 47.46 | 47.46 | 160.38 | 0.74 | 0.97 |
| ARwoFC | 0.00 | 50.55 | 84.34 | 568.15 | 68.44 | 24.75 | 174.89 | 0.68 | 0.93 | |
| ARwoFG | 0.00 | 54.45 | 93.73 | 782.06 | 42.69 | 42.69 | 129.45 | 0.78 | 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michel, S.; Wagner, C.; Nosenko, T.; Steiner, B.; Samad-Zamini, M.; Buerstmayr, M.; Mayer, K.; Buerstmayr, H. Merging Genomics and Transcriptomics for Predicting Fusarium Head Blight Resistance in Wheat. Genes 2021, 12, 114. https://doi.org/10.3390/genes12010114
Michel S, Wagner C, Nosenko T, Steiner B, Samad-Zamini M, Buerstmayr M, Mayer K, Buerstmayr H. Merging Genomics and Transcriptomics for Predicting Fusarium Head Blight Resistance in Wheat. Genes. 2021; 12(1):114. https://doi.org/10.3390/genes12010114
Chicago/Turabian StyleMichel, Sebastian, Christian Wagner, Tetyana Nosenko, Barbara Steiner, Mina Samad-Zamini, Maria Buerstmayr, Klaus Mayer, and Hermann Buerstmayr. 2021. "Merging Genomics and Transcriptomics for Predicting Fusarium Head Blight Resistance in Wheat" Genes 12, no. 1: 114. https://doi.org/10.3390/genes12010114
APA StyleMichel, S., Wagner, C., Nosenko, T., Steiner, B., Samad-Zamini, M., Buerstmayr, M., Mayer, K., & Buerstmayr, H. (2021). Merging Genomics and Transcriptomics for Predicting Fusarium Head Blight Resistance in Wheat. Genes, 12(1), 114. https://doi.org/10.3390/genes12010114





