Abstract
Eragrostis curvula presents mainly facultative genotypes that reproduce by diplosporous apomixis, retaining a percentage of sexual pistils that increase under drought and other stressful situations, indicating that some regulators activated by stress could be affecting the apomixis/sexual switch. Water stress experiments were performed in order to associate the increase in sexual embryo sacs with the differential expression of genes in a facultative apomictic cultivar using cytoembryology and RNA sequencing. The percentage of sexual embryo sacs increased from 4 to 24% and 501 out of the 201,011 transcripts were differentially expressed (DE) between control and stressed plants. DE transcripts were compared with previous transcriptomes where apomictic and sexual genotypes were contrasted. The results point as candidates to transcripts related to methylation, ubiquitination, hormone and signal transduction pathways, transcription regulation and cell wall biosynthesis, some acting as a general response to stress and some that are specific to the reproductive mode. We suggest that a DNA glycosylase EcROS1-like could be demethylating, thus de-repressing a gene or genes involved in the sexuality pathways. Many of the other DE transcripts could be part of a complex mechanism that regulates apomixis and sexuality in this grass, the ones in the intersection between control/stress and apo/sex being the strongest candidates.
1. Introduction
Apomixis refers to asexual propagation by seeds and it is a process composed of three components: apomeiosis, parthenogenesis and autonomous endosperm development or pseudogamy. Three hundred out of more than 400 species of angiosperms that reproduce by apomixis occur in the Poaceae, Asteraceae or Rosaceae [1]. However, some capacity for sexuality is usually maintained; thus, they benefit from using a very sophisticated combination of reproductive strategies, generating diversity and, concurrently, allowing the best fitted individuals to propagate clonally [2]. Due to its polyphyletic origin, there are numerous forms of apomixis: diplospory, apospory and adventitious embryogenesis [3]. The elucidation of the mechanism of apomixis is important not only for biological interest, but also for agricultural technology. In the agricultural industry, it is predicted that apomixis would decrease the cost of hybrid seed production significantly and increase the yield of existing inbred crops by converting them into high-yielding hybrids [4].
Although several comparative transcriptomic studies have already been performed in apomictic species, such as Pennisetum ciliare [5], Brachiaria brizantha [6], Poa pratensis [7], Panicum maximum [8], Paspalum simplex [9], Hieracium praealtum [10], Ranunculus auricomus [11], Boechera gunnisoniana [12] and Hypericum perforatum [13,14], and several candidate genes triggering specific components of apomixis are known, the function of their proteins is still not clear. Among the genes associated with the components of apomixis, found both in studies of apomictic species and in mutants that resemble apomixis, the following can be mentioned: SERK and APOSTART [15], DIF1 [16], BABY BOOM [17], APOLLO [18], DEMETER [19], MSII [20], RDR6 and SGS3 [21], DYAD/SWITCH1 [22,23], AGO9 [24], AGO104 [25], ORC [26], GID1 [27], FIE [28], AGAMOUS-LIKE 62 [29], PnTgs1-like [30] and DMC1 [31].
In organisms that can reproduce both sexually and asexually (facultative apomictic), stress plays an important role in determining which reproductive mode is used [32]. For example, in species with cyclical apomixis (like Daphnia pulex), reproduction is apomictic in one season, generally under favorable conditions, and sexual during stressful conditions [33]. Different species of fungi, algae and insects induce sexual reproduction under unfavorable conditions or in response to abiotic stress [34,35,36]. In plants, abiotic stress, such as drought or heat, can induce megaspore mother cells (MMCs) to undergo meiosis in the ovules of apomictic plants and produce genetically-reduced (sexual) embryo sacs [37,38,39,40]. Drought- and heat-stressed Boechera lignifera and B. gunnisoniana achieved major shifts from apomeiosis to meiosis in MMCs, whereas the non-stressed control plants exhibited 87–98% apomictic dyad formation, the heat- and water-stressed plants exhibited 75–80% sexual tetrad formation [38]. Differences in the photoperiod also induce increases in sexuality, such as in Themeda australis [41] and Ranunculus auricomus under long photoperiods [39] and in Paspalum cromyorrhizon [42] and Brachiaria brizantha [43] under short day conditions. The reproductive modes of other species also respond to environmental conditions; for instance, Gounaris et al. [44] detected a greater number of reduced embryo sacs under salt stress in Cenchrus ciliaris. Based on this, it is reasonable to expect that facultative apomicts tend to switch to sexual reproduction more often under stress conditions, and that such a stress-dependent switch facilitates the organism’s adaptation to a stressful environment [40].
Few studies associate the change in the frequency of apomictic/sexual embryo sacs under stress conditions with changes in gene expression. RNA-Seq studies conducted with immature pistils taken from drought-stressed and well-watered sexual and apomictic Boechera spp. plants show that this stress-induced switching includes global epigenetic-based changes in gene expression [45]. Gene ontology (GO) analyses of these differences in gene expression indicate that oxidative stress induces meiosis to occur instead of apomeiosis in apomictic Boechera [45]. Several authors, in different model plants, found that the genes that participate in stress pathways are related to the determination or regulation of apomixis [13,39,40].
Eragrostis curvula (weeping lovegrass), an African grass with cytotypes of different ploidy levels (e.g., 2x–8x) and displaying obligate and facultative apomixis and sexual reproduction [46], has become a model for the analysis of apomixis mechanisms, due to its particular diplosporous development (meiotic diplospory maintaining the same embryo: endosperm ploidy ratio as in sexual seeds). In recent years, the reproductive mode of this grass was studied extensively, providing information about the cytoembryological aspects of its apomictic–sexual development [47], differentially expressed (DE) transcripts [48,49,50,51], epigenetic aspects of apomictic regulation [52,53], mapping of the apomixis locus [54] and a high quality genome assembly [55]. Our group also demonstrated that under different internal and external stressful situations, including a change in ploidy, water stress, in vitro culture and intraspecific hybridization, the number of sexual embryo sacs increased in facultative apomictic plants of this grass [40,56]. Our group was able to observe that plants of the Tanganyika INTA cultivar produced fewer than 2% of sexual embryo sacs when growing in optimal conditions, but under different stress situations these plants showed an increase in the number of sexual embryo sacs [40]. This increase in the level of sexuality was associated with genetic and epigenetic changes, like methylation and molecular markers [40,56]. Evidence of epigenetic mechanisms controlling apomixis was also observed since differentially expressed patterns of RNA-directed DNA methylation (RdDM) genes [52] and microRNA between sexual and apomictic genotypes [53] were recently reported in this grass.
The aim of the present study was to identify genes that are differentially expressed in weeping lovegrass inflorescences of control and water-stressed facultative apomictic plants and to compare them with the differentially expressed genes between apomictic and sexual plants previously reported by our group [51]. This approach was taken based on previous findings about increases of sexual processes under stress conditions [40], in order to look for common pathways between stress and apomixis. It could give clues about the regulation of this intriguing reproductive mode.
2. Materials and Methods
2.1. Plant Materials
Plants of the tetraploid (2n = 4x = 40) facultative apomictic Don Walter cultivar were grown in 10 l pots in the greenhouse. Three plants coming from the same seed set (apomictic background) were divided asexually (two tillers each) and one tiller was assigned to the control treatment and the other one to the stress condition, totaling six plants. To avoid the noise represented by the genotype effect in a high heterozygous grass, we did not include a sexual or a full apomictic genotype as a control. For this reason we worked with clonal plants.
2.2. Stress Treatments
Plants were exposed to water stress conditions by water deprivation from three months before the onset of flowering until the end of the flowering season (September to March, 2016–2017). In order to maintain the biological functions of the plants, they were watered weekly with 50–80 mL per pot and a supplementary irrigation of an extra 100 mL per pot was carried out to induce the flowering close to the flowering season. Plants grown under normal conditions (300–500 mL water/week) were used as controls. As an indicator of the plant water status, the relative water content (RWC) was determined at the inflorescence collection time in leaves using the following formula: RWC (%) = (FM−DM)/(TM−DM) × 100, where FM, DM and TM are the fresh, dry, and turgid tissue weights, respectively [57].
2.3. Embryo Sacs Analyses
To analyze the effect of water stress on the reproductive mode, the different stages of megasporogenesis and megagametogenesis were observed under an optical microscope. Inflorescences from the control and treated plants were collected at the beginning of anthesis, when all embryo sac developmental stages were observable [47] and they were fixed in FAA (50% ethanol, 5% acetic acid, 10% formaldehyde in distilled water). Then, individual spikelets were dehydrated in a tertiary butyl alcohol series and embedded in Paraplast [58]. Samples were sectioned at 10 μm and stained with safranin-fast green. Observations were carried out with a Nikon Eclipse TE300 light transmission microscope. To assess the reproductive mode, the presence of meiosis or the number and position of nuclei in the embryo sac were observed according to Meier et al. [47]. More than three hundred spikelets were observed (41 from control plants and 271 from stressed ones).
2.4. RNA Extraction and Sequencing
Spikelets with basal flowers at the beginning of anthesis, containing embryo sacs at all developmental stages, were collected from control (DWC1, DWC2 and DWC3) and treated plants (DWS1, DWS2 and DWS3). In total, 30 mg of fresh tissue from each sample were ground to a fine powder using liquid nitrogen. The total RNA was extracted from the plant tissue as two fractions, small and large RNA, including RNA sequences smaller and larger than 200 bp, respectively, using a commercial NucleoSpin® miRNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. The large RNA fraction was sequenced in 150 bp reads in pair-end through an Illumina HiSeq1500 platform at INDEAR (Rosario, Argentina).
2.5. Bioinformatics Analyses
Quality assessments of the reads were performed using the FastQC software. Subsequently, the reads were filtered using the Trimmomatic software [59] with the following parameters: ILLUMINACLIP = TruSeq3-PE-2.fa:2:30:7:4:false, LEADING = 3, TRAILING = 3, SLIDINGWINDOW = 4 = 20, CROP = 150, HEADCROP = 13 and MINLEN = 36. The resulting paired-end reads were assembled using the Trinity software [60] with a KMER_SIZE:32. In order to remove redundant transcripts, a clustering with a sequence identity threshold of 0.9 was performed using the CD-HIT software [61,62]. A quality assessment of the assembly was made using the standard metrics provided by the downstream analysis of Trinity [63] and BUSCO [64,65].
Regarding the differential expression analysis, the transcript quantification (estimation of the abundance of each assembled transcript) was performed with RSEM software [66] using the trimmed paired and unpaired reads aligned by Bowtie2 as an input [67] with the parameters: fragment_length = 137 and fragment_std = 23. The differential expression analysis was carried out using the EDGE R-Cran package [68,69] and the DE transcripts were selected using a fold change (FC) = 2 and an e-value of 1e-3.
A functional annotation of the DE genes was performed with Blast2GO [70]. The distribution of level 2 and 3 GO terms—including biological process, cellular component and molecular function among the DE annotated transcripts—were plotted with Blast2GO. A comparative analysis of the GO terms containing the annotated down- and upregulated transcripts under stress conditions was performed and plotted on a bar chart. The KEGG pathways (Kyoto Encyclopedia of Genes and Genomes [71]) were also compared to detect differentially enriched pathways between the control and treated plants.
The Heatmap analysis was performed using the R Package pheatmap [68] using as an input the expression matrix used for the differential expression analysis.
2.6. Comparison with Previously Sequenced E. curvula Transcriptomes
The DE transcripts between the control and stressed plants and the DE transcripts between sexual and apomictic plants previously obtained [51] were compared in order to find a relationship between the genes/pathways related to stress/increase in sexuality and the genes/pathways involved in apomixis/sexuality using a unidirectional exonerate [72] alignment with a minimum identity of 90% and minimum coverage of 50% to match the sequences.
2.7. Search for Long Noncoding RNAs
Detection of long noncoding RNA (lncRNA) was carried out using the DE transcripts that could not be annotated with the Blast2GO software using the Magnoliopsida nonredundant protein database. The Coding Potential Calculator (CPC) software [73] was run using these transcripts as input, with the default parameters to detect the potential coding and lncRNA sequences. Finally, the lncRNA sequences annotated with the CPC software were searched in the dataset of DE transcripts between sexual and apomictic plants [51] in order to identify common transcript sequences.
2.8. Validation of Gene Expression by Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted as detailed above, under the same conditions and using the same RNA extraction kit used for the Illumina sequencing. The cDNA synthesis was performed using the ImProm II Reverse Transcription System (Promega) following the supplier’s instructions. The cDNA amplification was performed using specific primers designed according to the Integrated DNA technology (IDT) webpage (https://www.idtdna.com/scitools/Applications/RealTimePCR/). The primer pairs used in the qRT-PCR experiments are shown in Table S1. Real-time PCR reactions included 50 pmol of forward and reverse primers, 5 μL of cDNA diluted 100-fold and 10 μL of Real Mix (Biodynamics, Argentina). The amplification was carried out in a Rotor Gene 6000 thermocycler (Corbett Research, Australia). The expression level was normalized against the E. curvula UBICE gene (Table S1). The thermal cycling used for amplifications was as follows: 95 °C for 2 min, followed by 45 cycles at 94 °C for 10 s, then 15 s at the optimal annealing temperature for each primer pair and finally at 72 °C for 20 s using three biological and three technical replicates. The specificity of each reaction was verified through the dissociation curve profiles. To calculate the relative expression level and primer efficiency estimation, background-corrected raw fluorescence data were imported into LinRegPCR software version 11.0 [74,75]. The program uses a linear regression analysis to fit a straight line and estimate the PCR efficiency of each individual sample based on the slope of this line. The statistical analysis of the qRT-PCR fold change in the expression of genes among different treatments was analyzed using a Student’s t Test. A p-value of 0.05 was considered to be significant.
2.9. Data Availability
The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank
under the accession GIQX00000000. The version described in this paper
is the first version GIQX01000000.
3. Results
3.1. Percentage of Sexual vs. Apomictic Processes in Control and Water-Stressed Plants
The number of pistils and the percentages of sexual and apomictic embryo sacs from the control and water-stressed plants are shown in Table 1. The cytoembryology analyses showed that 4.97% of the embryo sacs were sexual (n = 161) in control plants, with an average RWC of 81.9%. Under water-stressed conditions, with an average RWC of 49.7%, the percentage of sexual embryo sacs increased to 23.84% (n = 172). The spikelets from stressed plants were smaller and had fewer flowers than the control ones. They also had a higher number of aborted seeds, so it was necessary to observe more spikelets to get an n = 172.

Table 1.
Percentage of sexual and apomictic embryo sacs in the control and water-stressed E. curvula plants (Don Walter cultivar) and the relative water content (RWC) for each treatment.
3.2. Sequencing and Assembly
Six RNA TruSeq HiSeq1500 Illumina libraries, corresponding to three biological replicates of control plants and three from water-stressed plants, were sequenced producing a total of 172,128,258 paired-end reads (2 × 150 bp). Then, after the quality analysis, performed with FastQC [76], the reads were trimmed resulting in 95,127,754 paired-end reads. These paired-end reads were de novo assembled (Trinity software) resulting in 305,798 transcripts. Finally, the redundant transcripts were removed and the remaining transcripts were clustered using the CDHIT software, resulting in a final number of 201,011.
Although an E. curvula genome assembly is available [55] and we could have used it as a reference genome, for this study we decided to de novo assemble the reads since the sequenced genome belongs to a sexual diploid genotype and the sequences corresponding to the region that determine apomixis might not be present, as was observed by Zappacosta et al. [54] using molecular markers linked to the trait. The assembly quality analysis, performed with Trinity stats, indicated an N50 of 1553 bp, an average contig length of 919.09 bp and a GC content of 46.38%. The percentage of aligned reads, determined by the Bowtie2 software, was 96.45% and the percentage of complete BUSCO genes in the final assembly was 87.2% (S = 54.6%, D = 32.6%), with 9.6% of fragmented and 3.2% of missing genes.
3.3. Differentially Expressed Transcript Analysis
A total of 501 DE transcripts were obtained with a fold change of two and an e-value of 1e-3. Out of these, 350 were downregulated while 151 were upregulated in the stressed plants. Table S2 shows all the information about the DE transcripts (ID and Blast, Blast2GO and KEGG analyses).
A principal component analysis (PCA) computed on the DE transcripts effectively separated the control from the treated samples, with the two first principal components explaining approximately 90% of the overall variance (Figure S1).
3.4. Gene Ontology Analysis
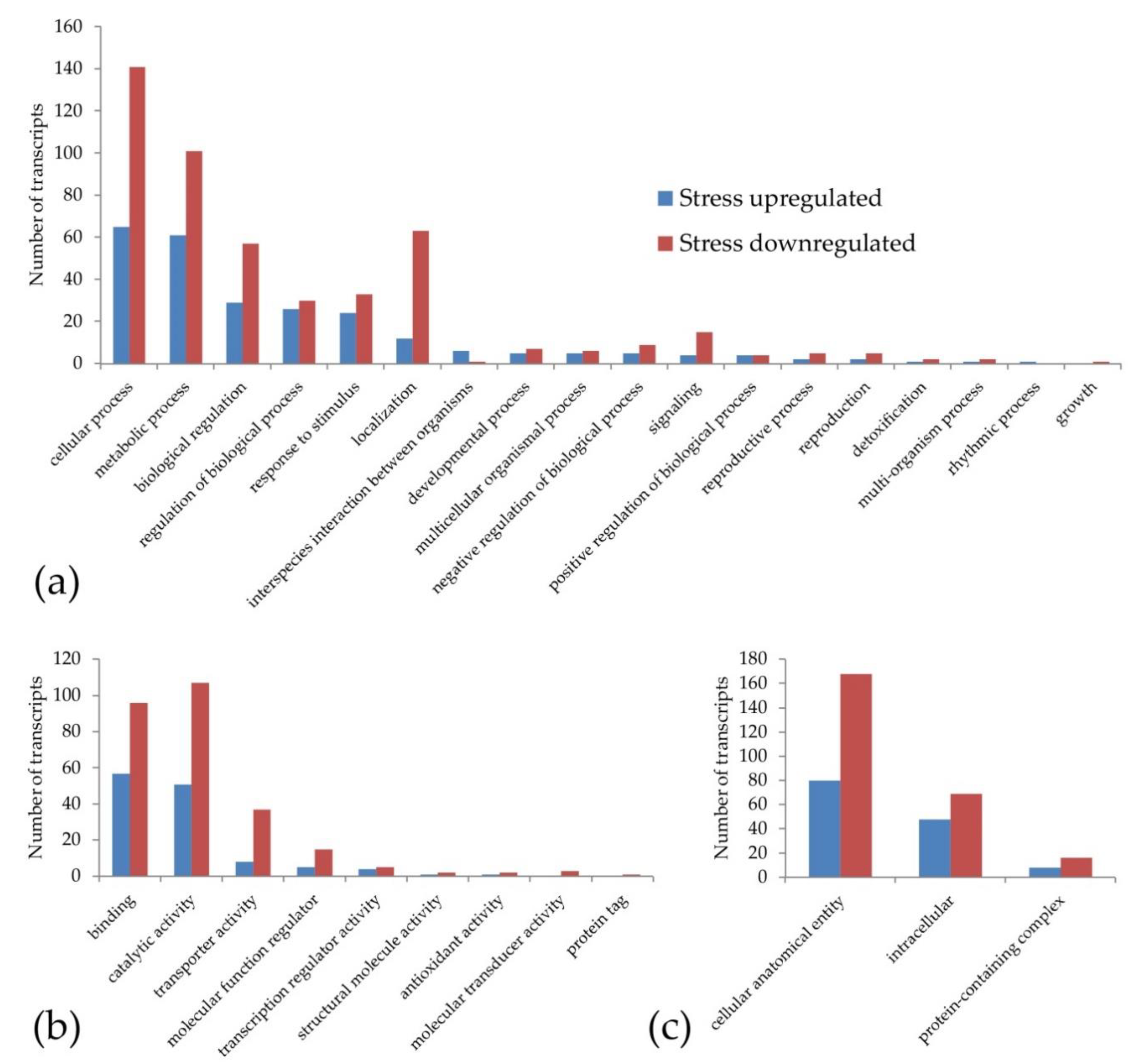
The gene ontology classification made in order to identify the pathways potentially associated with the increase in sexual pistils under stress conditions gave a result of 380 out of 501 DE annotated transcripts. This classification (Figure 1) shows that the transcripts upregulated and downregulated under stress treatments were included in the same main categories (except the rhythmic process and growth). However, the number of GO terms with downregulated transcripts was higher (264 downregulated vs. 116 upregulated under stress). This effect could be part of the general decrease in gene expression that happens under stress conditions due the lack of resources.
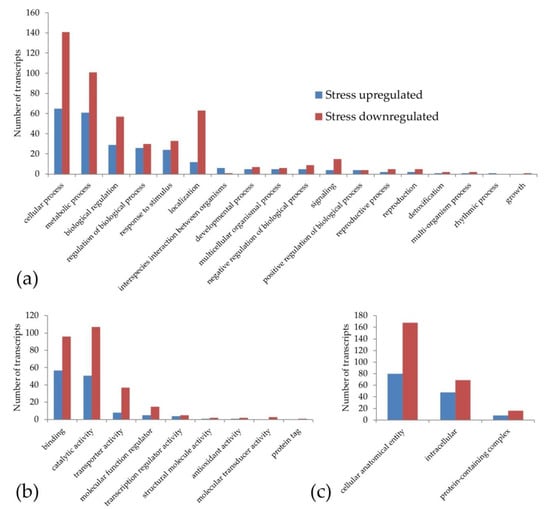
Figure 1.
Classification of the weeping lovegrass differentially expressed transcripts according to gene ontology: (a) biological process (b) molecular function and (c) cellular component. Each main category was classified at level 2. Blue bars show the transcripts that are upregulated and red bars represent the transcripts that are downregulated under stress conditions.
To carry out a more specific analysis, we searched for the presence of DE transcripts in the GO terms reproduction and reproductive processes. We found two upregulated DE transcripts with homology to a G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 GsSRK (DN35954_c0_g1_i7) and grassy tillers1 (DN35816_c1_g1_i1) and five downregulated DE transcripts under stress with homology to stromal processing peptidase (DN36893_c0_g1_i3), β-expansin (DN35576_c3_g4_i12, DN35576_c3_g4_i9), B3 domain-containing protein LFL1 (DN37977_c0_g1_i3) and indole-3-pyruvate monooxygenase YUCCA2 (DN36728_c1_g2_i3). Although some of these genes are related to stress responses, such as GsSRK, the overexpression of this gene in Arabidopsis exhibited more siliques at the adult developmental stage, among other traits [77]. All these DE transcripts are present in pathways that could participate in reproduction regulation, since they are involved in auxin pathways (YUCCA2, [9]), transcription regulation (grassy tillers1, [78]), embryogenesis (stromal processing peptidase, [79]), flowering time regulation (LFL1, [80]) and cell wall biosynthesis and pollen tube penetration through the stigma (β-expansin, [81]).
The GO terms only represented by upregulated transcripts at level 3 (Figure S2) were: chemical response, cell wall organization or biogenesis, response to biotic stimulus, response to external stimulus, response to other organisms, drug binding, cofactor binding, ligase activity, catalytic activity (acting on RNA), oxygen carrier activity, carbohydrate binding, envelope, RNA polymerase complex, thylakoid and Sm-like protein family complex. Although several of these GO terms may play a role in determining or regulating apomixis, we would like to highlight the ligase activity, where one of the differential transcripts shows homology with a putative E3 ubiquitin-protein ligase RING1a, an enzyme cited by other authors as a candidate to be involved in apomixis [82].
The GO level 3 terms that were only represented by downregulated transcripts (Figure S2) were: regulation of biological quality, regulation of molecular function, cell communication, signal transduction, response to abiotic stimulus, lyase activity, lipid binding, signaling receptor activity, isomerase activity, quaternary ammonium group binding, structural constituent of ribosome, membrane protein complex, extracellular space, external encapsulating structure and the microtubule associated complex. Among these GO terms, it can be highlighted that the membrane protein complex term contains two transcripts with homology to the AP-1 complex subunit sigma-1 and ENTH domain-containing protein, which were associated with SCD1-mediated vesicle transport and protein ubiquitination, a pathway cited by other authors as related to apomixis [83].
3.5. KEGG Pathway Classification
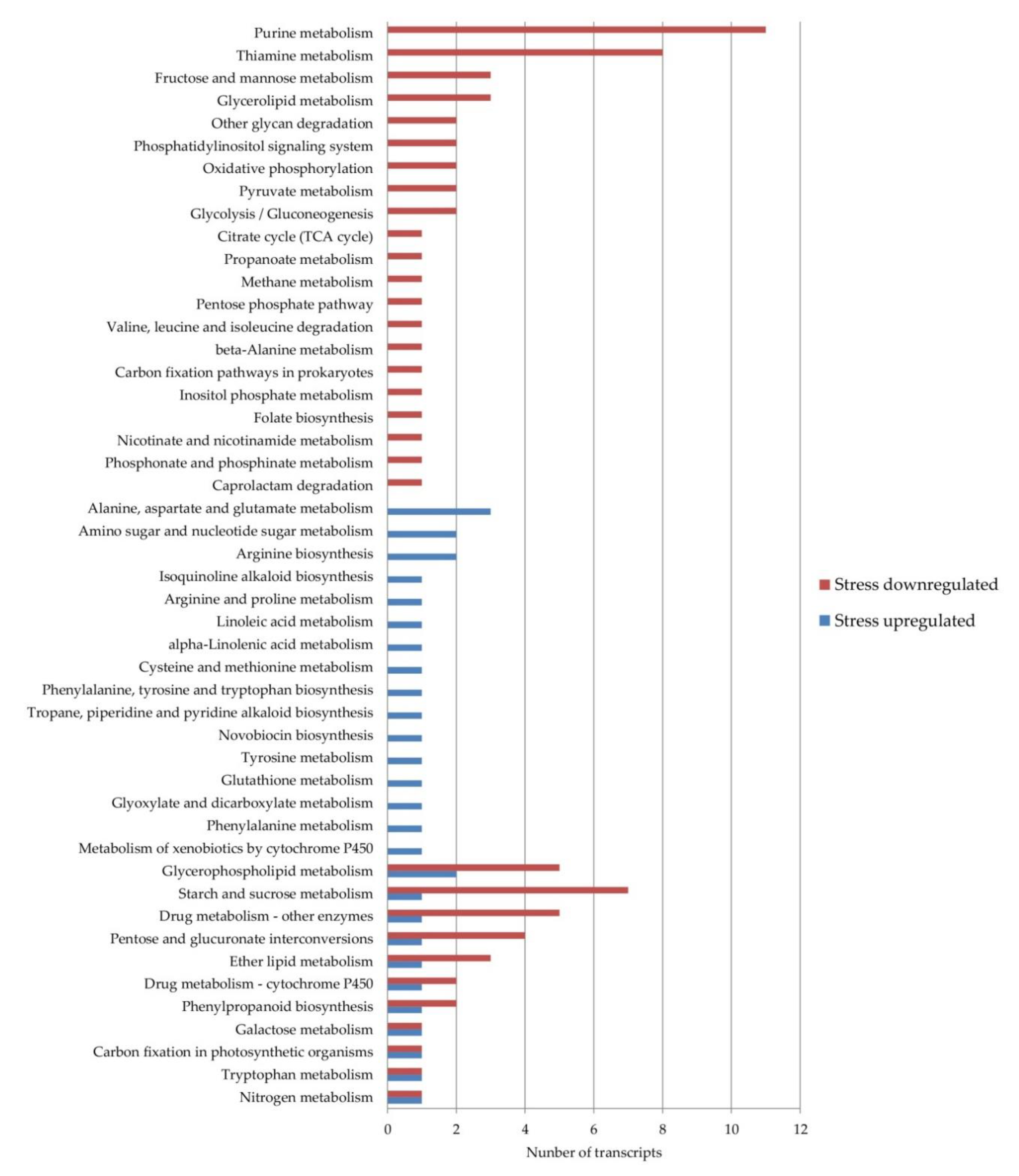
To identify the additional levels of regulation acting on the apomixis/sexuality switch under stress, the DE transcripts were analyzed using the KEGG database. Using the Blast2GO software, 13 transcripts that were upregulated and 37 that were downregulated under stress conditions were assigned to 48 different pathways (Figure 2).
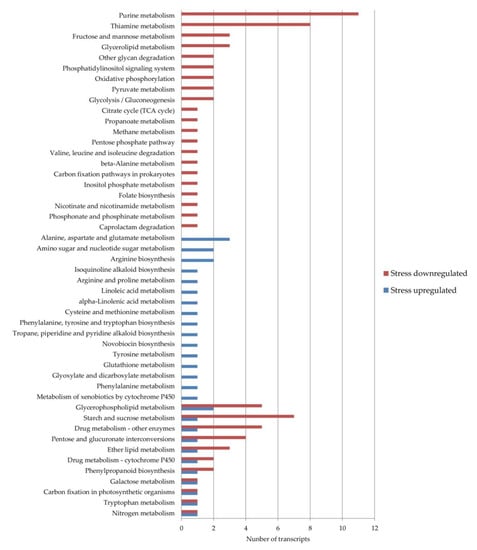
Figure 2.
Weeping lovegrass differentially expressed the transcripts present in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The blue bars show the number of transcripts upregulated and the red bars show the number of transcripts downregulated under stress conditions in each pathway.
In eleven pathways, DE transcripts from both categories, up- and downregulated, were present while 16 pathways were composed only by upregulated and 21 by downregulated transcripts under stress conditions. The pathways with the highest number of downregulated transcripts were in thiamine metabolism and purine metabolism. On the other hand, the pathways with the highest number of upregulated transcripts were the alanine, aspartate and glutamate metabolisms, arginine biosynthesis and the amino sugar and nucleotide sugar metabolisms. The upregulated transcripts included in more pathways were the enzymes aspartate aminotransferase cytoplasmic (ec:2.6.1.1—transaminase, DN38803_c1_g1_i6) and glutamine synthetase cytosolic isozyme 1–3 (ec:6.3.1.2—synthetase, DN38913_c0_g1_i4). The downregulated ones included the enzymes NADPH-dependent aldo–keto reductase (ec:1.1.1.2—dehydrogenase NADP+, DN38835_c1_g1_i3) and phosphoenolpyruvate carboxylase 4 (ec:4.1.1.31—carboxylase, DN19844_c0_g1_i1).
3.6. Comparison with Previous E. curvula Transcriptomes
In order to look for an association between the DE transcripts in plants under stress conditions and the increase in the number of sexual pistils, the results obtained in this study were compared with the DE transcripts obtained in a previous study (sexual vs. apomictic [51]), where two transcriptomes of weeping lovegrass, a fully sexual and a fully apomictic one, were sequenced.
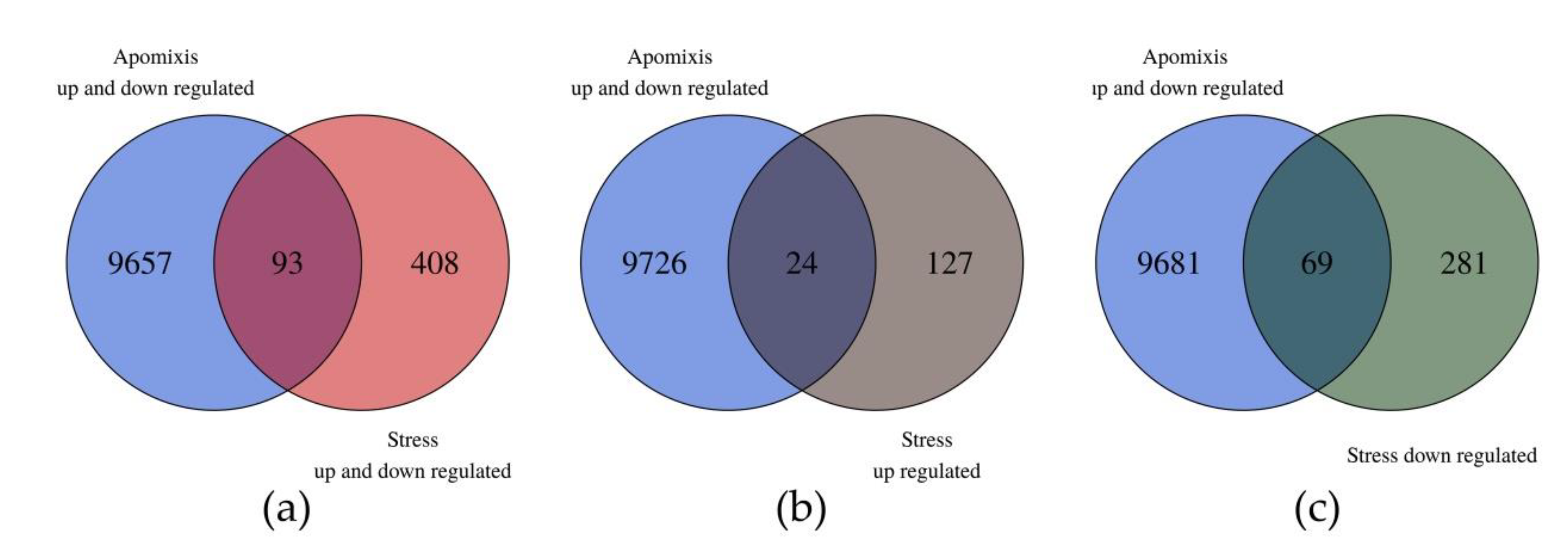
Ninety three out of the 501 DE transcripts showed homology in the unidirectional alignment with the 9750 DE transcripts reported by Garbus et al. [51]. Sixty nine out of the 93 DE transcripts belonged to the downregulated in the stress group and 24 to the upregulated one (Figure 3 and Table S2). From this analysis we can point out two interesting subgroups of transcripts that could be related to the increase in sexuality (Table 2 and Figure S3). The first one is composed of eight transcripts that were downregulated under stress conditions and upregulated in the apomictic genotype (in the apomixis/sexuality comparisons). The other group is composed of 20 transcripts that were upregulated under stress and downregulated in the apomictic genotype (Table 2).
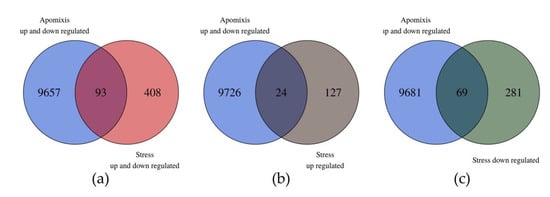
Figure 3.
Venn diagrams comparing the weeping lovegrass differentially expressed (DE) transcripts between control vs. stressed plants with the DE transcripts between apomictic vs. sexual plants. Data for the comparison apo/sex come from Garbus et al. [51]. (a) In the intersection between both groups of transcripts, there are 93 in common; (b) in the intersection are the common transcripts that are DE in apomictic plants (up- and downregulated) and downregulated under stress conditions; (c) in the intersection are the common transcripts that are DE in apomictic plants (up- and downregulated) and upregulated under stress conditions.

Table 2.
Differentially expressed transcripts between the control and stressed weeping lovegrass flowers showing homology with DE genes between apomixis/sexual flowers obtained previously in E. curvula by Garbus et al. [51]. In the description column the annotation provided by Blast2go was included.
Table 2 shows that there is more than one transcript for the same gene. It is important to remark that the β-expansin and grassy tillers1 genes were also annotated under the GO terms associated with reproduction. Another result to be highlighted from this analysis is a subtilisin (SBT5.3) downregulated under stress and overexpressed in apomictic plants. A member of this gene family, SBT1.4, was successfully validated by qRT-PCR. Subtilisins are proteases that are associated with the early stages of seed development [84] and reproductive mode [85,86].
3.7. Long Noncoding RNAs
From 501 DE transcripts blasted against the Magnoliopsida database, 452 were homologous with a coding sequence whereas 49 did not find a hit (Figure S2). Over these 49 transcripts introduced in the CPC software, 33 were predicted as noncoding with a high confidence. In total, 24 out of the 33 putative lncRNA were downregulated under stress conditions and nine were upregulated, representing 6% of the corresponding categories. Interestingly, 3 out of 33 of these RNAs were also found DE between apomictic and sexual plants in our previous work [51].
3.8. Analysis of Differentially Expressed Transcripts
The annotation of the DE transcripts that were not highlighted by the GO terms or KEGG pathways was also analyzed. Table 3 summarizes the annotated DE transcripts up- or downregulated under the stress treatment that have previously been mentioned by different authors in apomictic species or mutants. Among them, we can mention the transcription factor ethylene-responsive AINTEGUMENTA-like 5 (AIL5), belonging to the AP2 family, the same family as BABY BOOM (BBM), one of the few candidate genes for apomixis that has been confirmed to play a role in parthenogenesis [15]. Three DE transcripts were found associated with the brassinosteroid pathway (EXORDIUM, guanine nucleotide-binding protein α-1 subunit and serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B). This pathway has previously been mentioned by other authors as being associated with megagametogenesis in Poaceae [87] and apomixis induction [88].

Table 3.
Differentially expressed transcripts reported by different authors in different apomictic species compared with their sexual counterparts.
Another DE transcript upregulated under the stress treatment showed homology with MO25, a protein present in the apomixis-determining region in Hypericum perforatum [13] and associated with signal transduction (GO terms: intracellular signal transduction and positive regulation of protein serine/threonine kinase activity).
A very interesting candidate also related to apomixis that was found to be DE (upregulated under stress conditions) in the present study is a transcript with homology to the repressor of silencing ROS1A, a DNA-glycosylase that removes methylated cytosines and replaces them with unmethylated cytosines. If we postulate that sexuality is silenced in facultative apomictic plants, this gene is a very attractive candidate to de-repress this function by a demethylation pathway under stress situations, allowing an increase in the number of sexual pistils.
Among other genes previously related to apomixis and downregulated under stress conditions are the SNF1-related protein kinase regulatory subunit β-1 and F-box proteins (At2g14290, At3g07870, and At5g07610). Regarding genes previously related to apomixis and upregulated under stress, the transcription factor NAC10 should be mentioned.
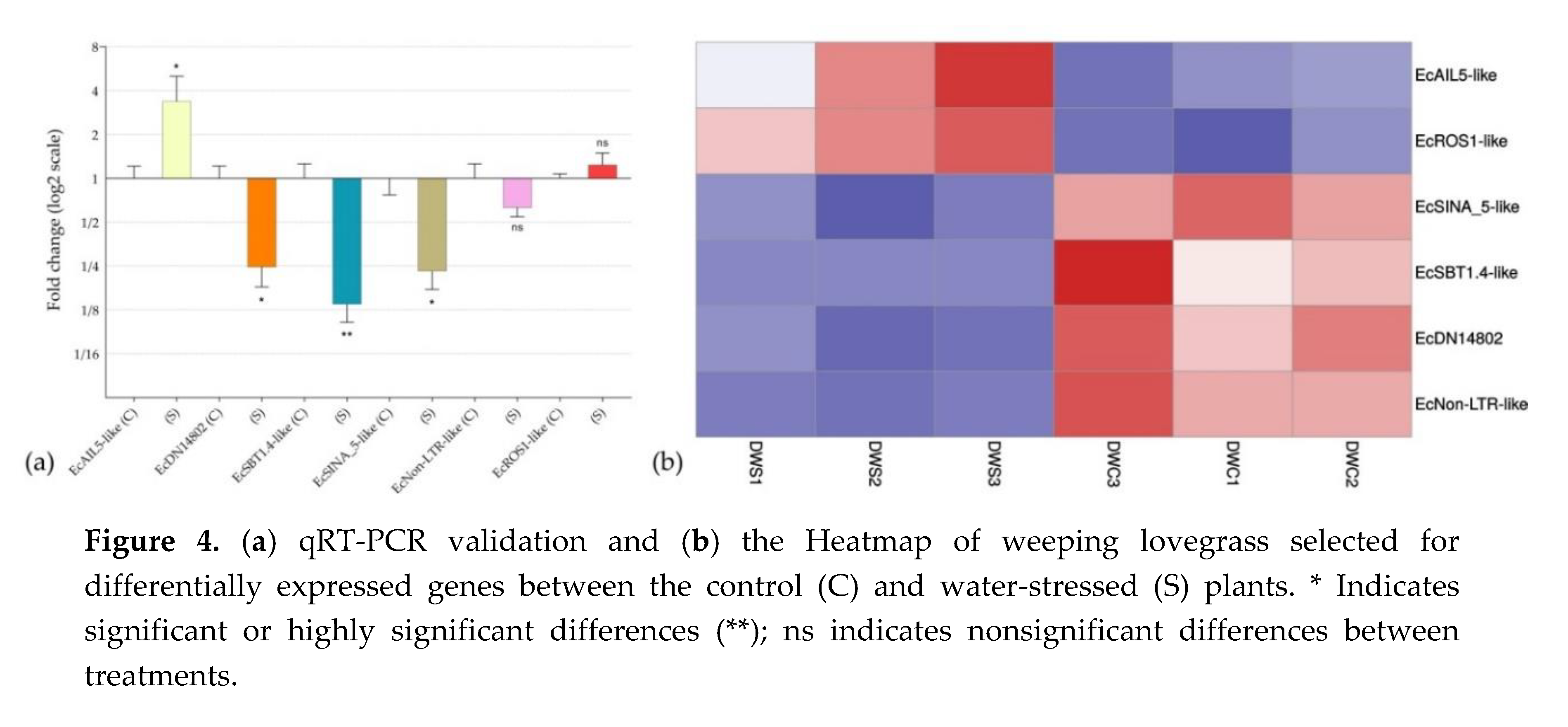
3.9. Validation by qRT-PCR
qRT-PCR assays were used to corroborate the in silico differential expression analysis of key genes (Figure 4a and Table S1), which were selected on the basis of their expression pattern and/or annotation. Heatmap (Figure 4b) was also used to show the significance of the in silico differential expression of the six selected genes.
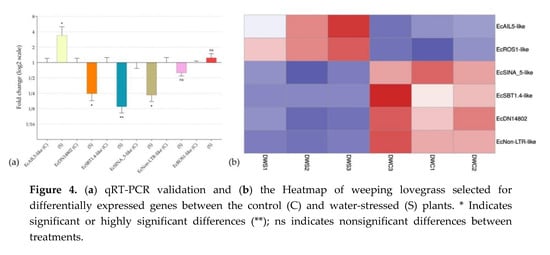
Figure 4.
(a) qRT-PCR validation and (b) the Heatmap of weeping lovegrass selected for differentially expressed genes between the control (C) and water-stressed (S) plants. * Indicates significant or highly significant differences (**); ns indicates nonsignificant differences between treatments.
EcAIL5-like, EcROS1-like and EcSINA_5-like, a transcription factor that belong to the AP2 family, a DNA glycosylase, and a putative E3 ubiquitin-protein ligase, respectively, were chosen for validation because they have previously been mentioned to be involved in pathways recently identified as having regulatory roles in apomixis.
An interesting transcript among those selected for qRT-PCR validation was EcDN14802, a transcript that was predicted as lncRNA, downregulated under stress conditions and not found in E. curvula sexual genotypes (OTA-S transcriptome and Victoria genome). These observations point out this transcript as a possible candidate related to apomictic processes. With the same criteria, the transcript EcNon-LTR-like was also selected. Both transcripts were found only in the apomictic genotype Don Walter.
EcSBT1.4-like is a stress downregulated protease that was selected for validation because it was previously found in E. curvula transcriptomes (Table 2) and because it was included in the GO terms reproductive process and reproduction.
Although EcNon-LTR-like and EcROS1-like were not statistically validated by qRT-PCR, they were very close and the Heatmap analysis showed a very consistent expression across all the three samples in each treatment.
4. Discussion
Our experimental design was aimed at finding a relationship between stress, the increase in sexual processes and the candidate genes involved using a facultative apomictic cultivar of E. curvula. Prior to the analysis, it is important to consider four aspects of the experimental design. Firstly is the fact that the experiments were conducted with clonal plants, so both the control and stressed plants have the same background. Secondly, the treatment involved water stress, hence some of the responses could be exclusively due to stress itself. Thirdly, the biological samples used consisted of whole flowers (spikelets), which comprise raquis, glumes, lemma, palea, ovary and anthers, therefore, the whole set of transcripts characterized here are derived from a variety of cell types, including somatic cells and male and female reproductive cells from premeiosis to anthesis. Finally, both treatments were conducted with a facultative apomictic cultivar, hence we expected to find mostly apomictic/sexual regulators, not apomictic triggers because they must be present in plants from both treatments. On the other hand, we are working with the knowledge that apomixis in E. curvula is determined by a genomic region [54] with an epigenetic component modulating the traits [40,52], and apomixis and sexuality co-exist in facultative plants [54], allowing us to hypothesize that sexuality is being repressed at different levels in apomictic plants. This repression is deregulated by different endogenous [56] and exogenous stresses [40], probably mediated by transcriptional and post transcriptional regulatory mechanisms.
In the present study, as in previous ones using this grass, it was shown that stress significantly increased the percentage of sexual processes, as was observed in the facultative apomictic Tanganyika and Don Walter cultivars, where the increase in sexual pistils ranged from 1.8 to 14.4% and from 4 to 22%, respectively [40,56]. Similar results were obtained in this study (4 to 24% increase) using plants of the Don Walter cultivar. This situation has also been observed in other plant species, such as Boechera, where Mateo de Arias [38] demonstrated that drought and heat stress caused a shift from apomeiosis to meiosis in female meiocytes when plants are stressed and the frequency of sexual embryo sacs was increased from an average of 10 to 30% in the drought-stress-treated plants, with similar percentages to the ones observed here in E. curvula.
Previous studies conducted in sexual and apomictic E. curvula flowers using different strategies, like expressed sequence tags (ESTs) [48,49], differential displays [50] and 454 sequencing [51], allowed us to detect that many of the DE transcripts are homologous to genes related with different environmental stresses. This fact, plus the observation that stress increases the frequency of sexual processes, encouraged us to look for an association between both findings. The present study using the Illumina platform represents a deeper and complete transcriptomic sequencing of spikelets from E. curvula that provides a great amount of information that can be used to look for DE genes potentially involved in the modulation of apomixis/sexuality. Despite the large number of assembled transcripts (201,011), few were DE between treatments (501). An important proportion of these transcripts could be annotated and classified under the GO terms and KEGG pathways. The nonannotated transcripts (49) were evaluated with the CPC software and 33 were predicted as long noncoding RNAs. The identification of these genes and the lncRNAs are a benchmark for understanding the common pathways between stress and apomixis.
The Blast2GO analysis at level 2 showed that the same GO terms are present in both treatments, having more downregulated than upregulated transcripts in each and this could be due to the overall stress response. An interesting finding was the presence of seven transcripts in the GO terms reproduction and reproductive process that are related to stress responses [77] and were also present in apo/sex comparisons made earlier in E. curvula [49], such as GsSRK. Other transcripts included in these terms are involved in auxin pathways (YUCCA2, [9]), transcription regulation (grassy tillers1, [78]), embryogenesis (stromal processing peptidase, [79]), flowering time regulation (LFL1, [80]), cell wall biosynthesis and the pollen tube penetration of the stigma (β-expansin, [81]) and could also be associated with apomixis in weeping lovegrass.
Based on homology or annotations, some of the DE transcripts are involved in pathways recently identified as having regulatory roles during apomixis [13,82,83]. One of these pathways is ubiquitination, in which a putative E3 ubiquitin-protein ligase RING1a and a sulfite exporter TauE/SafE family protein 3 were present, among others. Ubiquitination regulates nearly every aspect of cellular events in eukaryotes. It modifies intracellular proteins with the 76-amino acid polypeptide ubiquitin and destines them for proteolysis or activity alteration. The proteasomal degradation-mediated control of key cell cycle regulators and other targets influence reproductive fate decisions and germline development [93,94]. In this study, nine transcripts, representing seven genes that belong to the ubiquitination pathway, were found DE. Five out of these seven genes (E3 ubiquitin-protein ligase SINA-like 5, E3 ubiquitin-protein ligase SINA-like 10, U-box domain-containing protein 35, ubiquitin-NEDD8-like protein RUB2 and U-box domain-containing protein 15) were downregulated under stress conditions and two (putative E3 ubiquitin-protein ligase RING1a and sulfite exporter TauE/SafE family protein 3) were upregulated. This finding provides further evidence that tightly regulated protein degradation affecting cell cycle progression might be of crucial importance for governing the distinct specification and differentiation of apomictic and sexual germlines. Genes related to ubiquitin pathways were also observed in comparisons of apomictic and sexual weeping lovegrass [49,50,51] and Paspalum notatum flowers [95] and in ploidy related changes in the apomictic grass P. notatum [96].
Together with the ubiquitin pathway, F-box proteins were also mentioned by Zühl et al. [82] as differentially expressed and they might potentially play a role in the reproductive mode. Martelotto et al. [96] found transcripts homologous to F-box genes differentially expressed and related to ploidy changes in the apomictic species P. notatum. Here we found six downregulated F-box proteins and one upregulated. F-box proteins are part of Skp1–Cullin1–F-box protein (SCF) ligase complexes, acting in the polyubiquitination-mediated 26S proteasomal degradation [97]. Changes in this pathway due to the stress treatment could also be associated with the apomictic/sexual switch, as in ubiquitination.
Interestingly, when contrasted with previous data, four groups of DE transcripts were also found as DE in the comparison of apomixis vs. sexuality in weeping lovegrass [51]. In Table 2 we highlighted two groups. The first group is composed of eight transcripts that were downregulated under stress conditions and upregulated in the apomictic genotype (in the apo/sex comparisons). The second group is composed of 20 transcripts that were upregulated under stress and downregulated in the apomictic genotype. These 20 transcripts could be the ones that are associated to the increase in sexual embryo sacs. In the first group, transcripts presented homology with subtilisins, β-expansin, pollen allergen Cyn d 15, glycine-rich proteins, guanine nucleotide-binding proteins, pectine esterases inhibitors, cysteine proteases and a long noncoding RNA. These genes are mainly involved in cell wall modifications, proteolysis, signal transduction and a possible regulatory function (lncRNA). In the second group, we can mention transcription factors, such as the NAC domain containing protein, grassy tillers1 and HOX12; lipoxygenases; tryptamine hydroxycinnamoyl transferases and SNF1-type serine–threonine protein kinases. The latter gene belongs to the SnRK complex and another component of this pathway was found as DE in the present study (MARD1). The SnRK1 kinases control metabolism, growth and development, and stress tolerance by the direct phosphorylation of metabolic enzymes and regulatory proteins and by extensive transcriptional regulation. SnRK1 is also part of a more elaborate metabolic and stress signaling network, which includes the TOR kinase and ABA-signaling. The plant SnRK1–TOR system is heavily intertwined also with hormone signaling pathways, mainly auxins and brassinosteriods. Many components of these complex pathways were found as DE in the present study, and although they are related to stress responses, they may also be involved in the apo–sex ratio change. Mateo de Arias [38] also finds the DE components of these pathways and Gao [89] proposes that the differences in gene expression between apomictic and sexual plants might be triggered by the presence or absence of stress signaling and showed strong evidence that the TOR–brassinosteroids molecular stress response pathway is involved in the apomeiosis/meiosis switch. Recently, Carman et al. [91] patented components of these pathways as apomixis related, which reinforced the involvement of these genes, not only in the stress response, but also in the reproductive mode regulation.
Several genes related to hormones, such as auxins and brassinosteriods, could also be acting, since a number of genes involved in hormone perception and homeostasis, including cytokinins, auxins, and brassinosteroids, were DE in apomictic pistils [9,85,88]. Auxins and brassinosteroids are pathways that respond to stress and are considered to be key components for cell dedifferentiation (totipotence phenomenon) related to somatic embryogenesis [98], a situation that is similar to apomixis.
Another transcript that is overexpressed under stress conditions in the present study is EcROS1-like, a DNA glycosylase with a high homology to ROS1 (repressor of silencing 1) a DNA demethylase that is indispensable in both male and female gametophyte development [90]. The mutation of this gene in Arabidopsis showed DNA hypermethylation (an increased level of methylated cytosine) at nearly 5,000 loci involved in different pathways [99]. We suggest that EcROS1-like, as a specific or an unspecific stress response, could be demethylating a key target, thus de-repressing some gene or genes involved in sexuality pathways that are silenced in apomictic plants. Demethylation together with other pathways found here as DE could be part of the complex mechanism that regulates apomixis and sexuality in this grass, the ones in the intersection between control/stress and apo/sex being the strongest candidates.
In general, our results mainly point to the involvement of demethylation, protein degradation, transcriptional and post transcriptional regulatory mechanisms and regulation by plant hormones and signal transduction in the apomixis/sexuality switch regulation under stress situations.
5. Conclusions
These data, together with previous results [40], reinforce our previous hypothesis related to apomixis regulation in weeping lovegrass and its connections with stress. In this model, both pathways—the apomictic and the sexual one—co-exist in facultative apomictic plants, the sexuality mainly being repressed or expressed at very low levels under normal conditions. Stress situations can de-repress sexuality and the number of sexual embryo sacs increases in apomictic plants. We suggest that EcROS1-like can be demethylating, thus de-repressing some gene or genes involved in the sexuality pathways. Many of the other transcripts found as DE could be part of the complex mechanism that regulate apomixis and sexuality in this grass, the ones in the intersection between control/stress and apo/sex being the strongest candidates. Probably some of them are being upregulated under stress by this general demethylation response. Other related processes involved are ubiquitination, hormone and signal transduction pathways, transcription regulation and cell wall biosynthesis, some acting as a general response to stress and some that can be specific to the reproductive mode.
Finally, the availability of the sequence database reported here would make possible the characterization and validation of the numerous genes involved in apomixis and stress.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/9/969/s1, Figure S1: Principal component analysis (PCA) of the first two principal components representing 90.6% of the total variance using differentially expressed transcripts from control (DWC1, DWC2 and DWC3) and water-stressed (DWS1, DWS2 and DWS3) samples of E. curvula plants; Figure S2: Classification of the weeping lovegrass differentially expressed transcripts according to gene ontology (GO): (a) biological process (b) molecular function and (c) cellular component. Each main category was classified at level 3. Blue bars show the transcripts that are upregulated and red bars represent the transcripts that are downregulated under stress conditions; Figure S3: Venn diagrams comparing weeping lovegrass differentially expressed (DE) transcripts between control vs. stressed plants with the DE transcripts between apomictic vs. sexual plants. Data for the comparison apo/sex come from Garbus et al. [51]. (a): In the intersection are the common transcripts that are downregulated under stress conditions and downregulated in apomictic plants, (b): in the intersection are the common transcripts that are upregulated under stress conditions and downregulated in apomictic plants, (c): in the intersection are the common transcripts that are downregulated under stress conditions and upregulated in apomictic plants, (d): in the intersection are the common transcripts that are upregulated under stress conditions and upregulated in apomictic plants; Table S1: Primer pairs used for the qRT-PCR validation of transcripts expressed in control and water-stressed apomictic plants of E. curvula Don Walter cultivar, Table S2: DE transcripts information: transcript ID, Blast, Blast2GO and KEGG analyses results.
Author Contributions
Conceptualization, J.P.S., J.M.R., D.Z. and V.E.; methodology, J.P.S., D.Z. and J.C.; software, J.C. and C.A.G.; cytoembryological analysis, J.P.S., J.M.R. and J.G.; validation, J.P.S. and A.B.; writing—original draft preparation, J.P.S., D.Z., J.C.; writing—review and editing, J.P.S., D.Z., J.C. and V.E.; project administration, V.E.; funding acquisition, V.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, PICT Raíces 2014–1243 and PICT Raíces 2017–0879), Universidad Nacional del Sur (PGI 24/A199), Consejo Nacional de Investigaciones Científicas y Técnologicas (CONICET) and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No. 645674 (PROCROP).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Carman, J.G. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Hand, M.L.; Koltunow, A.M. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Koltunow, A.M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 1993, 5, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Carman, J.G. Apomixis for crop production: Status of technology development and commercialization implications. WJILDR 2004, 12, 29–48. [Google Scholar]
- Singh, M.; Burson, B.L.; Finlayson, S.A. Isolation of candidate genes for apomictic development in buffelgrass (Pennisetum ciliare). Plant Mol. Biol. 2007, 64, 673–682. [Google Scholar] [CrossRef]
- Rodrigues, J.C.; Cabral, G.B.; Dusi, D.M.; de Mello, L.V.; Rigden, D.J.; Carneiro, V.T. Identification of differentially expressed cDNA sequences in ovaries of sexual and apomictic plants of Brachiaria brizantha. Plant Mol. Biol. 2003, 53, 745–757. [Google Scholar] [CrossRef]
- Albertini, E.; Marconi, G.; Barcaccia, G.; Raggi, L.; Falcinelli, M. Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol. Biol. 2004, 56, 879–894. [Google Scholar] [CrossRef]
- Yamada-Akiyama, H.; Akiyama, Y.; Ebina, M.; Xu, Q.; Tsuruta, S.I.; Yazaki, J.; Kishimoto, N.; Kikuchi, S.; Takhara, M.; Takamizo, T.; et al. Analysis of expressed sequence tags in apomictic guineagrass (Panicum maximum). J. Plant Physiol. 2009, 166, 750–761. [Google Scholar] [CrossRef]
- Polegri, L.; Calderini, O.; Arcioni, S.; Pupilli, F. Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. J. Exp. Bot. 2010, 61, 1869–1883. [Google Scholar] [CrossRef]
- Okada, T.; Hu, Y.; Tucker, M.R.; Taylor, J.M.; Johnson, S.D.; Spriggs, A.; Tsuchiya, T.; Oelkers, K.; Rodrigues, J.C.M.; Koltunow, A.M. Enlarging cells initiating apomixis in Hieracium praealtum transition to an embryo sac program prior to entering mitosis. Plant Physiol. 2013, 163, 216–231. [Google Scholar] [CrossRef]
- Pellino, M.; Hojsgaard, D.; Schmutzer, T.; Scholz, U.; Hörandl, E.; Vogel, H.; Sharbel, T.F. Asexual genome evolution in the apomictic Ranunculus auricomus complex: Examining the effects of hybridization and mutation accumulation. Mol. Ecol. 2013, 22, 5908–5921. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Schmid, M.W.; Klostermeier, U.C.; Qi, W.; Guthoerl, D.; Sailer, C.; Waller, M.; Resenstiel, P.; Grossniklaus, U. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet. 2014, 10, e1004476. [Google Scholar] [CrossRef] [PubMed]
- Schallau, A.; Arzenton, F.; Johnston, A.J.; Hähnel, U.; Koszegi, D.; Blattner, F.R.; Altschmied, L.; Haberer, G.; Barcaccia, G.; Bäumlein, H. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010, 62, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Galla, G.; Basso, A.; Grisan, S.; Bellucci, M.; Pupilli, F.; Barcaccia, G. Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) accessions. Front. Plant Sci. 2019, 10, 654. [Google Scholar] [CrossRef] [PubMed]
- Albertini, E.; Marconi, G.; Reale, L.; Barcaccia, G.; Porceddu, A.; Ferranti, F.; Falcinelli, M. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiol. 2005, 138, 2185–2199. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.M.; Lister, C.; Page, T.; Fransz, P.; Findlay, K.; Jones, G.H.; Dickinson, H.G.; Dean, C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999, 19, 463–472. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Corral, J.M.; Vogel, H.; Aliyu, O.M.; Hensel, G.; Thiel, T.; Kumlehn, J.; Sharbel, T.F. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013, 163, 1660–1672. [Google Scholar] [CrossRef]
- Schoft, V.K.; Chumak, N.; Choi, Y.; Hannon, M.; Garcia-Aguilar, M.; Machlicova, A.; Slusarz, L.; Mosiolek, M.; Park, J.S.; Park, G.T.; et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 2011, 108, 8042–8047. [Google Scholar] [CrossRef]
- Guitton, A.E.; Berger, F. Loss of function of multicopy suppressor of IRA1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 2005, 15, 750–754. [Google Scholar] [CrossRef]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, I.; Ganesh, G.; Grossniklaus, U.; Subbiah, V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development 2000, 127, 197–207. [Google Scholar] [PubMed]
- Ravi, M.; Marimuthu, M.P.A.; Siddiqi, I. Gamete formation without meiosis in Arabidopsis. Nature 2008, 451, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628. [Google Scholar] [CrossRef]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Siena, L.A.; Ortiz, J.P.A.; Calderini, O.; Paolocci, F.; Cáceres, M.E.; Kaushal, P.; Grisan, S.; Pessino, S.C.; Pupilli, F. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J. Exp. Bot. 2016, 67, 1965–1978. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; de Alencar Dusi, D.M.; Irsigler, A.S.T.; Gomes, A.C.M.M.; Mendes, M.A.; Colombo, L.; de Campos Carneiro, V.T. GID1 expression is associated with ovule development of sexual and apomictic plants. Plant Cell Rep. 2018, 37, 293–306. [Google Scholar] [CrossRef]
- Ohad, N.; Yadegari, R.; Margossian, L.; Hannon, M.; Michaeli, D.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 1999, 11, 407–416. [Google Scholar] [CrossRef]
- Hehenberger, E.; Kradolfer, D.; Köhler, C. Endosperm cellularization defines an important developmental transition for embryo development. Development 2012, 139, 2031–2039. [Google Scholar] [CrossRef]
- Siena, L.A.; Ortiz, J.P.A.; Leblanc, O.; Pessino, S. PnTgs1-like expression during reproductive development supports a role for RNA methyltransferases in the aposporous pathway. BMC Plant Biol. 2014, 14, 297. [Google Scholar] [CrossRef]
- Zhao, L.; He, J.; Cai, H.; Lin, H.; Li, Y.; Liu, R.; Yang, Z.; Qin, Y. Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J. 2014, 80, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Nedelcu, A.; Marcu, O.; Michod, R. Sex as a response to oxidative stress: A twofold increase in cellular reactive oxygen species activates sex genes. Proc. R. Soc. B Biol. Sci. 2004, 271, 1591–1596. [Google Scholar] [CrossRef] [PubMed]
- Suomalainen, E.; Saura, A.; Lokki, J. Cytology and Evolution in Parthenogenesis; CRC Press, Inc.: Boca Raton, FL, USA, 1987. [Google Scholar]
- Berman, J.; Hadany, L. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 2012, 28, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Nedelcu, A.; Michod, R. Sex as a response to oxidative stress: The effect of antioxidants on sexual induction in a facultatively sexual lineage. Proc. R. Soc. B Biol. Sci. 2003, 270, S136–S139. [Google Scholar] [CrossRef]
- Hiruta, C.; Tochinai, S. How does the alteration of meiosis evolve to parthenogenesis?—Case study in a water flea, Daphnia pulex. In Meiosis-Molecular Mechanisms and Cytogenetic Diversity; Swan, A., Ed.; InTech: Rijeka, Croatia, 2012; pp. 109–122. [Google Scholar]
- Carman, J.G.; Jamison, M.; Elliott, E.; Dwivedi, K.K.; Naumova, T.N. Apospory appears to accelerate onset of meiosis and sexual embryo sac formation in sorghum ovules. BMC Plant Biol. 2011, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Mateo de Arias, M. Effects of Plant Stress on Facultative Apomixis in Boechera (Brassicaceae). Ph.D. Thesis, Utah State University, Logan, UT, USA, 2015. [Google Scholar]
- Klatt, S.; Hadacek, F.; Hodac, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef]
- Rodrigo, J.M.; Zappacosta, D.; Selva, J.P.; Garbus, I.; Albertini, E.; Echenique, V. Apomixis frequency under stress conditions in weeping lovegrass (Eragrostis curvula). PLoS ONE 2017, 12, e0175852. [Google Scholar] [CrossRef]
- Evans, L.; Knox, R. Environmental control of reproduction in Themeda australis. Aust. J. Bot. 1969, 17, 375. [Google Scholar] [CrossRef]
- Quarin, C.L. Seasonal changes in the incidence of apomixis of diploid, triploid, and tetraploid plants of Paspalum cromyorrhizon. Euphytica 1986, 35, 515–522. [Google Scholar] [CrossRef]
- Lutts, S.; Ndikumana, J.; Louant, B.P. Male and female sporogenesis and gametogenesis in apomictic Brachiaria brizantha, Brachiaria decumbens and F1 hybrids with sexual colchicine induced tetraploid Brachiaria ruziziensis. Euphytica 1994, 78, 19–25. [Google Scholar] [CrossRef]
- Gounaris, E.; Sherwood, R.; Gounaris, I.; Hamilton, R.; Gustine, D. Inorganic salts modify embryo sac development in sexual and aposporous Cenchrus ciliaris. Sex. Plant Reprod. 1991, 4, 188–192. [Google Scholar] [CrossRef]
- Schilling, M.P. Hybridization, Population Genetic Structure and Gene Expression in the Genus Boechera. Ph.D. Thesis, Utah State University, Logan, UT, USA, 2016. [Google Scholar]
- Voigt, P.W.; Bashaw, E.C. Facultative apomixis in Eragrostis curvula. Crop Sci. 1976, 16, 803–806. [Google Scholar] [CrossRef]
- Meier, M.; Zappacosta, D.; Selva, J.P.; Pessino, S.; Echenique, V. Evaluation of different methods for assessing the reproductive mode of weeping lovegrass plants, Eragrostis curvula (Schrad.) Nees. Aust. J. Bot. 2011, 59, 253–261. [Google Scholar] [CrossRef]
- Cervigni, G.; Paniego, N.; Díaz, M.; Selva, J.P.; Zappacosta, D.; Zanazzi, D.; Landerreche, I.; Martelotto, L.; Felitti, S.; Pessino, S.; et al. Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol. Biol. 2008, 67, 1–10. [Google Scholar] [CrossRef]
- Cervigni, G.D.; Paniego, N.; Pessino, S.; Selva, J.P.; Díaz, M.; Spangenberg, G.; Echenique, V. Gene expression in diplosporous and sexual Eragrostis curvula genotypes with differing ploidy levels. Plant Mol. Biol. 2008, 67, 11–23. [Google Scholar] [CrossRef]
- Selva, J.P.; Pessino, S.; Meier, M.; Echenique, V. Identification of candidate genes related to polyploidy and/or apomixis in Eragrostis curvula. Amer. J. Plant Sci. 2012, 3, 403–416. [Google Scholar] [CrossRef]
- Garbus, I.; Romero, J.; Selva, J.P.; Pasten, M.C.; Chinestra, C.; Carballo, J.; Zappacosta, D.; Echenique, V. De novo transcriptome sequencing and assembly from apomictic and sexual Eragrostis curvula genotypes. PLoS ONE 2017, 12, e0185595. [Google Scholar] [CrossRef]
- Selva, J.P.; Siena, L.; Rodrigo, J.M.; Garbus, I.; Zappacosta, D.; Romero, J.; Ortiz, J.P.; Pessino, S.; Leblanc, O.; Echenique, V. Temporal and spatial expression of genes involved in DNA methylation during reproductive development of sexual and apomictic Eragrostis curvula. Sci. Rep. 2017, 7, 15092. [Google Scholar] [CrossRef]
- Garbus, I.; Selva, J.P.; Pasten, M.C.; Bellido, A.M.; Carballo, J.; Albertini, E.; Echenique, V. Characterization and discovery of miRNA and miRNA targets from apomictic and sexual genotypes of Eragrostis curvula. BMC Genom. 2019, 20, 839. [Google Scholar] [CrossRef]
- Zappacosta, D.; Gallardo, J.; Carballo, J.; Meier, M.; Rodrigo, J.M.; Gallo, C.; Selva, J.P.; Stein, J.; Ortiz, J.P.; Albertini, E.; et al. A high-density linkage map of the forage grass Eragrostis curvula and localization of the diplospory locus. Front. Plant Sci. 2019, 10, 918. [Google Scholar] [CrossRef]
- Carballo, J.; Santos, B.; Zappacosta, D.; Garbus, I.; Selva, J.P.; Gallo, C.; Díaz, A.; Albertini, E.; Caccamo, M.; Echenique, V. A high-quality genome of Eragrostis curvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci. Rep. 2019, 9, 10250. [Google Scholar] [CrossRef]
- Zappacosta, D.; Ochogavía, A.; Rodrigo, J.M.; Romero, J.; Meier, M.; Garbus, I.; Pessino, S.C.; Echenique, V. Increased apomixis expression concurrent with genetic and epigenetic variation in a newly synthesized Eragrostis curvula polyploid. Sci. Rep. 2014, 4, 4423. [Google Scholar] [CrossRef]
- Fritz, G. Measurement of water status by water content methods. In An Introductory Plant Physiology; Noggle, G., Fritz, G., Eds.; Prentice-Hall, Inc.: Englewood Cliffs, NJ, USA, 1976; pp. 445–447. [Google Scholar]
- Johansen, D. Plant Microtechnique; Mc Graw-Hill Book Company Inc.: New York, NY, USA, 1940. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. published online ahead of print, 6 December 2017. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, J.D.; Chen, Y.; Smyth, K.G. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Slater, G.S.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, J.B.; Moornman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Lekanne Deprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 April 2019).
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef]
- Wills, D.M.; Whipple, C.J.; Takuno, S.; Kursel, L.E.; Shannon, L.M.; Ross-Ibarra, J.; Doebley, J.F. From many, one: Genetic control of prolificacy during maize domestication. PLoS Genet. 2013, 9, e1003604. [Google Scholar] [CrossRef]
- Trösch, R.; Jarvis, P. The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS ONE 2011, 6, e23039. [Google Scholar] [CrossRef]
- Peng, L.T.; Shi, Z.Y.; Li, L.; Shen, G.Z.; Zhang, J.L. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. J. Plant Physiol. 2008, 165, 876–885. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef]
- Zühl, L.; Volkert, C.; Ibberson, D.; Schmidt, A. Differential activity of F-box genes and E3 ligases distinguishes sexual versus apomictic germline specification in Boechera. J. Exp. Bot. 2019, 70, 5643–5657. [Google Scholar] [CrossRef]
- Bocchini, M.; Galla, G.; Pupilli, F.; Bellucci, M.; Barcaccia, G.; Ortiz, J.P.A.; Pessino, S.C.; Albertini, E. The vesicle trafficking regulator PN_SCD1 is demethylated and overexpressed in florets of apomictic Paspalum notatum genotypes. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- D’erfurth, I.; Le Signor, C.; Aubert, G.; Sanchez, M.; Vernoud, V.; Darchy, B.; Lherminier, J.; Bourion, V.; Bouteiller, N.; Bendahmane, A.; et al. A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol. 2012, 196, 738–751. [Google Scholar] [CrossRef]
- Galla, G.; Zenoni, S.; Avesani, L.; Altschmied, L.; Rizzo, P.; Sharbel, T.F.; Barcaccia, G. Pistil transcriptome analysis to disclose genes and gene products related to aposporous apomixis in Hypericum perforatum L. Front. Plant Sci. 2017, 8, 79. [Google Scholar] [CrossRef]
- Bräuning, S.; Catanach, A.; Lord, J.M.; Bicknell, R.; Macknight, R.C. Comparative transcriptome analysis of the wild-type model apomict Hieracium praealtum and its loss of parthenogenesis (lop) mutant. BMC Plant Biol. 2018, 18, 206. [Google Scholar] [CrossRef]
- Gruszka, D. Exploring the brassinosteroid signaling in monocots reveals novel components of the pathway and implications for plant breeding. Int. J. Mol. Sci. 2020, 21, 354. [Google Scholar] [CrossRef]
- Sherwood, D.A. A Simple Metabolic Switch May Activate Apomixis in Arabidopsis thaliana. Ph.D. Thesis, Utah State University, Logan, UT, USA, 2018. [Google Scholar]
- Gao, L. Pharmacologically Induced Meiosis Apomeiosis Interconversions in Boechera, Arabidopsis and Vigna. Ph.D. Thesis, Utah State University, Logan, UT, USA, 2018. [Google Scholar]
- Ono, A.; Yamaguchi, K.; Fukada-Tanaka, S.; Terada, R.; Mitsui, T.; Iida, S. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012, 71, 564–574. [Google Scholar] [CrossRef]
- Carman, J.G.; Sherwood, D.; Gao, L. Methods of Inducing Apomictic or Sexual Reproduction. U.S. Patent Application No. 16/273,132, 9 April 2020. [Google Scholar]
- De Storme, N.; Geelen, D. The impact of environmental stress on male reproductive development in plants: Biological processes and molecular mechanisms. Plant Cell Environ. 2014, 37, 1–18. [Google Scholar] [CrossRef]
- Dieterle, M.; Thomann, A.; Renou, J.P.; Parmentier, Y.; Cognat, V.; Lemonnier, G.; Muller, R.; Shen, W.H.; Kretsch, T.; Genschik, P. Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 2005, 41, 386–399. [Google Scholar] [CrossRef]
- Thomann, A.; Brukhin, V.; Dieterle, M.; Gheyeselinck, J.; Vantard, M.; Grossniklaus, U.; Genschik, P. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 2005, 43, 437–448. [Google Scholar] [CrossRef]
- Laspina, N.V.; Vega, T.; Seijo, J.G.; González, A.M.; Martelotto, L.G.; Stein, J.; Podio, M.; Ortiz, J.P.A.; Echenique, V.; Quarin, C.L.; et al. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol. Biol. 2008, 67, 615–628. [Google Scholar] [CrossRef]
- Martelotto, L.G.; Ortiz, J.P.A.; Stein, J.; Espinoza, F.; Quarin, C.L.; Pessino, S.C. A comprehensive analysis of gene expression alterations in a newly synthesized Paspalum notatum autotetraploid. Plant Sci. 2005, 169, 211–220. [Google Scholar] [CrossRef]
- Skowyra, D.; Craig, K.L.; Tyers, M.; Elledge, S.J.; Harper, J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 1997, 91, 209–219. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C.; Wang, A. Molecular regulation of plant somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 2013, 49, 631–642. [Google Scholar] [CrossRef]
- Qian, W.; Miki, D.; Zhang, H.; Liu, Y.; Zhang, X.; Tang, K.; Kan, Y.; Li, X.; Li, S.; Zhu, X.; et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012, 336, 1445–1448. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).