Genetic Dissection of Apomixis in Dandelions Identifies a Dominant Parthenogenesis Locus and Highlights the Complexity of Autonomous Endosperm Formation
Abstract
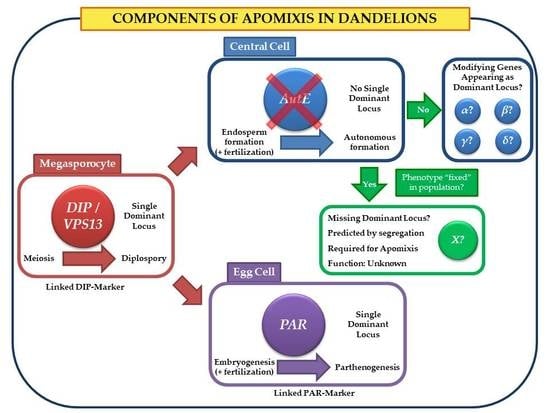
1. Introduction
2. Materials and Methods
2.1. The Cross between a Sexual Diploid and an Apomictic Triploid
2.2. Apomixis Phenotyping
2.3. Microsatellite Genotyping
2.4. Nomarski DIC Microscopy Phenotyping
2.5. Seed Flow Cytometry
3. Results
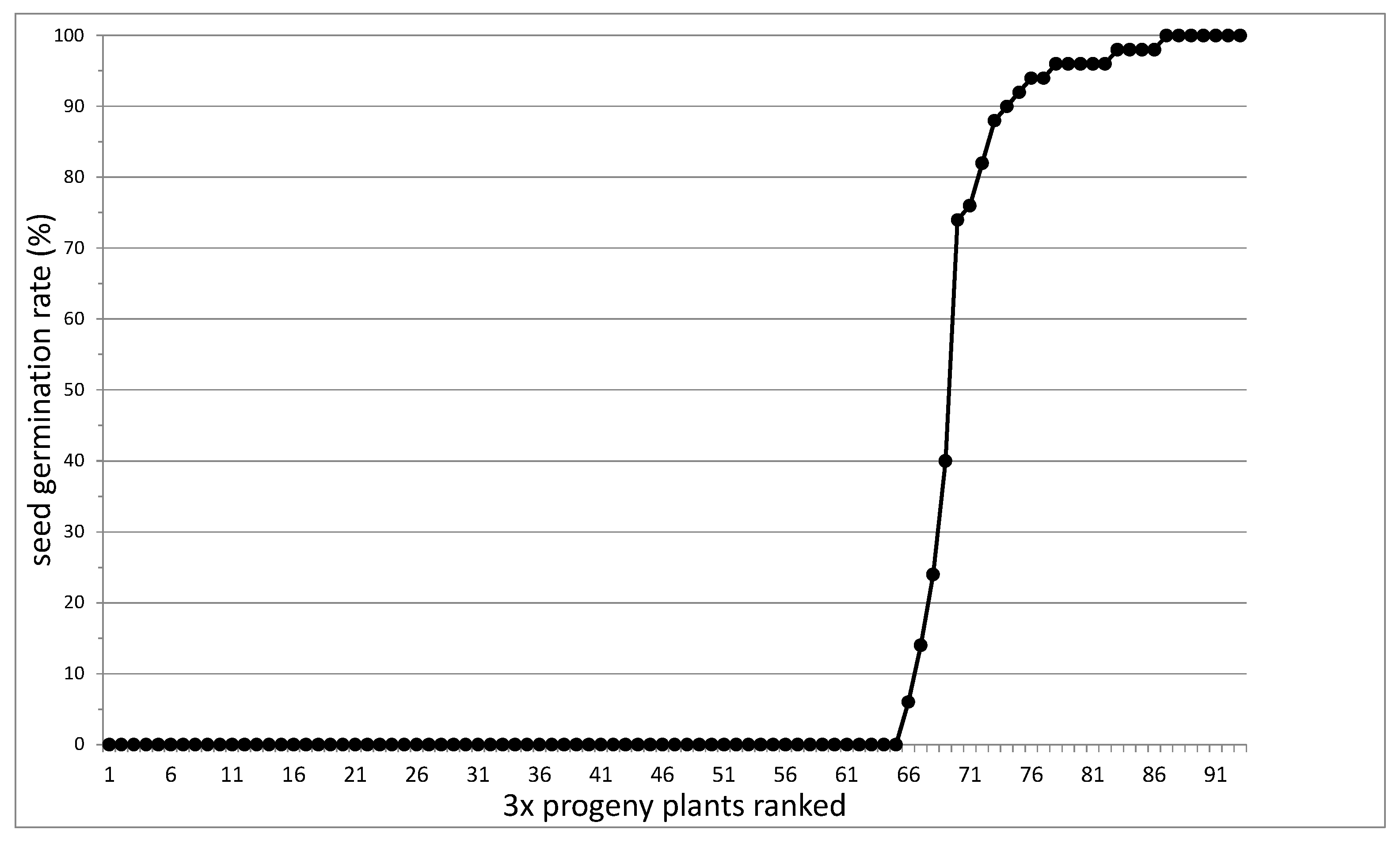
3.1. Segregation of Apomixis as a Whole
3.2. Association between SSR Markers and Apomixis
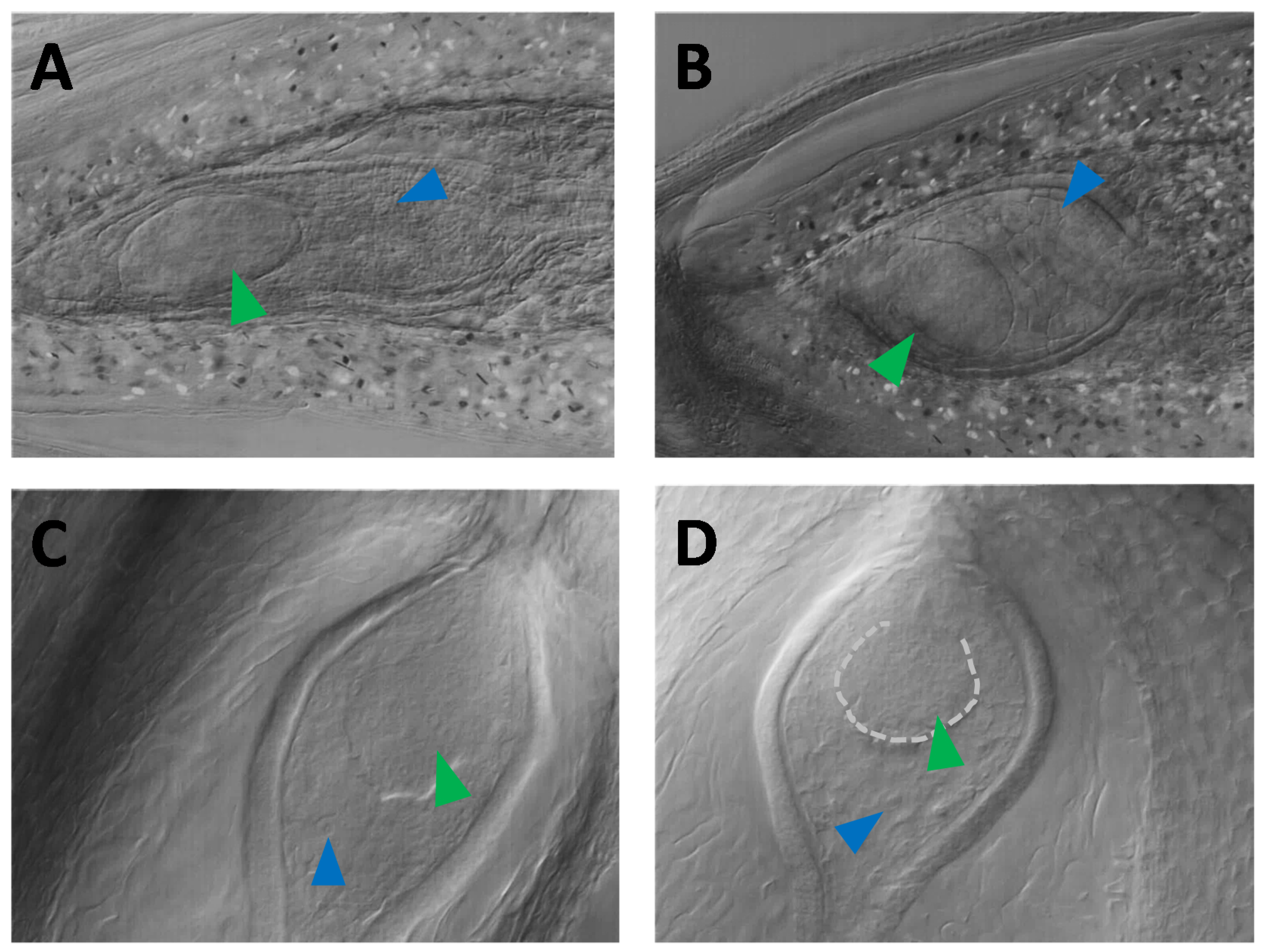
3.3. Nomarski DIC Microscopy Phenotyping of Parthenogenesis and Autonomous Endosperm
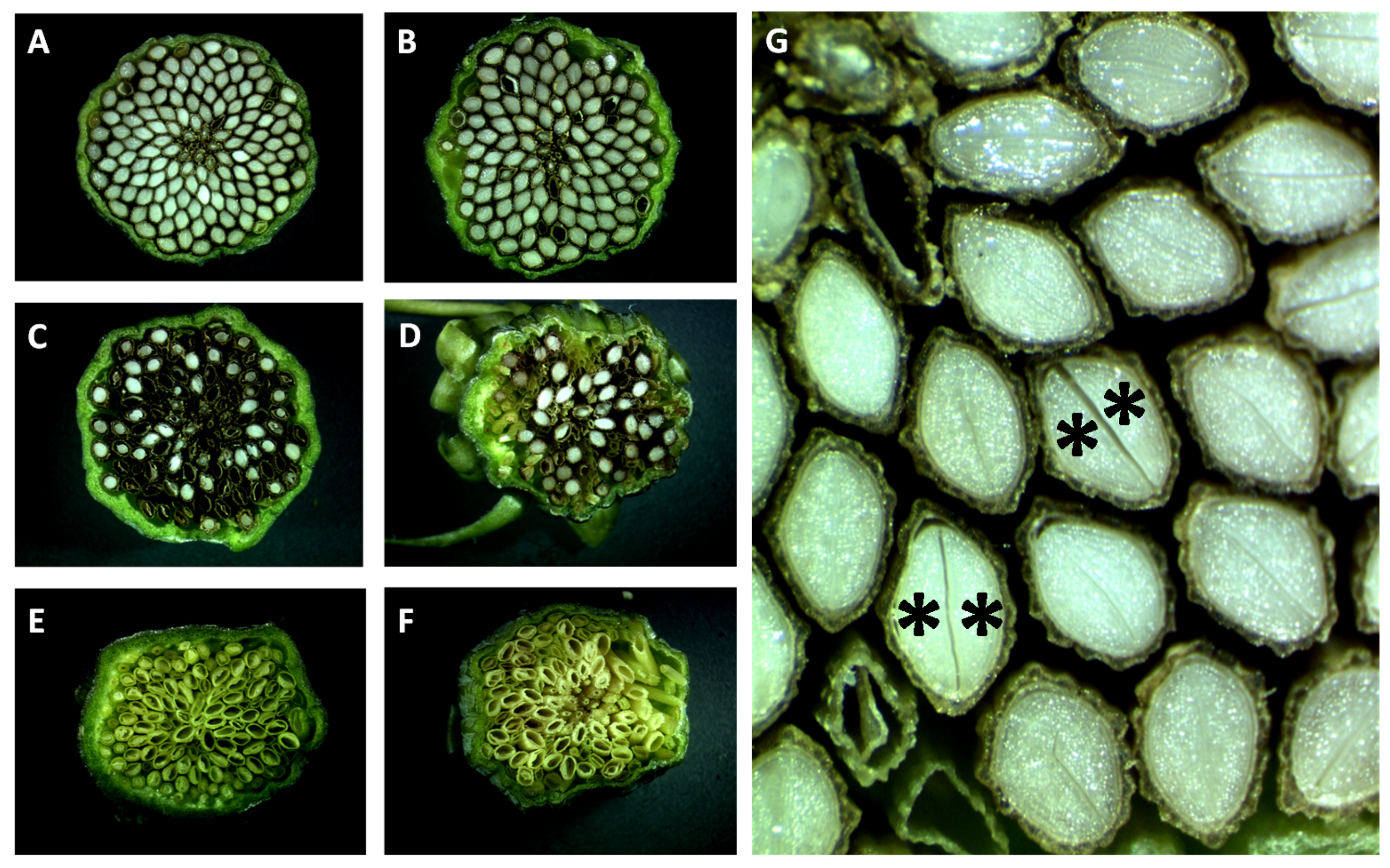
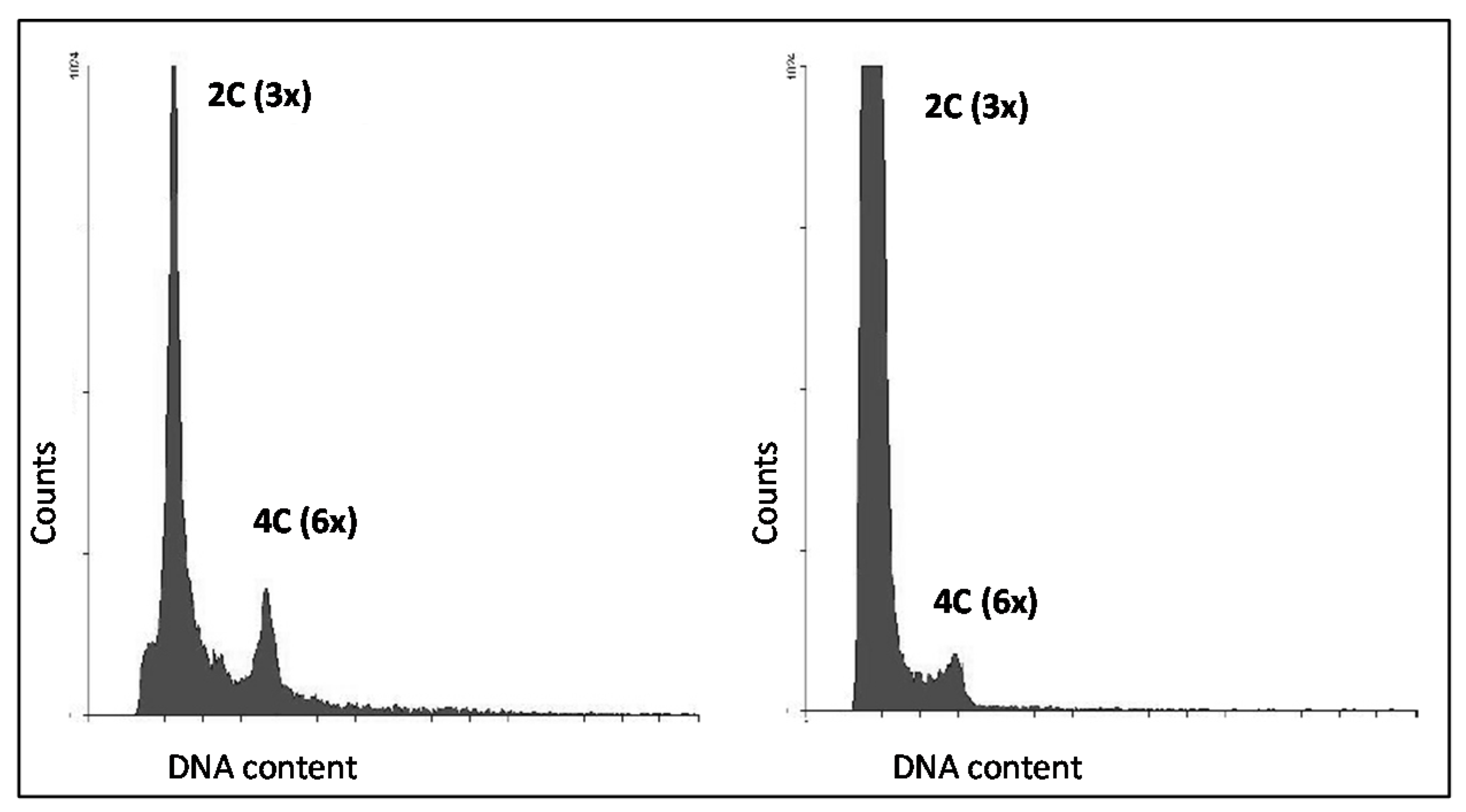
3.4. Seed Flow Cytometry to Assess Autonomous Endosperm Formation
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nogler, G.A. Gametophytic apomixis. In Embryology of Angiosperms; Johri, B.A., Ed.; Springer: Berlin, Germany, 1984; pp. 475–518. [Google Scholar]
- Grossniklaus, U.; Nogler, G.A.; Van Dijk, P.J. How to avoid sex: The genetic control of gametophytic apomixis. Plant Cell 2001, 13, 15–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jefferson, R.A. Apomixis: A social revolution for agriculture? Biotechnol. Dev. Monit. 1994, 19, 14–16. [Google Scholar]
- Vielle-Calzada, J.-P.; Crane, C.F.; Stelly, D.M. Apomixis: The asexual revolution. Science 1996, 274, 1322–1323. [Google Scholar] [CrossRef]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis technology development—Virgin births in farmers’ fields? Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, P.J.; Rigola, D.; Schauer, S.E. Plant Breeding: Surprisingly less sex is better. Curr. Biol. 2016, 26, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Hands, P.; Rabiger, D.S.; Koltunow, A. Mechanisms of endosperm initiation. Plant Reprod. 2016, 29, 215–225. [Google Scholar] [CrossRef]
- Noyes, R.D. Apomixis in the Asteraceae: Diamonds in the rough. Funct. Plant Sci. Biotechnol. 2007, 1, 207–222. [Google Scholar]
- Ozias-Akins, P.; Van Dijk, P.J. Mendelian genetics of apomixis in plants. Ann. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef]
- Hand, M.L.; Koltunow, A.M.G. The genetic control of apomixis: Asexual seed formation. Genetics 2014, 197, 441–450. [Google Scholar] [CrossRef]
- Noyes, R.D.; Rieseberg, L.H. Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 2000, 155, 379–390. [Google Scholar]
- Noyes, R.D.; Baker, R.; Mai, B. Mendelian segregation for two-factor apomixis in Erigeron annuus (Asteraceae). Heredity 2007, 98, 92–98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Catanach, A.S.; Erasmuson, S.K.; Podivinsky, E.; Jordan, B.R.; Bicknell, R. Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. USA 2006, 103, 18650–18655. [Google Scholar] [CrossRef] [PubMed]
- Koltunow, A.M.G.; Johnson, S.D.; Rodrigues, J.C.M.; Okada, T.; Hu, Y.; Tsuchiya, T.; Wilson, S.; Fletcher, P.; Ito, K.; Suzuki, G.; et al. Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J. 2011, 66, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.T.; Johnson, S.D.; Eichmann, J.; Koltunow, A.M.G. Genetic analyses of the inheritance and expressivity of autonomous endosperm formation in Hieracium with different modes of embryo sac and seed formation. Ann. Bot. 2017, 119, 1001–1010. [Google Scholar] [CrossRef]
- Vijverberg, K.; Ozias-Akins, P.; Schranz, M.E. Identifying and Engineering Genes for Parthenogenesis in Plants. Front Plant Sci. 2019, 10, 128. [Google Scholar] [CrossRef]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. Induction of parthenogenesis by a BBM-like gene. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef]
- Guitton, A.-E.; Berger, F. Loss of Function of MULTICOPY SUPPRESSOR OF IRA 1 Produces Nonviable Parthenogenetic Embryos in Arabidopsis. Curr. Biol. 2005, 15, 750–754. [Google Scholar] [CrossRef]
- Fenby, N.; Pu, H.; Pennell, R.; Praekelt, U.; Day, R.; Scott, R. An uncoupling screen for autonomous embryo mutants in Arabidopsis thaliana. Sex Plant Reprod. 2010, 4, 255–264. [Google Scholar] [CrossRef]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef]
- Chaudhury, A.M.; Ming, L.; Miller, C.; Craig, S.; Dennis, E.S.; Peacock, W.J. Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1997, 94, 4223–4228. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Vielle-Calzada, J.P.; Hoeppner, M.A.; Gagliano, W.B. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Bilodeau, P.; Koltunow, A.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A.M. Genes controlling fertilization independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.; Tucker, M.R.; Johnson, S.D.; Hrmova, M.; Koltunow, A.M. Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell 2008, 20, 2372–2386. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.C.M.; Okada, T.; Johnson, S.D.; Koltunow, A.M. A MULTICOPY SUPPRESSOR OF IRA1 (MSI1) homologue is not associated with the switch to autonomous seed development in apomictic (asexual) Hieracium plants. Plant Sci. 2010, 179, 590–597. [Google Scholar] [CrossRef]
- Van Dijk, P.J.; Bakx-Schotman, J.M.T. Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum) is controlled by a sex-specific dominant locus. Genetics 2004, 166, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Vijverberg, K.; Van der Hulst, R.; Lindhout, P.; Van Dijk, P.J. A genetic linkage map of the diplosporous chromosomal region in Taraxacum (common dandelion; Asteraceae). Theor. Appl. Genet. 2004, 108, 725–732. [Google Scholar] [CrossRef]
- Vašut, R.; Vijverberg, K.; Van Dijk, P.J.; de Jong, H. Fluorescent In Situ Hybridization shows DIPLOSPOROUS located on one of the NOR-chromosomes in apomictic dandelions in the absence of a large hemizygous chromosomal region. Genome 2015, 57, 609–620. [Google Scholar] [CrossRef]
- Vijverberg, K.; Milanovic-Ivanovic, S.; Bakx-Schotman, J.M.T.; Van Dijk, P.J. Genetic fine-mapping of DIPLOSPOROUS in Taraxacum (dandelion, Asteraceae) indicates a duplicated DIP-gene. BMC Plant Biol. 2010, 10, 154. [Google Scholar] [CrossRef]
- Van Dijk, P.J.; Rigola, D.; Prins, M.W.; van Tunen, A.J. Diplospory Gene. WO2017/039452 A1, 9 March 2017. [Google Scholar]
- Van Dijk, P.J.; Tas, I.C.Q.; Falque, M.; Bakx-Schotman, J.M.T. Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 1999, 83, 715–721. [Google Scholar] [CrossRef]
- Van Dijk, P.J.; Van Baarlen, P.; De Jong, J.H. The occurrence of phenotypically complementary apomixis-recombinants in crosses between sexual and apomictic dandelions (Taraxacum officinale). Sex. Plant Reprod. 2003, 16, 71–76. [Google Scholar] [CrossRef]
- Mártonfiová, L.; Majeský, L.; Mártonfi, P. Polyploid progeny from crosses between diploid sexuals and tetraploid apomictic pollen donors in Taraxacum sect. Ruderalia. Acta Biol. Crac. Ser. Bot. 2007, 49, 47–54. [Google Scholar]
- Van Dijk, P.J.; De Jong, J.H.; Vijverberg, K.; Biere, A. An apomixis-gene’s view on dandelions. In Lost Sex, the Evolutionary Biology of Asexual Reproduction; Schön, I., Van Dijk, P.J., Martens, K., Eds.; Springer: Berlin, Germany, 2009; pp. 475–493. [Google Scholar]
- Van der Hulst, R.G.M.; Meirmans, P.; Van Tienderen, P.H.; Van Damme, J.M.M. Nuclear-cytoplasmic male-sterility in diploid dandelions. Heredity 2004, 93, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Tas, I.C.Q.; Van Dijk, P.J. Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity 1999, 83, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Falque, M.; Keurentjes, J.; Bakx-Schotman, J.M.T.; Van Dijk, P.J. Development and characterization of microsatellite markers in the sexual-apomictic complex Taraxacum officinale (dandelion). Theor. Appl. Genet. 1998, 97, 283–292. [Google Scholar] [CrossRef]
- Vašut, R.J.; Van Dijk, P.J.; Falque, M.; Travnicek, B.; De Jong, J.H. Development and characterisation of nine new microsatellite markers in Taraxacum (Asteraceae). Mol. Ecol. Notes 2004, 4, 645–648. [Google Scholar] [CrossRef]
- Van Baarlen, P.; De Jong, J.H.; Van Dijk, P.J. Comparative cyto-embryological investigations of sexual and apomictic dandelions (Taraxacum) and their apomictic hybrids. Sex. Plant Reprod. 2002, 15, 31–38. [Google Scholar] [CrossRef]
- Matzk, F.; Meister, A.; Schubert, I. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 2000, 21, 97–108. [Google Scholar] [CrossRef]
- Ulrich, I.; Ulrich, W. High-resolution flow cytometry of nuclear DNA in higher plants. Protoplasma 1991, 165, 212–215. [Google Scholar] [CrossRef]
- Hojsgaard, D. Apomixis Technology: Separating the Wheat from the Chaff. Genes 2020, 11, 411. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Conner, J.A. Clonal reproduction through seeds in sight for crops. Trends Genet. 2020, 36, 215–226. [Google Scholar] [CrossRef]
- Fagerlind, F. Makrosporogenese und Embryosackbildung bei agamospermischen Taraxacum-Biotypen. Svensk. Bot. Tidskr. 1947, 41, 365–390. [Google Scholar]
- Cooper, D.C.; Brink, R.A. The endosperm-embryo relationship in the autonomous apomict, Taraxacum officinale. Bot. Gaz. 1949, 111, 139–152. [Google Scholar] [CrossRef]
- Nowack, M.K.; Grini, P.E.; Jakoby, M.J.; Lafos, M.; Koncz, C.; Schnittger, A. A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Gen. 2006, 38, 63–67. [Google Scholar] [CrossRef] [PubMed]



| Marker | Repeat Motif | Forward Primer Sequence | Reverse Primer Sequence | TA | Size | Reference |
|---|---|---|---|---|---|---|
| MSTA31 | (CT)17 | CCTCAAAGCCCGAACTT | ACGACCCCAACTGATTTTTAC | 51.0 | 240 | [37] |
| MSTA44B | (CT)19 | AGTTTCTCTAAAATGGGAAGAT | TGTCAGGTATATTCAAAAGATTC | 51.0 | 191 | [37] |
| MSTA53 | (TC)12(GT)8 | CAATTATTATGGTCTCGTCCTT | CCAGTTGAAGCAAAAACAGT | 55.0 | 203 | [37] |
| MSTA64 | (TC)4TT(TC)2TT(TC)2TT(TC)5A(TC)4-(A)16 | TGCTTTTTGAACGACAGTG | TTTGCTTGGTTATTAGTGAACAT | 55.0 | 191 | [37] |
| MSTA67 | (TC)22T(CA)12 | TTCGGATATGACCCTTCACT | GACATCTTGCACCTAAAAACAAT | 56.0 | 219 | [37] |
| MSTA73 | (TC)21CTG(TC)8 | CCGCATGAGGTTGTCT | TGGGCTGTTTAATAGAACTTA | 53.0 | 216 | [37] |
| MSTA74 | (CT)10 | GAGGTCTTTTTATTCGGTTTT | GGATGCCTTTACAGTTACAAT | 49.0 | 223 | [37] |
| MSTA78 | (CT)9 | TGATTGATTCTGCCCTAAACC | TGCCAAGACATCCGAAAAG | 52.0 | 151 | [37] |
| MSTA85 | (CT)20 | TGCATGTTCGTTCTACTGGT | ACGTAATAAAATTGGAAGTCAGG | 55.0 | 196 | [37] |
| MSTA101 | (CCT)2TCT(TC)16 | GCATGGGGGTCGAGGGGTAT | CCGCGATGGACTTATTCTTGGTTG | 57.8 | 198 | [38] |
| MSTA105 | (TC)23 | CACCGTTCAAAAATAAAGATAAAA | AGAATAGCTCCGTCAAGTAGG | 54.3 | 203 | [38] |
| MSTA131 | (AT)7 | TACCCTGCAAACATTACTCTTCTG | GTTGGCCTGTTAATACTTGATACG | 55.0 | 181 | [38] |
| SSR Locus | Allele 1 | Allele 2 | Allele 3 | Chi-Square | d.f. | p-Value | Significance |
|---|---|---|---|---|---|---|---|
| MSTA44B | b | d | e | ||||
| apomicts | 20 | 27 | 11 | ||||
| non-apomicts | 47 | 31 | 50 | 11.36 | 2 | 0.00 | p < 0.05 |
| MSTA53 | a | b | c | ||||
| apomicts | 14 | 20 | 16 | ||||
| non-apomicts | 44 | 44 | 38 | 0.81 | 2 | 0.67 | n.s. |
| MSTA64 | a | b | c | ||||
| apomicts | 17 | 14 | 17 | ||||
| non-apomicts | 37 | 36 | 41 | 0.55 | 2 | 0.93 | n.s. |
| MSTA67 | a | d | e | ||||
| apomicts | 24 | 17 | 18 | ||||
| non-apomicts | 38 | 46 | 46 | 2.43 | 2 | 0.30 | n.s. |
| MSTA73 | a | a | c | ||||
| apomicts | 19 | 9 | |||||
| non-apomicts | 38 | 19 | 0.01 | 1 | 0.91 | n.s. | |
| MSTA74 | b | c | e | ||||
| apomicts | 22 | 17 | 15 | ||||
| non-apomicts | 44 | 34 | 46 | 1.45 | 2 | 0.48 | n.s. |
| MSTA78 | a | c | d | ||||
| apomicts | 29 | 18 | 11 | ||||
| non-apomicts | 33 | 46 | 43 | 9.98 | 2 | 0.01 | p < 0.05 |
| MSTA85 | a | b | b | ||||
| apomicts | 20 | 36 | |||||
| non-apomicts | 32 | 90 | 1.67 | 1 | 0.20 | n.s. | |
| MSTA101 | a | c | d | ||||
| apomicts | 27 | 12 | 15 | ||||
| non-apomicts | 51 | 36 | 25 | 1.84 | 2 | 0.40 | n.s. |
| MSTA105 | b | c | d | ||||
| apomicts | 17 | 18 | 21 | ||||
| non-apomicts | 36 | 44 | 42 | 0.28 | 2 | 0.87 | n.s. |
| MSTA131 | a | c | d | ||||
| apomicts | 13 | 21 | 24 | ||||
| non-apomicts | 44 | 41 | 45 | 2.49 | 2 | 0.29 | n.s. |
| Ploidy | Germin. % | Apomixis | Dip Marker (MST78-a) | Par Marker (MST44B-d) | DIC Microscopy | ||||
|---|---|---|---|---|---|---|---|---|---|
| PAR | AUT | FCSS AUT | AUT Combined | ||||||
| TJX 320 | 2 × | no | ab | ac | non | non | non | – | |
| 68 | 3 × | 98 | yes | acd | bde | yes | yes | yes | + |
| 2 | 3 × | 96 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 16 | 3 × | 100 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 18 | 3 × | 96 | yes | ad | bd | n.d. | n.d. | n.d. | + |
| 23 | 3 × | 14 | yes | ac | be | n.d. | ? | n.d. | + |
| 24 | 3 × | 100 | yes | ad | bd | n.d. | ? | n.d. | + |
| 31 | 3 × | 100 | yes | ad | bd | n.d. | n.d. | n.d. | + |
| 34 | 3 × | 98 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 37 | 3 × | 40 | yes | ac | bd | + | + | n.d. | + |
| 48 | 3 × | 78 | yes | ac | bd | + | + | n.d. | + |
| 50 | 3 × | 98 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 69 | 3 × | n.q. | yes | ad | de | + | + | n.d. | + |
| 73 | 3 × | 100 | yes | ac | de | + | + | n.d. | + |
| 76 | 3 × | 94 | yes | ad | bd | n.d. | n.d. | n.d. | + |
| 98 | 3 × | 100 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 99 | 3 × | 74 | yes | ad | bd | + | + | n.d. | + |
| 113 | 3 × | 90 | yes | ad | bd | n.d. | n.d. | n.d. | + |
| 115 | 3 × | 96 | yes | ad | de | n.d. | n.d. | n.d. | + |
| 127 | 3 × | 82 | yes | ad | bd | + | + | n.d. | + |
| 132 | 3 × | 94 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 133 | 3 × | 98 | yes | ac | be | n.d. | n.d. | n.d. | + |
| 136 | 3 × | 96 | yes | ac | de | n.d. | n.d. | n.d. | + |
| 144 | 3 × | 88 | yes | ac | de | n.d. | n.d. | n.d. | + |
| 154 | 3 × | 100 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 158 | 3 × | 96 | yes | ac | de | n.d. | n.d. | n.d. | + |
| 159 | 3 × | 24 | yes | ac | de | n.d. | n.d. | n.d. | + |
| 165 | 3 × | 98 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 183 | 3 × | 92 | yes | ac | bd | n.d. | n.d. | n.d. | + |
| 194 | 3 × | 6 | yes | ad | de | n.d. | n.d. | n.d. | + |
| 201 | 3 × | 100 | yes | ad | de | n.d. | n.d. | n.d. | + |
| 163 | 3 × | 0 | no | ad | de | + | + | + | + |
| 70 | 3 × | 0 | no | ac | bd | + | ? | + | + |
| 22 | 3 × | 0 | no | ac | de | + | ? | n.d. | ? |
| 56 | 3 × | 0 | no | ac | de | + | ? | n.d. | ? |
| 148 | 3 × | 0 | no | ac | de | ? | + | + | + |
| 95 | 3 × | 0 | no | ac | bd | ? | ? | + | + |
| 114 | 3 × | 0 | no | ac | de | n.d. | n.d. | + | + |
| 123 | 3 × | 0 | no | ad | de | ? | + | n.d. | + |
| 195 | 3 × | 0 | no | ac | de | ? | ? | + | + |
| 175 | 3 × | 0 | no | ac | bd | n.d. | n.d. | – | – |
| 193 | 3 × | 0 | no | ac | de | n.d. | n.d. | – | – |
| 30 | 3 × | 0 | no | ad | be | ? | + | + | + |
| 45 | 3 × | 0 | no | ad | be | ? | + | + | + |
| 139 | 3 × | 0 | no | ad | be | ? | ? | + | + |
| 170 | 3 × | 0 | no | ad | be | ? | + | + | + |
| 181 | 3 × | 0 | no | ac | be | ? | + | + | + |
| 185 | 3 × | 0 | no | ac | be | ? | + | + | + |
| 190 | 3 × | 0 | no | ac | be | ? | + | + | + |
| 177 | 3 × | 0 | no | ac | be | ? | + | n.d. | + |
| 68 | 3 × | 0 | no | ad | be | n.d. | n.d. | + | + |
| 78 | 3 × | 0 | no | ad | be | n.d. | n.d. | + | + |
| 79 | 3 × | 0 | no | ac | be | ? | ? | + | + |
| 86 | 3 × | 0 | no | ad | be | n.d. | n.d. | + | + |
| 117 | 3 × | 0 | no | ad | be | n.d. | n.d. | + | + |
| 140 | 3 × | 0 | no | ad | be | ? | ? | + | + |
| 149 | 3 × | 0 | no | ac | be | ? | ? | + | + |
| 176 | 3 × | 0 | no | ad | be | ? | ? | + | + |
| 207 | 3 × | 0 | no | ad | be | ? | ? | + | + |
| 65 | 3 × | 0 | no | ac | be | ? | ? | – | – |
| 112 | 3 × | 0 | no | ac | be | ? | ? | n.d. | ? |
| 157 | 3 × | 0 | no | ad | bc | ? | ? | n.d. | ? |
| 33 | 3 × | 0 | no | ad | be | ? | ? | n.d. | ? |
| 43 | 3 × | 0 | no | ac | be | ? | ? | n.d. | ? |
| 208 | 3 × | 0 | no | ac | be | ? | ? | n.d. | ? |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Dijk, P.J.; Op den Camp, R.; Schauer, S.E. Genetic Dissection of Apomixis in Dandelions Identifies a Dominant Parthenogenesis Locus and Highlights the Complexity of Autonomous Endosperm Formation. Genes 2020, 11, 961. https://doi.org/10.3390/genes11090961
Van Dijk PJ, Op den Camp R, Schauer SE. Genetic Dissection of Apomixis in Dandelions Identifies a Dominant Parthenogenesis Locus and Highlights the Complexity of Autonomous Endosperm Formation. Genes. 2020; 11(9):961. https://doi.org/10.3390/genes11090961
Chicago/Turabian StyleVan Dijk, Peter J., Rik Op den Camp, and Stephen E. Schauer. 2020. "Genetic Dissection of Apomixis in Dandelions Identifies a Dominant Parthenogenesis Locus and Highlights the Complexity of Autonomous Endosperm Formation" Genes 11, no. 9: 961. https://doi.org/10.3390/genes11090961
APA StyleVan Dijk, P. J., Op den Camp, R., & Schauer, S. E. (2020). Genetic Dissection of Apomixis in Dandelions Identifies a Dominant Parthenogenesis Locus and Highlights the Complexity of Autonomous Endosperm Formation. Genes, 11(9), 961. https://doi.org/10.3390/genes11090961