miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
2.2. Animals
2.3. Primer Design and Synthesis
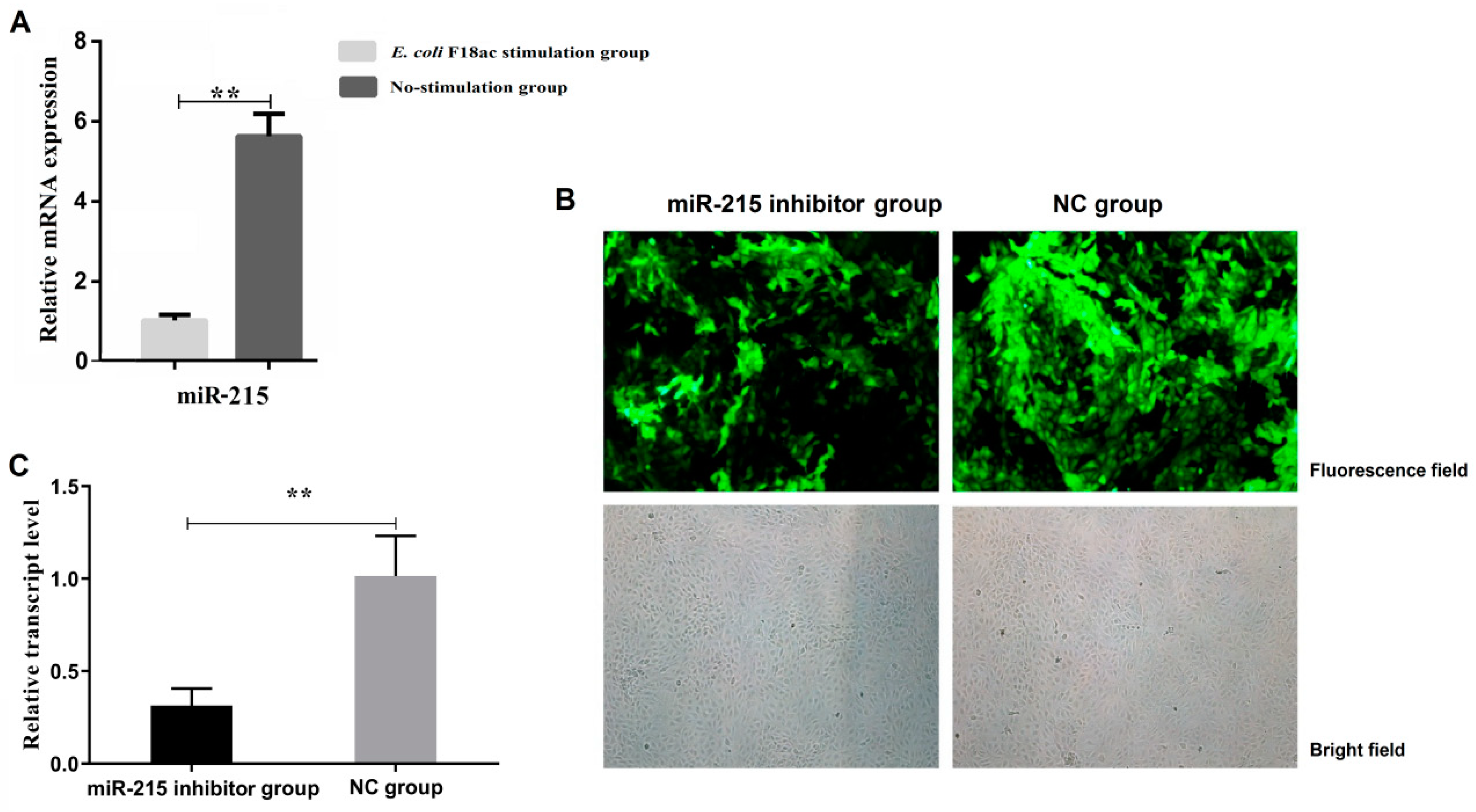
2.4. Expression Changes of miR-215 in IPEC-J2 Cells Stimulated by E. coli F18ac
2.5. miR-215 Inhibitor Lentivirus Transfected IPEC-J2 Cell Line
2.6. RNA-Seq Analysis
2.6.1. Library Construction and Transcriptome Sequencing
2.6.2. Bioinformatics Analysis and Quantitative Verification
2.7. Prediction and Verification of Porcine miR-215 Target Genes
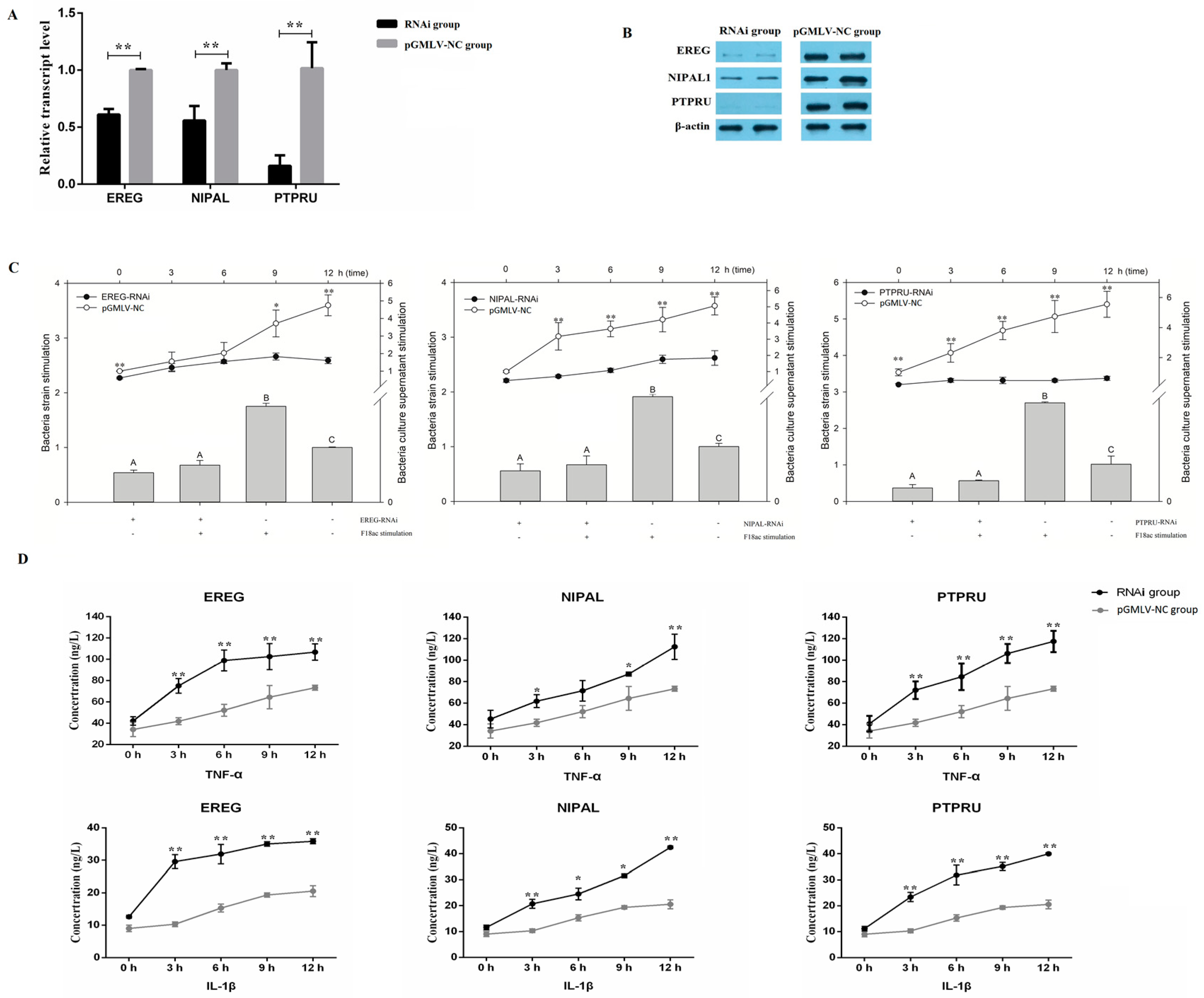
2.8. RNAi and E. coli F18ac Stimulation Experiment
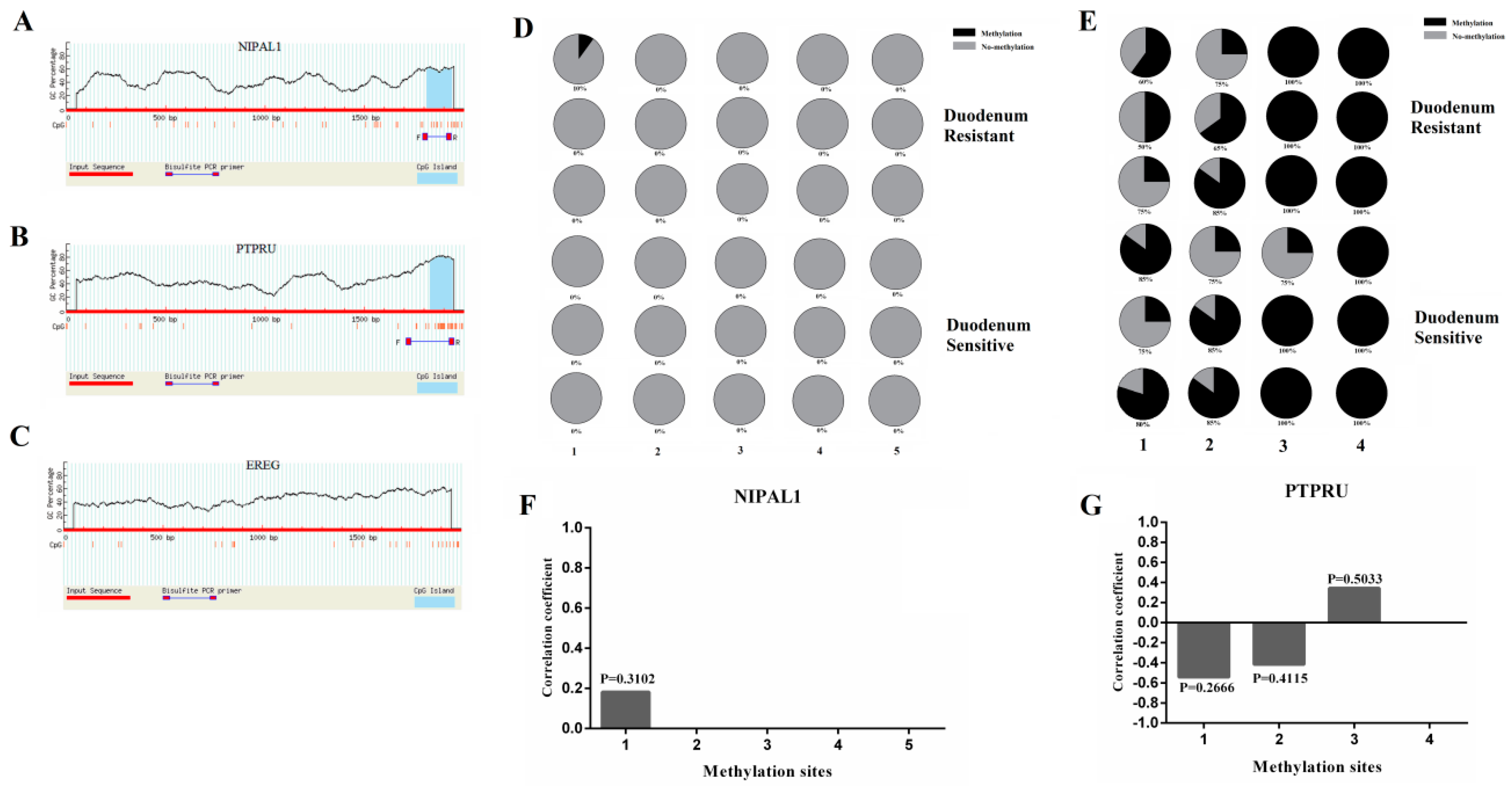
2.9. Promoter Regions Prediction and Core Promoter Identification of miR-215 Target Genes
2.10. Promoter Regions Prediction and Core Promoter Identification of miR-215 Target Genes
2.11. Construction of Mutant Vectors of Core Promoter Regions and Detection of Dual Luciferase Activity
2.12. qPCR and Western Blot Analysis
2.13. Statistical Analysis
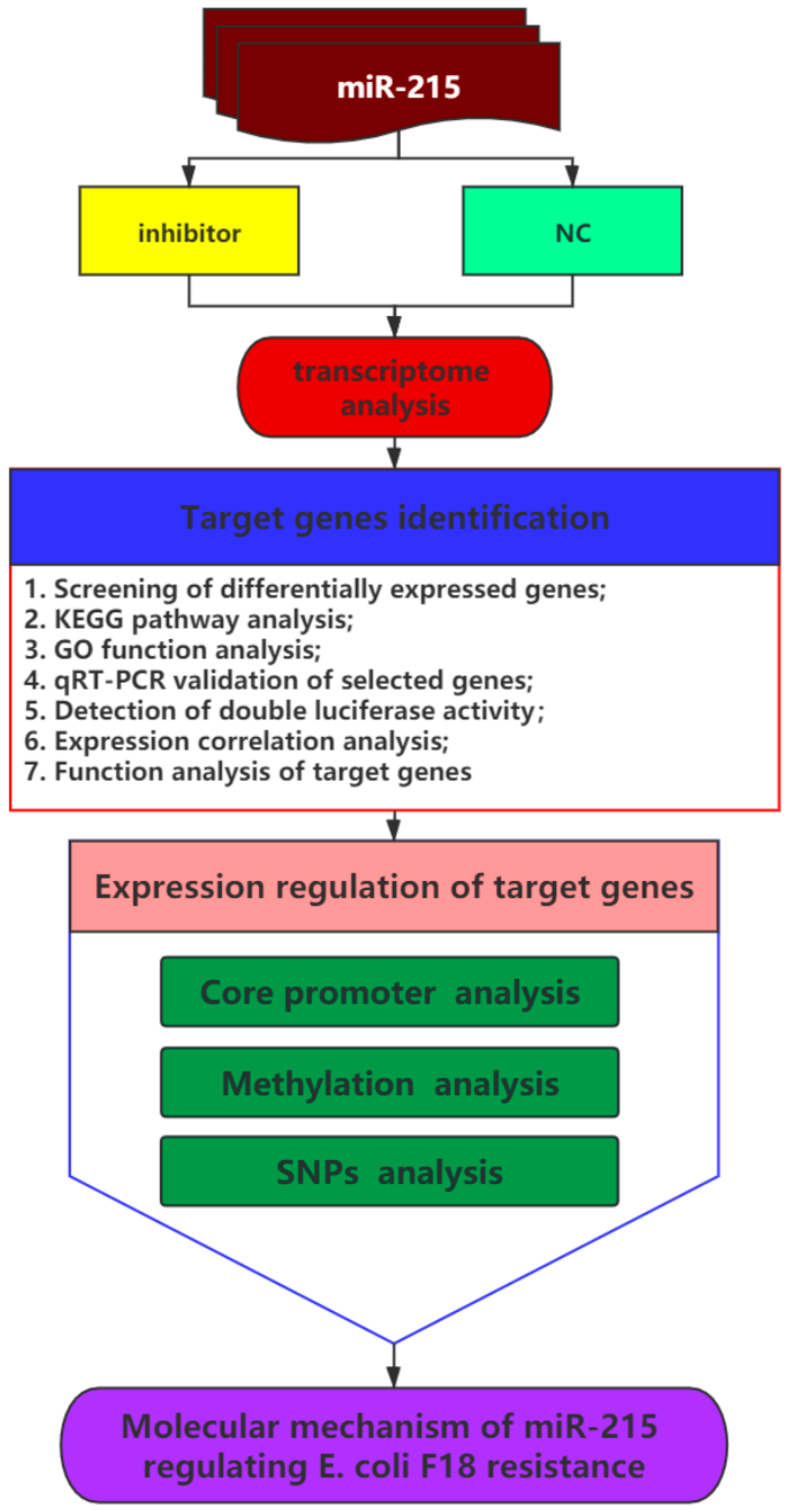
3. Results
3.1. Expression Changes of miR-215 in IPEC-J2 Cells Stimulated by E. coli F18ac and Construction of Cell Line with miR-215 Stable Interference
3.2. RNA-Seq Results of IPEC-J2 Cells between miR-215 Inhibitor Group and NC Group
3.3. Prediction and Verification of miR-215 Target Genes
3.4. The Regulation of miR-215 Target Genes Expression Levels in the Process of E. coli Infection of IPEC-J2 Cells
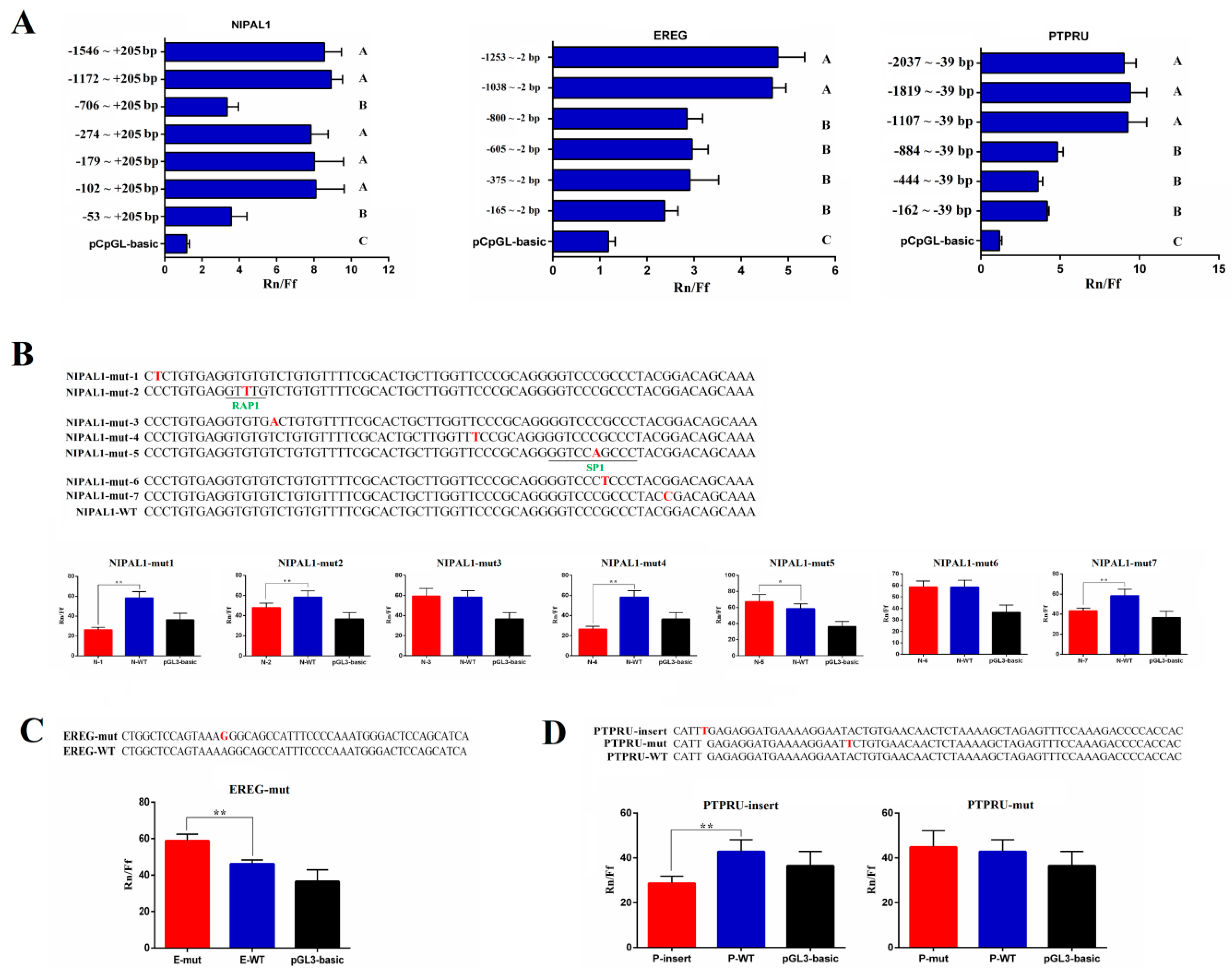
3.5. Detection of Promoter Methylation in Target Genes of miRNA-215 and Effect of SNPs in Core Promoter Region on Promoter Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bai, L.; Liang, R.; Yang, Y.; Hou, X.; Wang, Z.; Zhu, S.; Wang, C.; Tang, Z.; Li, K. MicroRNA-21 regulates PI3K/Akt/mTOR signaling by targeting TGFβI during skeletal muscle development in pigs. PLoS ONE 2015, 10, e0119396. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776. [Google Scholar] [CrossRef]
- Wienholds, E.; Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 2005, 579, 5911–5922. [Google Scholar] [CrossRef]
- Liu, C.; Tang, D.G. MicroRNA regulation of cancer stem cells. Cancer Res. 2011, 71, 5950–5954. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, L.; Liao, S.; Xu, Z.; Zhou, Y. The porcine microRNA transcriptome response to transmissible gastroenteritis virus infection. PLoS ONE 2015, 10, e0120377. [Google Scholar] [CrossRef]
- Loveday, E.K.; Svinti, V.; Diederich, S.; Pasick, J.; Jean, F. Temporal-and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J. Virol. 2012, 86, 6109–6122. [Google Scholar] [CrossRef]
- Xia, B.; Song, H.; Chen, Y.; Xia, X.; Sun, H. Efficient inhibition of porcine reproductive and respiratory syndrome virus replication by artificial microRNAs targeting the untranslated regions. Arch. Virol. 2013, 158, 55–61. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Pawlina-Tyszko, K.; Żukowski, K.; Tyra, M.; Derebecka, N.; Wesoły, J.; Szmatola, T.; Piórkowska, K. Identification of molecular mechanisms related to pig fatness at the transcriptome and miRNAome levels. Genes 2020, 11, 600. [Google Scholar] [CrossRef]
- Braun, C.J.; Zhang, X.; Savelyeva, I.; Wolff, S.; Moll, U.M.; Schepeler, T.; Ørntoft, T.F.; Andersen, C.L.; Dobbelstein, M. P53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008, 68, 10094–10104. [Google Scholar] [CrossRef] [PubMed]
- Georges, S.A.; Biery, M.C.; Kim, S.Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008, 68, 10105–10112. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Wang, Y.; Titmus, M.A.; Botchkina, G.; Formentini, A.; Kornmann, M.; Ju, J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. Cancer 2010, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- White, N.M.; Khella, H.W.; Grigull, J.; Adzovic, S.; Youssef, Y.M.; Honey, R.J.; Stewart, R.; Pace, K.T.; Bjarnason, G.A.; Jewett, M.A.S.; et al. MiRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br. J. Cancer 2011, 105, 1741–1749. [Google Scholar] [CrossRef]
- Liu, F.; You, X.; Chi, X.; Wang, T.; Ye, L.; Niu, J.; Zhang, X. Hepatitis B virus X protein mutant HBxΔ127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem. Biophys. Res. Commun. 2014, 444, 128–134. [Google Scholar] [CrossRef]
- Liang, H.; Li, Y.; Luo, R.; Shen, F.J. MicroRNA-215 is a potential prognostic marker for cervical cancer. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014, 34, 207–212. [Google Scholar] [CrossRef]
- Chen, C.; Deng, B.; Qiao, M.; Zheng, R.; Chai, J.; Ding, Y.; Peng, J.; Jiang, S. Solexa sequencing identification of conserved and novel microRNAs in backfat of large white and Chinese meishan pigs. PLoS ONE 2012, 7, e31426. [Google Scholar] [CrossRef]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Brameld, J.M.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, S.L.; Liu, Y.; Liu, Y.L.; Wang, W.J. Analysis of microRNA expression profiles in weaned pig skeletal muscle after lipopolysaccharide challenge. Int. J. Mol. Sci. 2015, 16, 22438–22455. [Google Scholar] [CrossRef]
- Sharbati, S.; Friedländer, M.R.; Sharbati, J.; Hoeke, L.; Chen, W.; Keller, A.; Stähler, P.F.; Rajewsky, N.; Einspanier, R. Deciphering the porcine intestinal microRNA transcriptome. BMC Genom. 2010, 11, 275. [Google Scholar] [CrossRef]
- Ye, L.; Su, X.; Wu, Z.; Zheng, X.; Wang, J.; Zi, C.; Zhu, G.; Wu, S.; Bao, W. Analysis of differential miRNA expression in the duodenum of Escherichia coli F18-sensitive and-resistant weaned piglets. PLoS ONE 2012, 7, e43741. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, J.; Zhao, Q.; Zi, C.; Wu, Z.; Su, X.; Huo, Y.; Zhu, G.; Wu, S.; Bao, W. Genetic variation in exon 10 of the BPI gene is associated with Escherichia coli F18 susceptibility in Sutai piglets. Gene 2013, 523, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Wang, H.; Zhu, G.; Wu, S.; Bao, W. Porcine CD14 gene silencing partially inhibited the bacterial immune response mediated by TLR4 signaling pathway. Gene 2017, 628, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.D.; Brenner, S.E.; Dudoit, S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010, 38, e131. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Dai, C.; Yang, L.; Jin, J.; Wang, H.; Wu, S.; Bao, W. Regulation and molecular mechanism of TLR5 on resistance to escherichia coli F18 in weaned piglets. Animals 2019, 9, 735. [Google Scholar] [CrossRef]
- Toyoda, H.; Komurasaki, T.; Uchida, D.; Morimoto, S. Distribution of mRNA for human epiregulin, a differentially expressed member of the epidermal growth factor family. Biochem. J. 1997, 326, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Adeyinka, A.; Nui, Y.; Cherlet, T.; Snell, L.; Watson, P.H.; Murphy, L.C. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin. Cancer Res. 2002, 8, 1747–1753. [Google Scholar] [PubMed]
- Yun, J.; Song, S.H.; Park, J.; Park, J.; Kim, H.P.; Yoon, Y.K.; Han, S.W.; Oh, D.Y.; Im, S.A.; Bang, Y.J.; et al. Gene silencing of EREG mediated by DNA methylation and histone modification in human gastric cancers. Lab. Investig. 2012, 92, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Sunaga, N.; Kaira, K. Epiregulin as a therapeutic target in non-small-cell lung cancer. Lung Cancer 2015, 6, 91. [Google Scholar] [CrossRef]
- Qu, X.; Sandmann, T.; Frierson, H.J.; Fu, L.; Fuentes, E.; Walter, K.; Okrah, K.; Rumpel, C.; Moskaluk, C.; Lu, S.; et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene 2016, 35, 6403–6415. [Google Scholar] [CrossRef]
- Liu, S.; Ye, D.; Xu, D.; Liao, Y.; Zhang, L.; Liu, L.; Yu, W.; Wang, Y.; He, Y.; Hu, J.; et al. Autocrine epiregulin activates EGFR pathway for lung metastasis via EMT in salivary adenoid cystic carcinoma. Oncotarget 2016, 7, 25251. [Google Scholar] [CrossRef]
- Yamashita, Y.; Kawashima, I.; Yanai, Y.; Nishibori, M.; Richards, J.S.; Shimada, M. Hormone-induced expression of tumor necrosis factor α-converting enzyme/A disintegrin and metalloprotease-17 impacts porcine cumulus cell oocyte complex expansion and meiotic maturation via ligand activation of the epidermal growth factor receptor. Endocrinology 2007, 148, 6164–6175. [Google Scholar] [CrossRef]
- Xiang, Z.F.; Zhang, J.Z.; Li, X.B.; Xie, H.B.; Wang, Q.H. Comparison of gene expression between cumulus oocyte complexes and naked oocytes by suppression subtractive hybridization in swine. Asian Australas. J. Anim. Sci. 2010, 23, 17–24. [Google Scholar] [CrossRef]
- Okwueze, M.I.; Cardwell, N.L.; Pollins, A.C.; Nanney, L.B. Modulation of porcine wound repair with a transfected ErbB3 gene and relevant EGF-like ligands. J. Investig. Dermatol. 2007, 127, 1030–1041. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, Y.; Li, K.; Liu, J.; Wang, H.; Sun, B.; Xiong, Z.; Jiang, H.; Zheng, J.; Hu, Z. Protein tyrosine phosphatase receptor U (PTPRU) is required for glioma growth and motility. Carcinogenesis 2014, 35, 1901–1910. [Google Scholar] [CrossRef]
- Jacobs, F.M.; van der Linden, A.J.; Wang, Y.; von Oerthel, L.; Sul, H.S.; Burbach, J.P.H.; Smidt, M.P. Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development 2009, 136, 2363–2373. [Google Scholar] [CrossRef] [PubMed]
- Le, W.; Zhang, L.; Xie, W.; Li, S.; Dani, J.A. Pitx3 deficiency produces decreased dopamine signaling and induces motor deficits in Pitx3 (−/−) mice. Neurobiol. Aging 2015, 36, 3314–3320. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yan, H.X.; Yang, W.; Zhang, R.; Chen, L.; Tang, L.; Zhai, B.; Liu, S.Q.; Cao, H.F.; Man, X.B.; Wu, H.P.; et al. Protein-tyrosine phosphatase PCP-2 inhibits β-catenin signaling and increases E-cadherin-dependent cell adhesion. J. Biol. Chem. 2006, 281, 15423–15433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, Z.; Xiong, Z.; Zheng, J.; Hu, Z.; Qiu, J. Knockdown of protein tyrosine phosphatase receptor U inhibits growth and motility of gastric cancer cells. Int. J. Clin. Exp. Pathol. 2014, 7, 5750. [Google Scholar]
- Sasahira, T.; Nishiguchi, Y.; Kurihara-Shimomura, M.; Nakashima, C.; Kuniyasu, H.; Kirita, T. NIPA-like domain containing 1 is a novel tumor-promoting factor in oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2018, 144, 875–882. [Google Scholar] [CrossRef]
- Cingiz, M.Ö.; Biricik, G.; Diri, B. A combinatorial approach to construct core and generic gene co-expression networks of colon cancer. In Proceedings of the 2017 IEEE International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Gdynia, Poland, 3–5 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 102–106. [Google Scholar] [CrossRef]
- Nakayama, A.; Nakaoka, H.; Yamamoto, K.; Sakiyama, M.; Shaukat, A.; Toyoda, Y.; Okada, Y.; Kamatani, Y.; Nakamura, T.; Takada, T.; et al. GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Ann. Rheum. Dis. 2017, 76, 869–877. [Google Scholar] [CrossRef]
- Polan, M.B.; Pastore, M.T.; Steingass, K.; Hashimoto, S.; Thrush, D.L.; Pyatt, R.; Reshmi, S.; Gastier-Foster, J.M.; Astbury, C.; McBride, K.L. Neurodevelopmental disorders among individuals with duplication of 4p13 to 4p12 containing a GABA A receptor subunit gene cluster. Eur. J. Hum. Genet. 2014, 22, 105–109. [Google Scholar] [CrossRef]
- Thuong, N.T.; Hawn, T.R.; Chau, T.T.; Bang, N.D.; Yen, N.T.; Thwaites, G.E.; Teo, Y.Y.; Seielstad, M.; Hibberd, M.; Lan, N.T.; et al. Epiregulin (EREG) variation is associated with susceptibility to tuberculosis. Genes Immun. 2012, 13, 275–281. [Google Scholar] [CrossRef][Green Version]
- Jonker, D.J.; Karapetis, C.; Harbison, C.; Callaghan, C.J.; Tu, D.; Simes, R.J.; Xu, L.; Moore, M.J.; Zalcberg, J.R.; Khambata-Ford, S. High epiregulin (EREG) gene expression plus K-ras wild-type (WT) status as predictors of cetuximab benefit in the treatment of advanced colorectal cancer (ACRC): Results from NCIC CTG CO. 17—A phase III trial of cetuximab versus best supportive care (BSC). J. Clin. Oncol. 2009, 27, 4016. [Google Scholar] [CrossRef]
- Zhou, R.; Zhou, X.; Yin, Z.; Guo, J.; Hu, T.; Jiang, S.; Liu, L.; Dong, X.; Zhang, S.; Wu, G. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci. Rep. 2016, 6, 35714. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, L.; Xia, Y.; Yang, Q.; Li, X.; Tang, G.; Jiang, Y.; Wang, J.; Li, M.; Zhu, L. DNA methylation landscape of fat deposits and fatty acid composition in obese and lean pigs. Sci. Rep. 2016, 6, 35063. [Google Scholar] [CrossRef]
- Wang, J.; Cao, M.; Yang, M.; Lin, Y.; Che, L.; Fang, Z.; Xu, S.; Feng, B.; Li, J.; Wu, D. Intra-uterine undernutrition amplifies age-associated glucose intolerance in pigs via altered DNA methylation at muscle GLUT4 promoter. Br. J. Nutr. 2016, 116, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Ning, C.; Zheng, X.; Fu, J.; Wang, A.; Zhang, Q.; Liu, J. Genome-wide DNA methylation and transcriptome analyses reveal genes involved in immune responses of pig peripheral blood mononuclear cells to poly I: C. Sci. Rep. 2017, 7, 9709. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Ma, Y.; Ma, P.; Wang, S.; Fan, Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells 2013, 31, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, J.B.; Xue, Y.; Peng, Y.L.; Chen, G.; Fang, M.Y. Differential expression of CYB 5A in Chinese and European pig breeds due to genetic variations in the promoter region. Anim. Genet. 2015, 46, 16–22. [Google Scholar] [CrossRef]
- Sweeney, T.; Halloran, A.M.; Hamill, R.M.; Davey, G.C.; Gil, M.; Southwood, O.I.; Ryan, M.T. Novel variation in the FABP3 promoter and its association with fatness traits in pigs. Meat. Sci. 2015, 100, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Chu, W.; Shi, H.; Yu, S.G.; Han, H.Y.; Gu, S.H.; Chen, J. Genetic variants in ABCA1 promoter affect transcription activity and plasma HDL level in pigs. Gene 2015, 555, 414–420. [Google Scholar] [CrossRef]
- Mencıía, M.; Moqtaderi, Z.; Geisberg, J.V.; Kuras, L.; Struhl, K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 2002, 9, 823–833. [Google Scholar] [CrossRef]
- Tan, N.Y.; Khachigian, L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell Biol. 2009, 29, 2483–2488. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, C.-H.; Wang, F.; Wang, S.-Q.; Wu, Z.-C.; Wu, S.-L.; Bao, W.-B. miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets. Genes 2020, 11, 1053. https://doi.org/10.3390/genes11091053
Dai C-H, Wang F, Wang S-Q, Wu Z-C, Wu S-L, Bao W-B. miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets. Genes. 2020; 11(9):1053. https://doi.org/10.3390/genes11091053
Chicago/Turabian StyleDai, Chao-Hui, Fang Wang, Shi-Qin Wang, Zheng-Chang Wu, Sheng-Long Wu, and Wen-Bin Bao. 2020. "miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets" Genes 11, no. 9: 1053. https://doi.org/10.3390/genes11091053
APA StyleDai, C.-H., Wang, F., Wang, S.-Q., Wu, Z.-C., Wu, S.-L., & Bao, W.-B. (2020). miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets. Genes, 11(9), 1053. https://doi.org/10.3390/genes11091053





