PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Site Selection and In Vitro Cleavage Assay
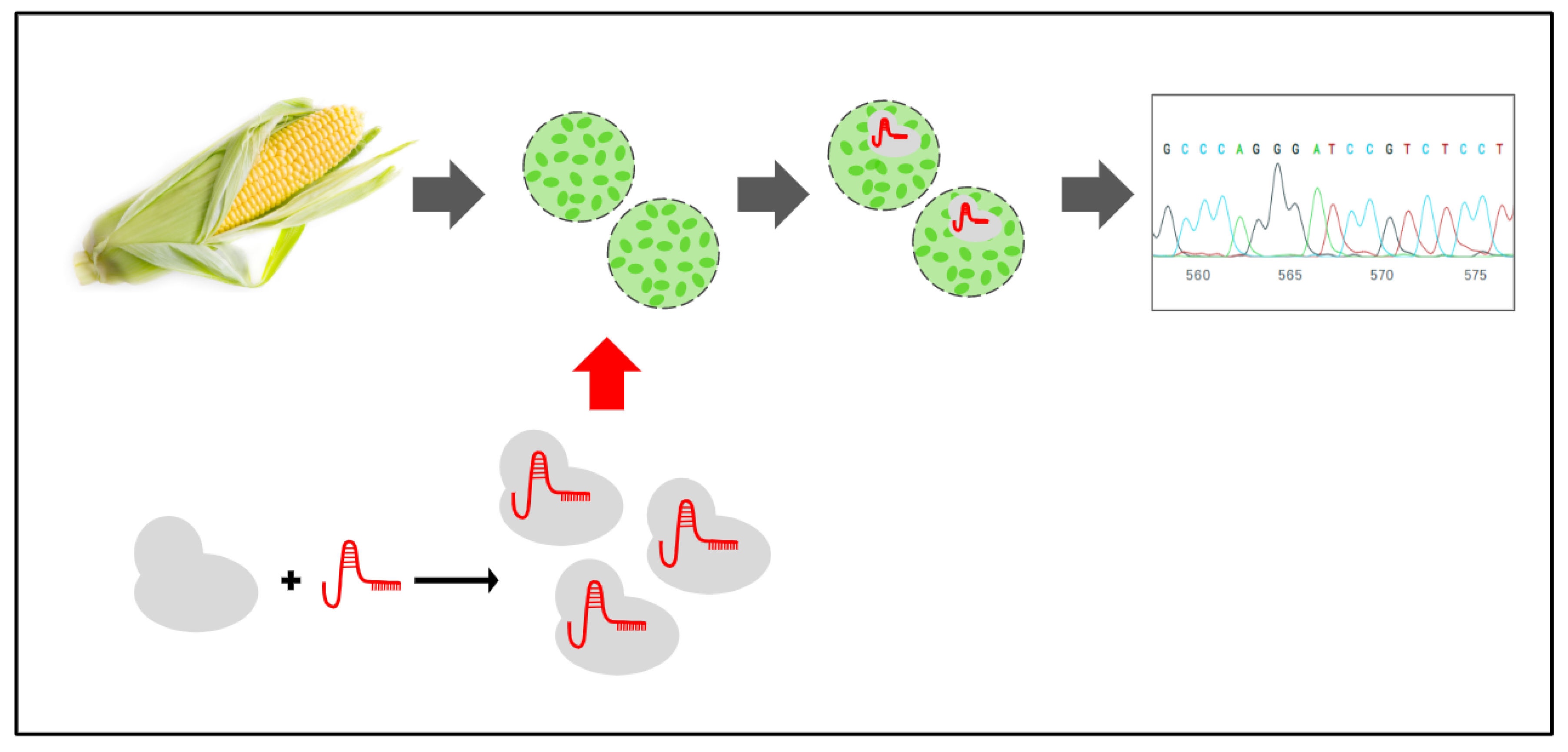
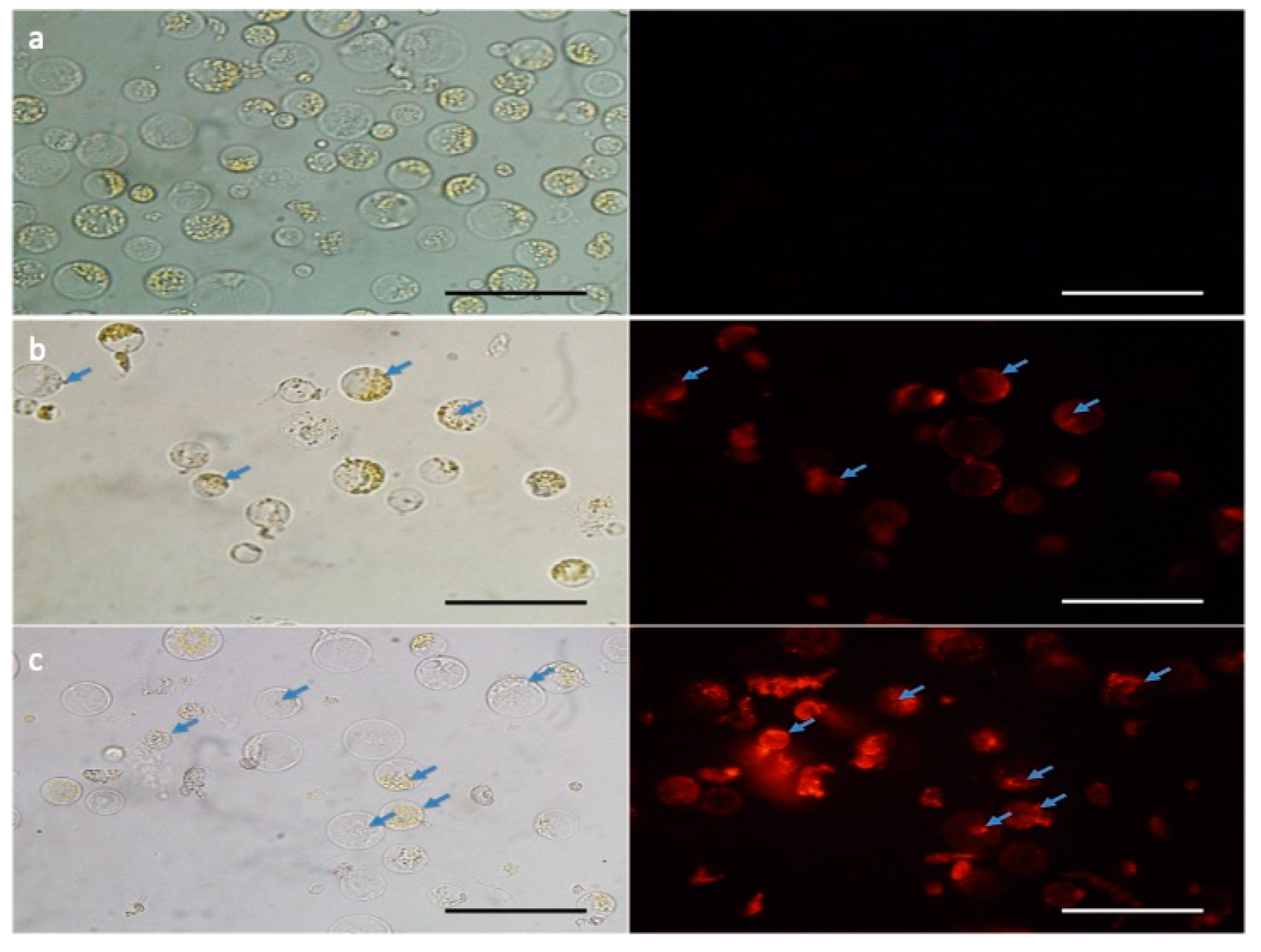
2.2. Maize Protoplast Isolation and Fluorescent Transfection Assay
2.3. Maize Protoplast Transformation
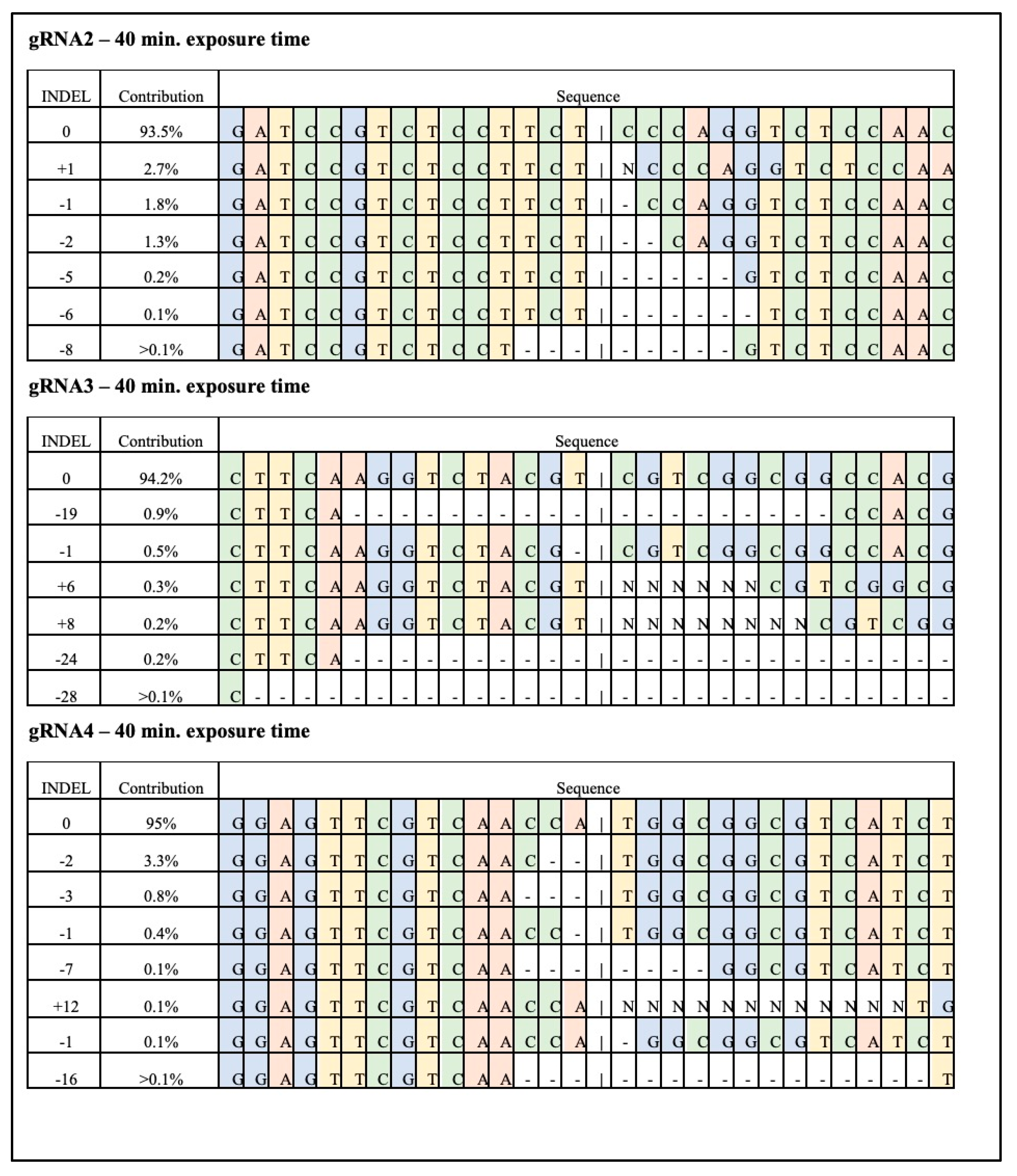
2.4. Gene-Editing Efficiency Analysis by Sanger Sequencing
3. Results
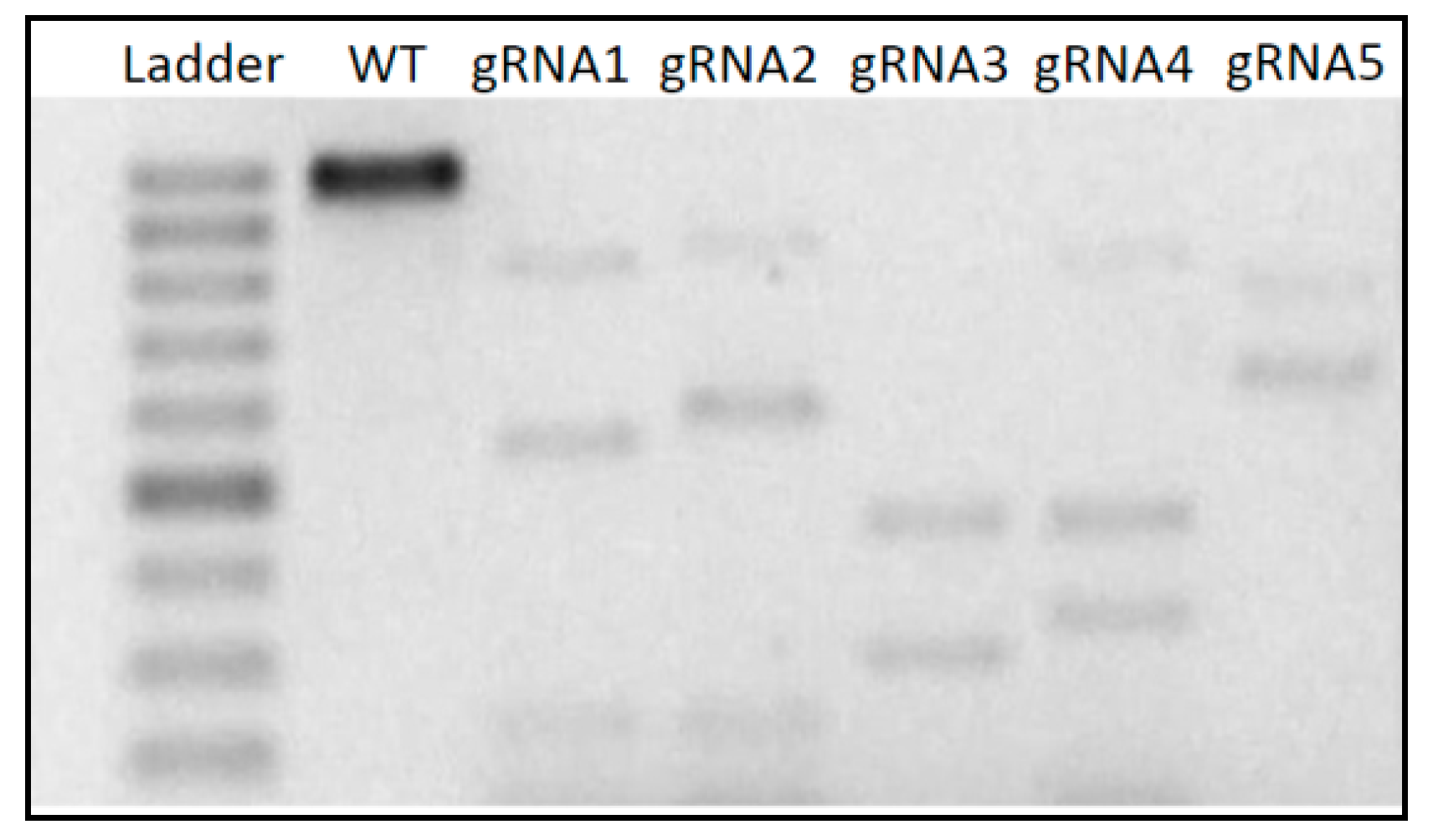
3.1. In Vitro Cleavage Assay
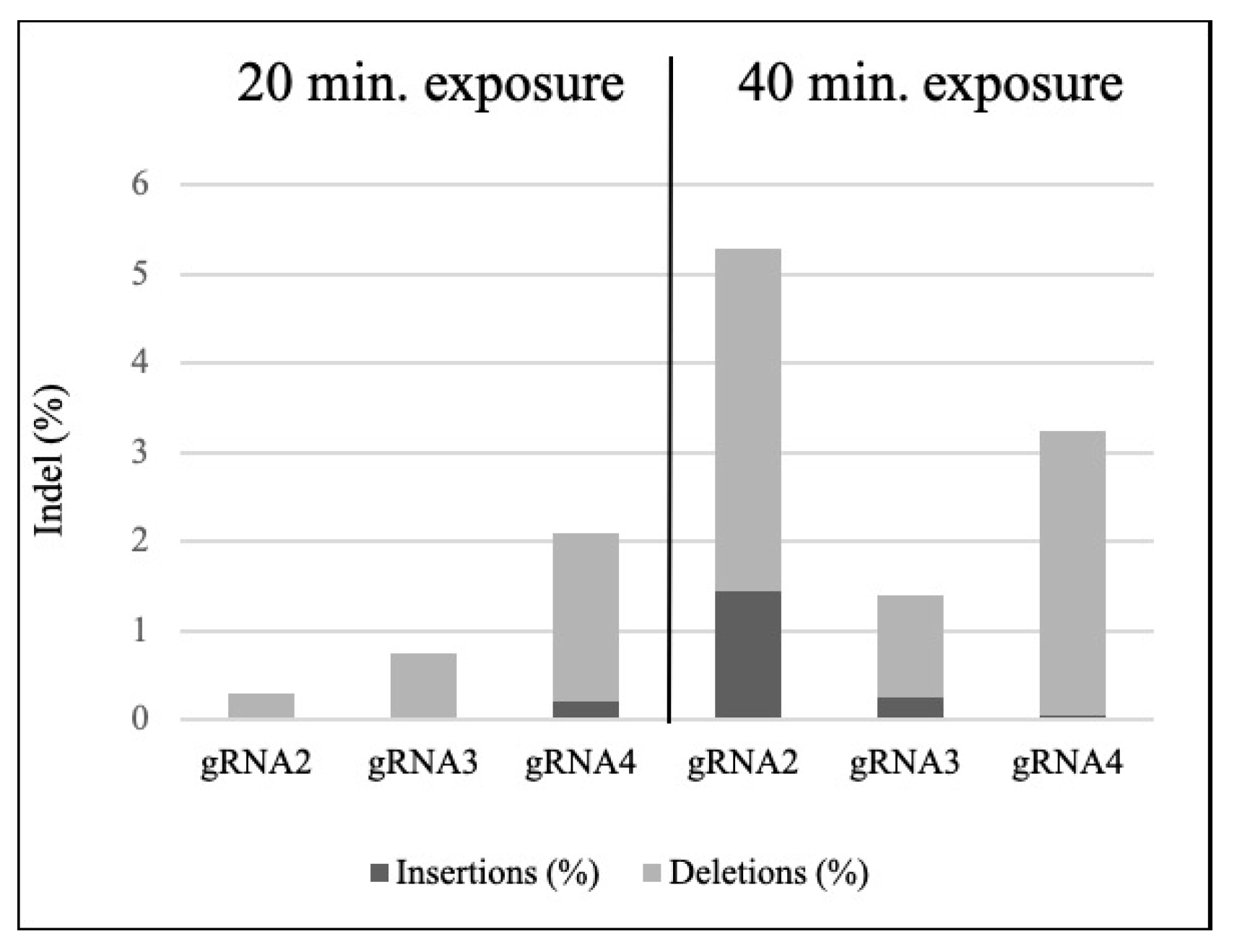
3.2. Targeted Mutagenesis in Maize Using CRISPR-Cas9 Ribonucleoproteins
4. Discussion
4.1. Ribonucleoprotein Delivery in Plants
4.2. CRISPR Delivery Methods in Maize
4.3. Analytical Platforms for Gene-Editing Detection
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Grobman, A.; Bonavia, D. Origin, Domestication, and Evolution of Maize; Cambridge University Press (CUP): Cambridge, UK, 2013; pp. 329–486. [Google Scholar]
- Li, H.; Wang, M.; Li, W.; He, L.; Zhou, Y.; Zhu, J.; Che, R.; Warburton, M.L.; Yang, X.; Yan, J. Genetic variants and underlying mechanisms influencing variance heterogeneity in maize. Plant J. 2020, 103, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Doudna, J. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-free genome editing: Past, present and future. Front. Plant Sci. 2019, 9. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, Y.; Zhu, F.; Hong, Y.; Ge, R. Efficient genome editing using CRISPR/Cas9 ribonucleoprotein approach in cultured Medaka fish cells. Biol. Open 2018, 7, bio035170. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N. DNA-free genome editing methods for targeted crop improvement. Plant Cell Rep. 2016, 35, 1469–1474. [Google Scholar] [CrossRef]
- Agarwal, A.; Yadava, P.; Kumar, K.; Singh, I.; Kaul, T.; Pattanayak, A.; Agrawal, P.K. Insights into maize genome editing via CRISPR/Cas9. Physiol. Mol. Biol. Plants 2018, 24, 175–183. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalan, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Kanchiswamy, C.N. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Ohlsson, P.; González, M.N.; Samuelsson, M.; Hofvander, P.; Olsson, P. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Cigan, A.M. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef]
- Liang, Z.; Zong, Y.; Gao, C. An efficient targeted mutagenesis system using CRISPR/Cas in monocotyledons. Curr. Protoc. Plant Biol. 2016, 1, 329–344. [Google Scholar] [CrossRef]
- Char, S.N.; Neelakandan, A.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K.; et al. An agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2016, 15, 257–268. [Google Scholar] [CrossRef]
- Feng, C.; Su, H.; Bai, H.; Wang, R.; Liu, Y.; Guo, X.; Liu, C.; Zhang, J.; Yuan, J.; Birchler, J.A.; et al. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 2018, 16, 1848–1857. [Google Scholar] [CrossRef]
- Dong, L.; Qi, X.; Zhu, J.; Liu, C.; Zhang, X.; Cheng, B.; Mao, L.; Xie, C. Supersweet and waxy: Meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019, 17, 1853–1855. [Google Scholar] [CrossRef]
- Sheen, J. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 1991, 3, 225–245. [Google Scholar]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Hsiau, T.; Conant, D.; Rossi, N.; Maures, T.; Waite, K.; Yang, J.; Joshi, S.; Kelso, R.; Holden, K.; Enzmann, B.L.; et al. Inference of crispr edits from sanger trace data. bioRxiv 2019, 251082. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-S.; Hsu, C.; Yang, L.; Lee, L.; Fu, J.; Cheng, Q.; Wu, F.; Hsiao, H.C.-W.; Zhang, Y.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Zhu, C.; Zischewski, J.; Perez, L.; Bassié, L.; Nadi, R.; Forni, G.; Lade, S.B.; Soto, E.; Jin, X.; et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 2016, 14, 2203–2216. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.; Kim, J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Braatz, J.; Harloff, H.-J.; Mascher, M.; Stein, N.; Himmelbach, A.; Harloff, H.-J. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef]
- Sandhya, D.; Jogam, P.; Allini, V.R.; Abbagani, S.; Alok, A. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 2020, 18, 1–11. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-free genome editing of brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 Ribonucleoprotein complexes. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 2019, 5, 363. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J. Genet. Genomics 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Xing, H.; Dong, L.; Wang, Z.; Zhang, H.; Han, C.; Liu, B.; Wang, X.; Chen, Q. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016, 16, 58. [Google Scholar] [CrossRef]
- Zhu, J.; Song, N.; Sun, S.; Yang, W.; Zhao, H.; Song, W.; Lai, J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genomics 2016, 43, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xu, Q.; Liu, Y.; Zhang, J.; Ren, D.; Wang, G.; Liu, Y. Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.L.; et al. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Doll, N.M.; Gilles, L.M.; Gérentes, M.F.; Richard, C.; Just, J.; Fierlej, Y.; Borrelli, V.G.M.; Gendrot, G.; Ingram, C.G.; Rogowsky, P.M. Single and multiple gene knockouts by CRISPR–Cas9 in maize. Plant Cell Rep. 2019, 38, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, F.; Liu, L.; Char, S.N.; Ding, Y.; Je, B.I.; Schmelz, E.; Yang, B.; Jackson, D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci. USA 2019, 117, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jian, L.; Xu, J.; Zhang, Q.; Zhang, M.; Jin, M.; Peng, Y.; Yan, J.; Han, B.; Liu, J.; et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 2020, 32, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Gao, H.; Mutti, J.; Young, J.K.; Yang, M.; Schroder, M.; Lenderts, B.; Wang, L.; Peterson, D.; Clair, G.S.; Jones, S.; et al. Complex trait loci in maize enabled by CRISPR-Cas9 mediated gene insertion. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Barone, P.; Wu, E.; Lenderts, B.; Anand, A.; Gordon-Kamm, W.; Svitashev, S.; Kumar, S. Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol. Plant 2020, 13. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Chen, R.; Yang, L.; Fan, K.; Liu, Y.; Wang, G.; Ren, Z.; Liu, Y. Generation of transgene-free Semidwarf maize plants by gene editing of gibberellin-oxidase20-3 using CRISPR/Cas9. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Park, J.; Choe, S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res. 2019, 28, 61–64. [Google Scholar] [CrossRef]
- Vu, G.T.H.; Cao, X.H.; Fauser, F.; Reiss, B.; Puchta, H.; Schubert, I. Endogenous sequence patterns predispose the repair modes of CRISPR /Cas9-induced DNA double-stranded breaks in Arabidopsis thaliana. Plant J. 2017, 92, 57–67. [Google Scholar] [CrossRef]
- Agapito-Tenfen, S.Z.; Okoli, A.S.; Bernstein, M.J.; Wikmark, O.-G.; Myhr, A.I. Revisiting risk governance of GM plants: The need to consider new and emerging gene-editing techniques. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
| Primer | Sequence (5′–3′) | Amplicon Size (bp) |
| ZmIPK-F | GAAGAAGCAGCAGAGCTTCA | 876 |
| ZmIPK-R | CAGAAGAAATCCGTGAGGACAG | |
| crRNA | Sequence (5′–3′) | Cleavage Fragments (bp) |
| crRNA1 | AGCTCGACCACGCCGCCGAC | 279 | 597 |
| crRNA2 * | GGGATCCGTCTCCTTCTCCC | 617 | 259 |
| crRNA3 * | ATCTTCAAGGTCTACGTCGT | 525 | 351 |
| crRNA4 * | CAGGAGTTCGTCAACCATGG | 498 |378 |
| crRNA5 | ACAAGCTCTACGGAGACGAC | 141 | 735 |
| Sample | Incubation Time (min) | % of Indel | Model Fit (R2) | KO Score | Mutation Range (bp) | Greater Contribution (bp) |
|---|---|---|---|---|---|---|
| Cas9 only | 20 | 0 | 0 | 0 | 0 | |
| gRNA2 only | 20 | 0 | 0 | 0 | 0 | |
| gRNA3 only | 20 | 0 | 0 | 0 | 0 | |
| gRNA4 only | 20 | 0 | 0 | 0 | 0 | |
| Cas9 + gRNA2 rep1 | 20 | 0 | 0.99 | 0 | 0 | 0 |
| Cas9 + gRNA2 rep2 | 20 | 1 | 1 | 1 | −4 to −2 | −2 |
| Cas9 + gRNA3 rep1 | 20 | 0 | 1 | 0 | −7 to −1 | −2 |
| Cas9 + gRNA3 rep2 | 20 | 1 | 1 | 1 | −7 to−1 | −2 |
| Cas9 + gRNA4 rep1 | 20 | 3 | 0.99 | 2 | −7 to +12 | −2 |
| Cas9 + gRNA4 rep2 | 20 | 1 | 0.99 | 1 | −2 to +3 | −2 |
| Cas9 + gRNA2 rep1 | 40 | 4 | 0.99 | 4 | −7 to +1 | −1 |
| Cas9 + gRNA2 rep2 | 40 | 6 | 1 | 6 | -8 to +1 | +1 |
| Cas9 + gRNA3 rep1 | 40 | 1 | 1 | 1 | −19 to−1 | −1 |
| Cas9 + gRNA3 rep2 | 40 | 2 | 0.96 | 2 | −28 to +8 | −19 |
| Cas9 + gRNA4 rep1 | 40 | 2 | 0.99 | 2 | −3 to −2 | −2 |
| Cas9 + gRNA4 rep2 | 40 | 5 | 1 | 4 | −16 to +12 | −2 |
| Reference | Plant Species | Plant Material | Transfection Method | Gene-Editing Efficiency |
|---|---|---|---|---|
| RNP delivered in plants | ||||
| [12] | Arabidopsis thaliana, Lactuca sativa, Nicotiana attenuata, Oryza sativa | Protoplasts | PEG-mediated | 5.7–40.0% |
| [13] | Malus domestica, Vitis vinifera | Protoplasts | PEG-mediated | 0.1–6.9% |
| [14] | Petunia x hybrid | Protoplasts | PEG-mediated | 2.4–21.0% |
| [31] | Triticum aestivum | Protoplasts, immature embryos | PEG-mediated, Biolistics | 0.2–45.3% |
| [15] | Solanum tuberosum | Protoplasts | PEG-mediated | 1.0–25.0% |
| [32] | Brassica oleracea, Brassica rapa | Protoplasts | PEG-mediated | 0.1–24.5% |
| [33] | Oryza sativa | Zygotes | PEG-mediated | 14.0–64.0% |
| Other delivery methods in maize | ||||
| [34] | Zea mays | Protoplasts | Vector via PEG-mediated | 13.1% |
| Immature embryos | Agrobacterium-mediated | 16.4–19.1% | ||
| [35] | Zea mays | Protoplasts | Vector via PEG-mediated | N.A |
| [24] | Zea mays | Immature embryos | Vector via biolistics | 1.3–4.6% |
| [17] | Zea mays | Protoplasts | Vector via biolistics | 80.0–90.0% |
| [36] | Zea mays | Immature embryos | Agrobacterium-mediated | 57.1–71.4% |
| [19] | Zea mays | Protoplasts | Vector via PEG-mediated | 2.8–27.0% |
| Immature embryos | Agrobacterium-mediated | 19.0–31.0% | ||
| [37] | Zea mays | Protoplasts | Vector via PEG-mediated | 4.0–11.9% |
| Immature embryos | Agrobacterium-mediated | 65.8–86.9% | ||
| [16] | Zea mays | Immature embryos | RNP via Biolistics | 0.01–0.7% |
| [18] | Zea mays | Immature embryos | Agrobacterium-mediated | 12.0–74.0% |
| [38] | Zea mays | Immature embryos | Vector via Biolistics | 60.0–98.0% |
| [39] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [19] | Zea mays | Immature embryos | Agrobacterium-mediated | 5.0–100% |
| [20] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [40] | Zea mays | Immature embryos | Agrobacterium-mediated | 90.0–100% |
| [41] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [42] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [43] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [44] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [45] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| [46] | Zea mays | Immature embryos | Vector via biolistics | N.A |
| [47] | Zea mays | Immature embryos | Agrobacterium-mediated | 25.0–100% |
| [48] | Zea mays | Immature embryos | Agrobacterium-mediated | N.A |
| Present study | Zea mays | Protoplasts | RNA via PEG-mediated | 0.85–5.85% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sant’Ana, R.R.A.; Caprestano, C.A.; Nodari, R.O.; Agapito-Tenfen, S.Z. PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts. Genes 2020, 11, 1029. https://doi.org/10.3390/genes11091029
Sant’Ana RRA, Caprestano CA, Nodari RO, Agapito-Tenfen SZ. PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts. Genes. 2020; 11(9):1029. https://doi.org/10.3390/genes11091029
Chicago/Turabian StyleSant’Ana, Rodrigo Ribeiro Arnt, Clarissa Alves Caprestano, Rubens Onofre Nodari, and Sarah Zanon Agapito-Tenfen. 2020. "PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts" Genes 11, no. 9: 1029. https://doi.org/10.3390/genes11091029
APA StyleSant’Ana, R. R. A., Caprestano, C. A., Nodari, R. O., & Agapito-Tenfen, S. Z. (2020). PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts. Genes, 11(9), 1029. https://doi.org/10.3390/genes11091029








