Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Data
2.2. Single-Gene Transcriptomic Quantifiers
2.2.1. Biological Replicas, Profiling Redundancy and Average Expression Level
2.2.2. Expression Variation
2.2.3. Expression Coordination
2.3. Gene Commanding Height (GCH) and Gene Master Regulator (GMR)
2.4. Expression Regulation
2.5. Quantifiers of the Functional Genomic Fabrics
3. Results
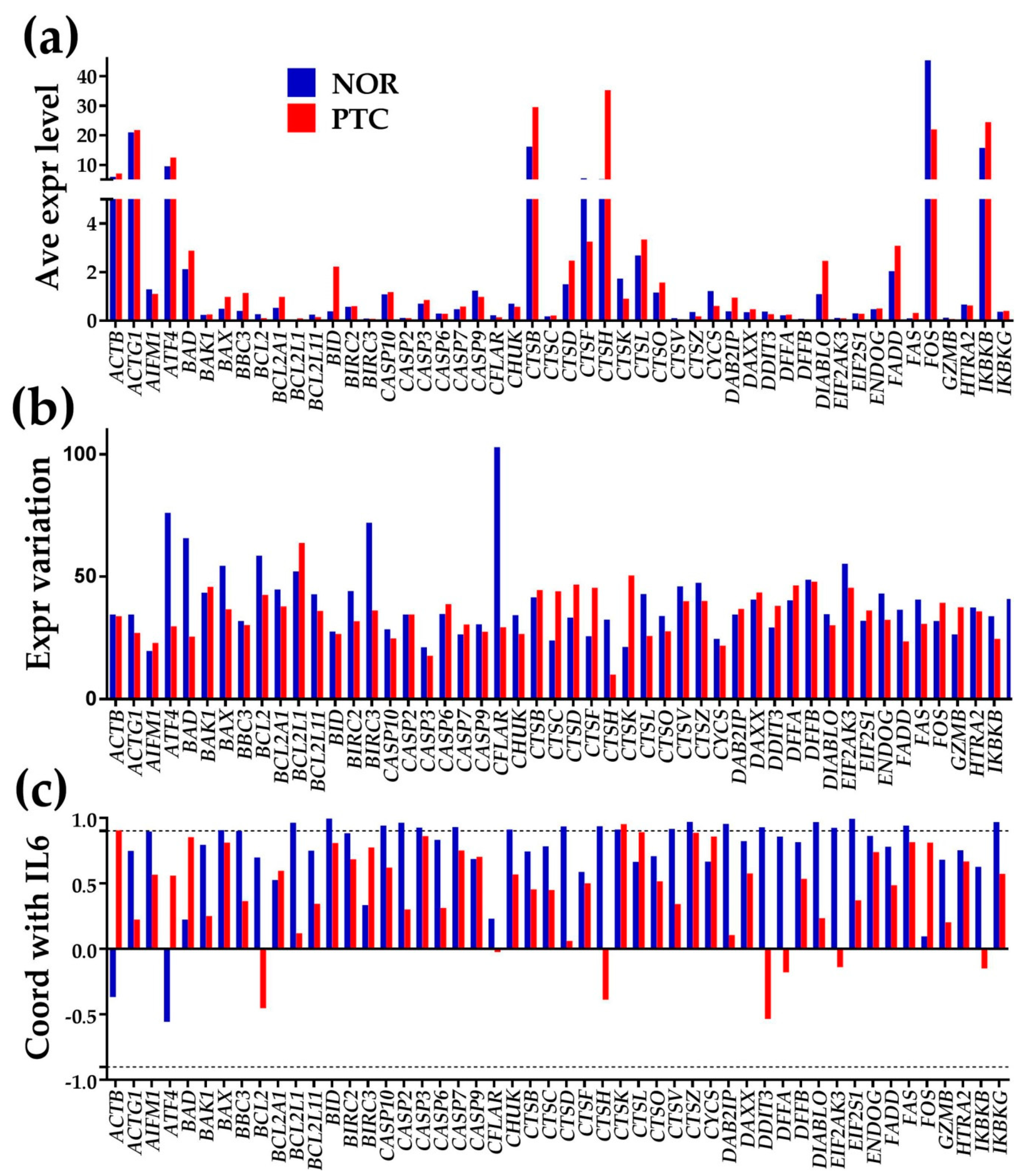
3.1. Overall Results
3.2. Three Independent Measures for Each Gene
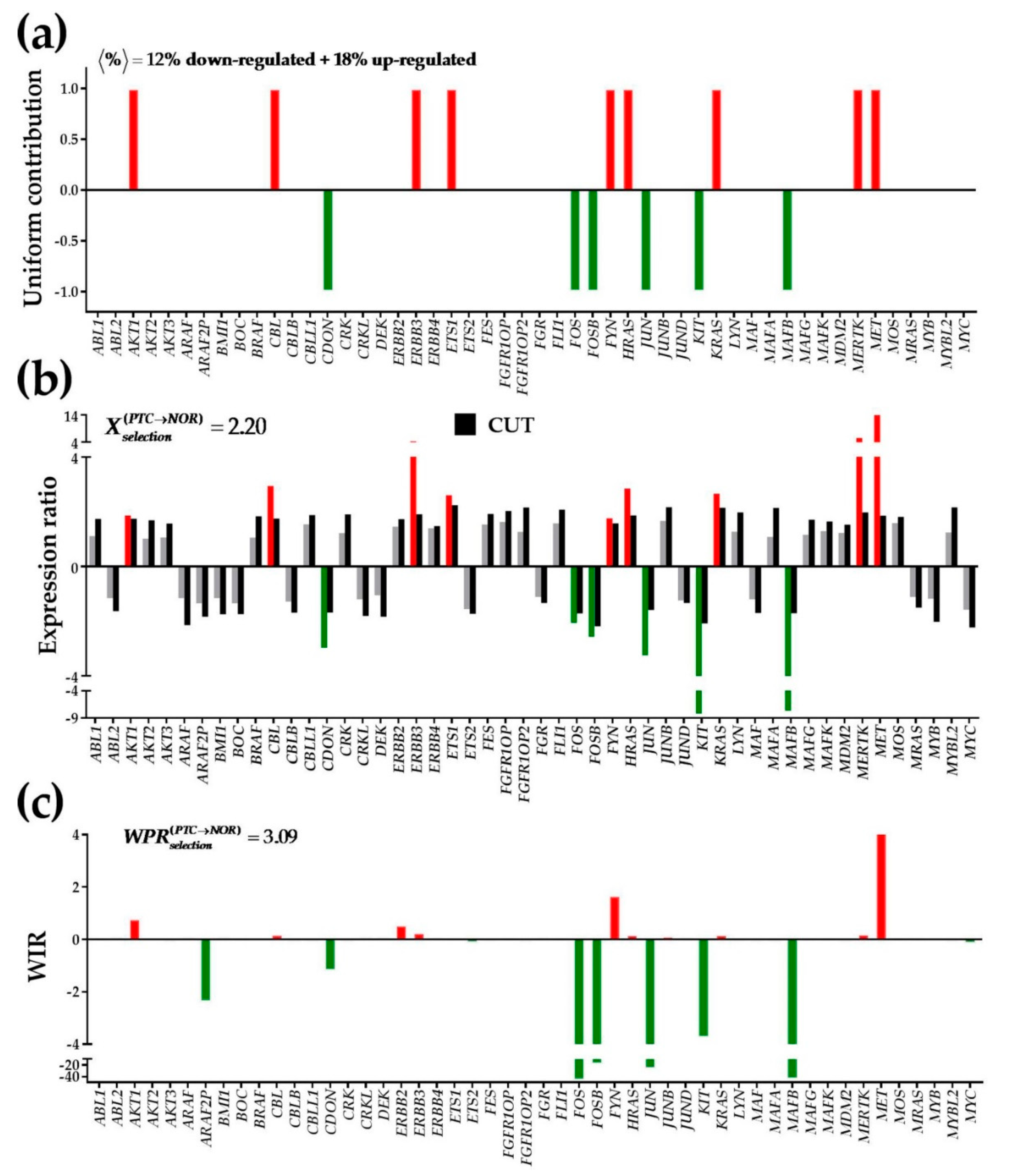
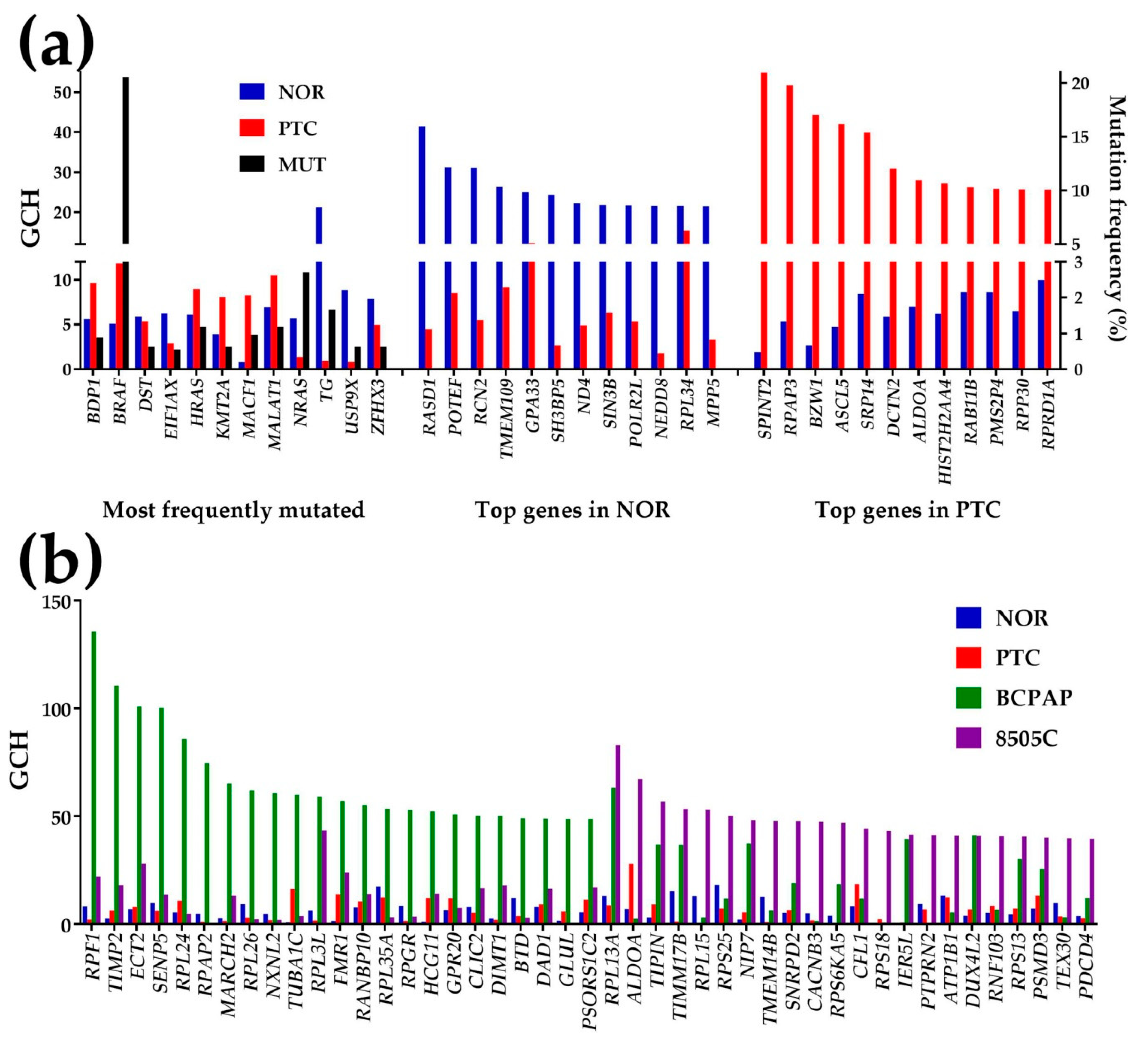
3.3. Expression Regulation
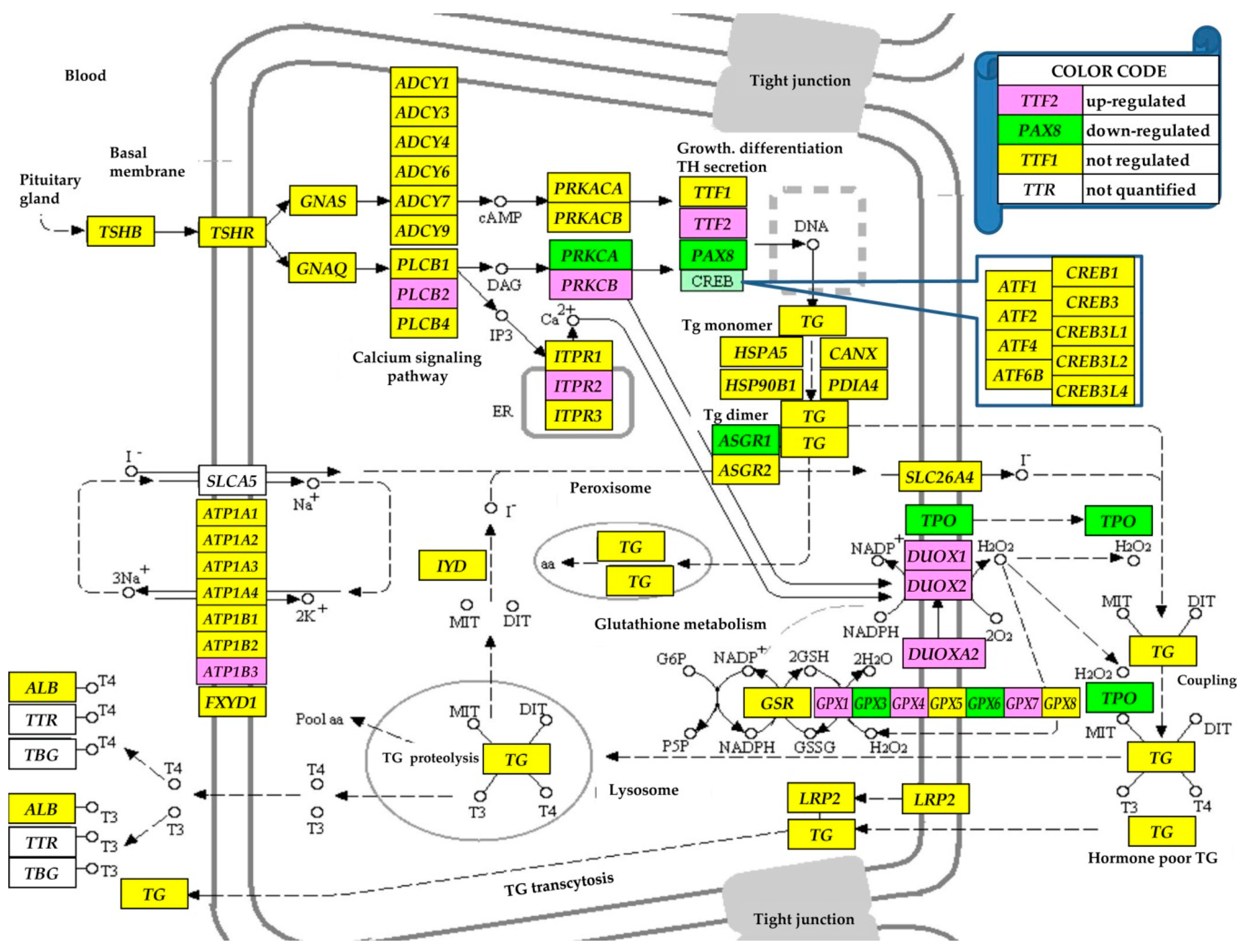
3.4. Regulation of the Thyroid Hormone Synthesis
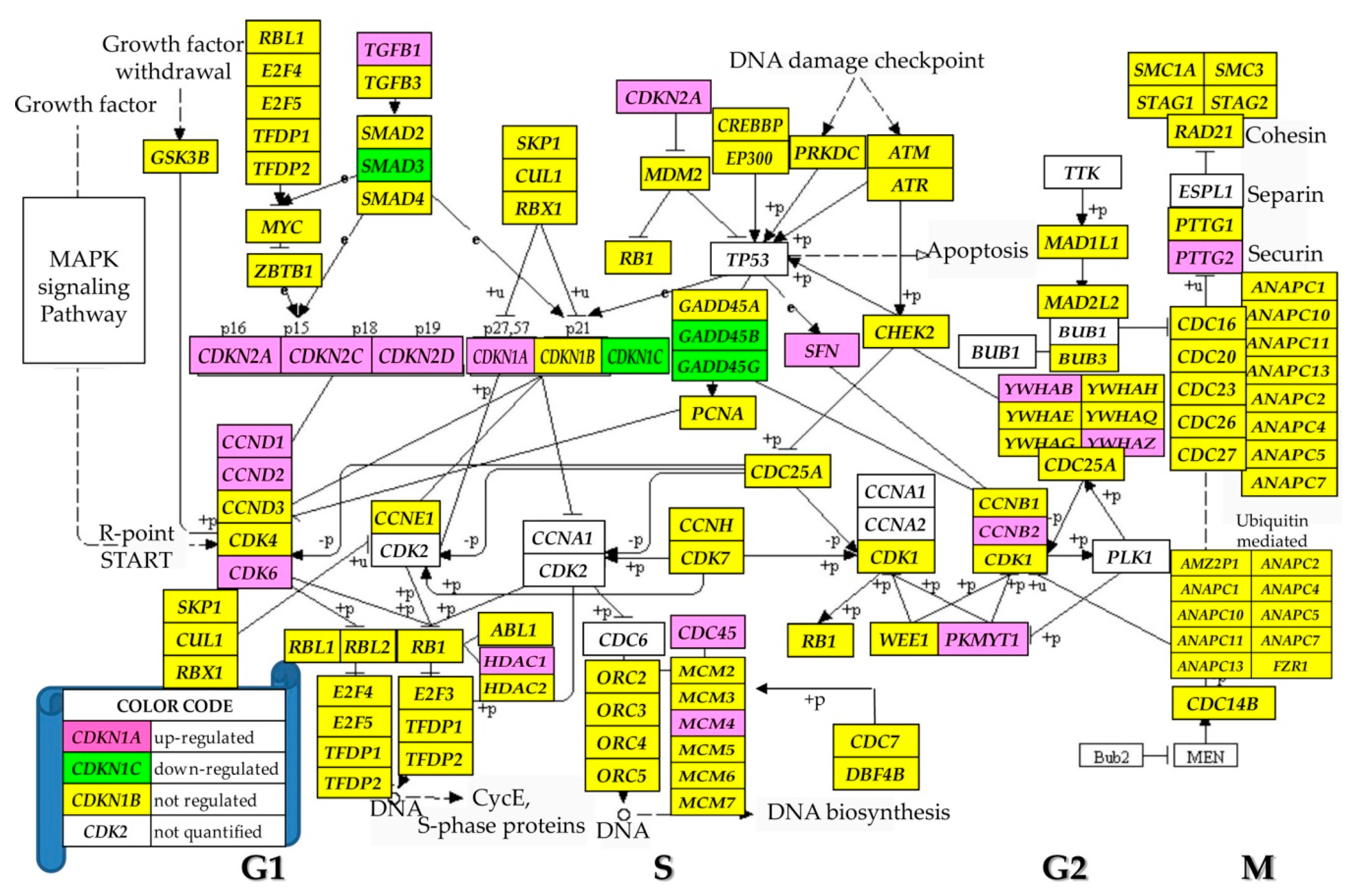
3.5. Regulation of the Cell-Cycle Pathway
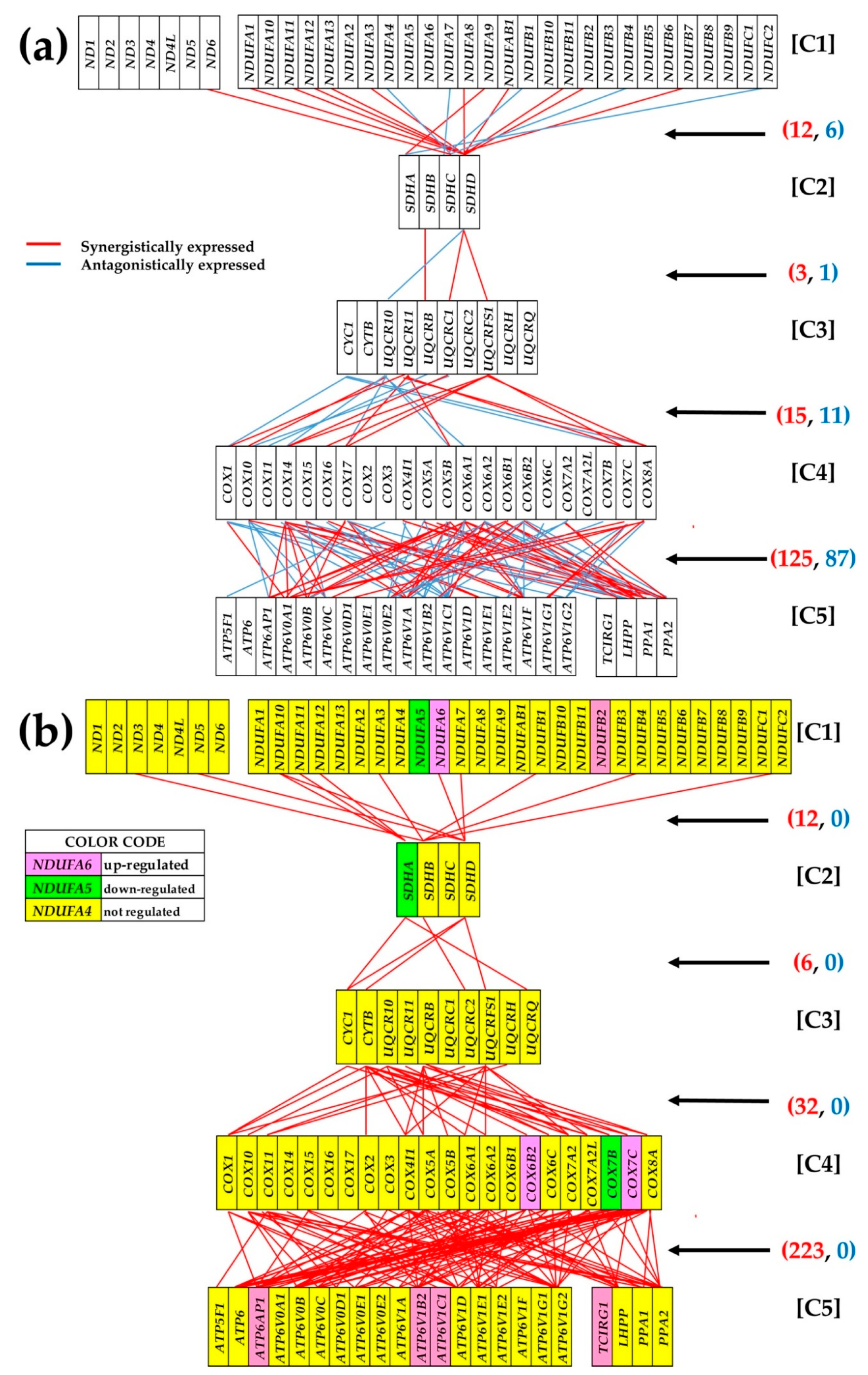
3.6. Remodeling of the Oxidative Phosphorylation Pathway
3.7. Gene Hierarchy
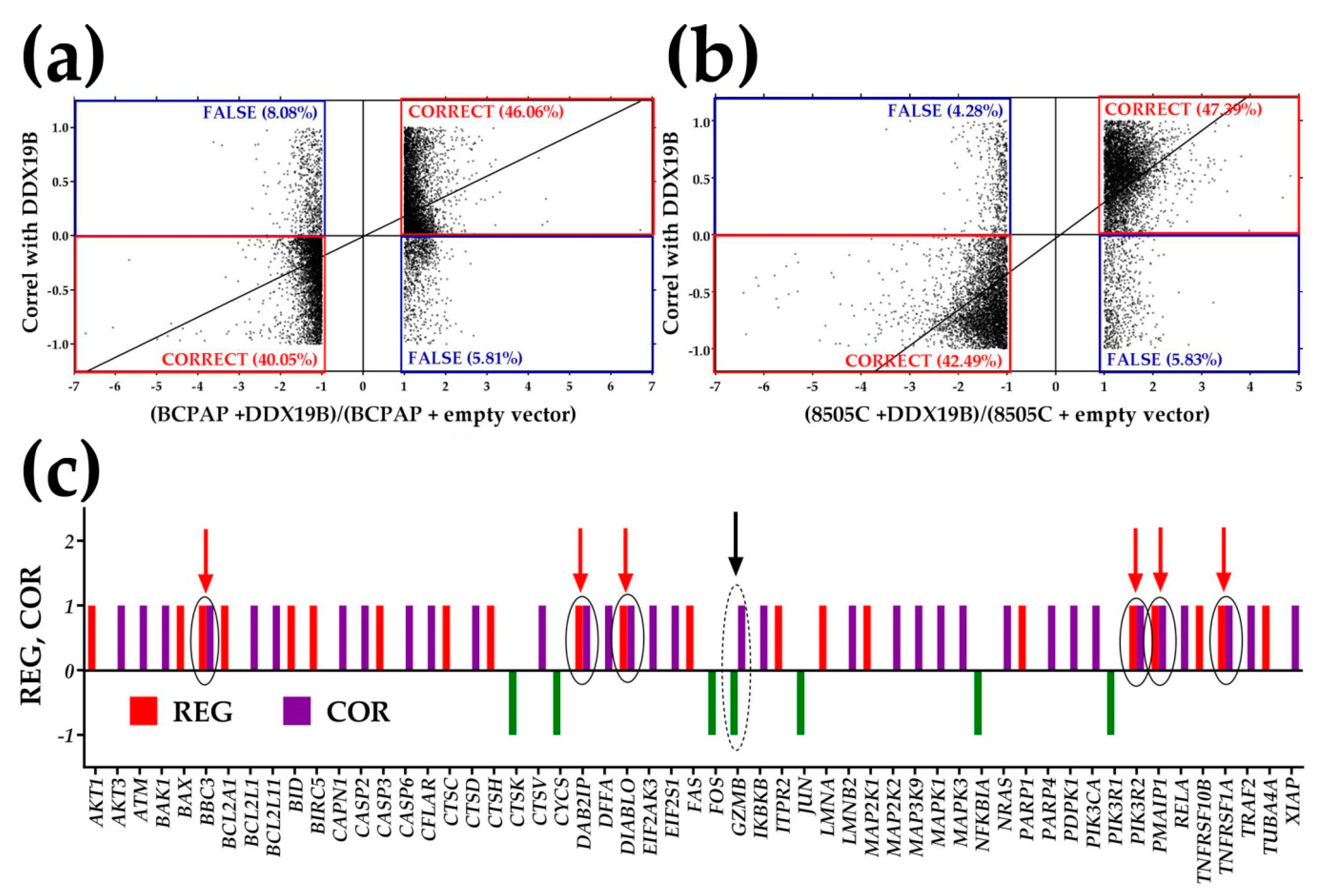
3.8. The Gene Master Regulator at Play
4. Discussion
- Each cell phenotype from the tumor is governed by a different gene hierarchy and a distinct organization of its transcriptome;
- As selected from the most altered genes in a large population of cancer patients, the biomarkers have low GCH and therefore little therapeutic value;
- The GMR of the cancer nodule is the most legitimate target of the gene therapy because it is the most influential gene for cancer cells while having very little role in the surrounding normal cells;
- SPINT2 was identified as the GMR of the PTC nodule of the profiled tumor and a gene with very low GCH score in NOR;
- The up-regulation of the synergistically expressed apoptosis genes in untreated PTC following the experimental SPINT2 overexpression was identified as a potential mechanism of selectively killing the cancer cells.
5. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Cancer Org Portal. Available online: https://www.cancer.org/cancer/thyroid-cancer (accessed on 26 July 2020).
- Thyroid Cancer Portal. Available online: https://www.thyroid.org/thyroid-cancer/ (accessed on 26 July 2020).
- Cancer Gov. Available online: https://portal.gdc.cancer.gov (accessed on 26 July 2020).
- 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Alexander, E.K.; Kennedy, G.C.; Baloch, Z.W.; Cibas, E.S.; Chudova, D.; Diggans, J.; Friedman, L.; Kloos, R.T.; LiVolsi, V.A.; Mandel, S.J.; et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N. Engl. J. Med. 2012, 367, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.I.; Junit, S.M.; Ng, K.L.; Jayapalan, J.J.; Karikalan, B.; Hashim, O.H. Papillary Thyroid Cancer: Genetic Alterations and Molecular Biomrker Investigations. Int. J. Med. Sci. 2019, 16, 450–460. [Google Scholar] [CrossRef]
- Foundation Medicine. Available online: https://www.foundationmedicine.com/genomic-testing (accessed on 12 July 2020).
- Iacobas, D.A.; Tuli, N.; Iacobas, S.; Rasamny, J.K.; Moscatello, A.; Geliebter, J.; Tiwari, R.K. Gene master regulators of papillary and anaplastic thyroid cancer phenotypes. Oncotarget 2018, 9, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, S.; Ede, N.; Iacobas, D.A. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes 2019, 10, 560. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A. Commentary on “The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. J. Cancer Immunol. 2019, 1, 31–33. [Google Scholar] [CrossRef]
- Iacobas, D.A. The Genomic Fabric Perspective on the Transcriptome between Universal Quantifiers and Personalized Genomic Medicine. Biol. Theory 2016, 11, 123–137. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gds/?term=iacobas (accessed on 26 July 2020).
- Zhang, H.; Xing, Z.; Mani, S.K.; Bancel, B.; Durantel, D.; Zoulim, F.; Tran, E.J.; Merle, P.; Andrisani, O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 2016, 64, 1033–1048. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, C.; Cao, J.; Xiong, M. NEMP1 Promotes Tamoxifen Resistance in Breast Cancer Cells. Biochem. Genet. 2019, 57, 813–826. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Z.; Li, Q.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Dai, Y.; Jiao, Y.; Zhang, Z.; et al. Prognostic significance of the PANK family expression in acute myeloid leukemia. Ann. Transl. Med. 2019, 7, 261. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Stout, R.; Spray, D.C. Cellular environment remodels the genomic fabrics of functional pathways in astrocytes. Genes 2020, 11, 520. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Lee, P.R.; Cohen, J.E.; Fields, R.D. Coordinated Activity of Transcriptional Networks Responding to the Pattern of Action Potential Firing in Neurons. Genes 2019, 10, 754. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A.; Iacobas, S.; Spray, D.C. Connexin43 and the brain transcriptome of the newborn mice. Genomics 2007, 89, 113–123. [Google Scholar] [CrossRef][Green Version]
- Mathew, R.; Huang, J.; Iacobas, S.; Iacobas, D.A. Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways. Genes 2020, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A.; Iacobas, S.; Tanowitz, H.B.; de Carvalho, A.C.; Spray, D.C. Functional genomic fabrics are remodeled in a mouse model of Chagasic cardiomyopathy and restored following cell therapy. Microbes Infect. 2018, 20, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/ (accessed on 21 June 2020).
- Liu, Y.C.; Yeh, C.T.; Lin, K.H. Molecular Functions of Thyroid Hormone Signaling in Regulation of Cancer Progression and Anti-Apoptosis. Int. J. Mol. Sci. 2019, 20, 4986. [Google Scholar] [CrossRef]
- Bai, J.W.; Wei, M.; Li, J.W.; Zhang, G.J. Notch Signaling Pathway and Endocrine Resistance in Breast Cancer. Front. Pharmacol. 2020, 11, 924. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef]
- Kobawala, T.P.; Trivedi, T.I.; Gajjar, K.K.; Patel, D.H.; Patel, G.H.; Ghosh, N.R. Significance of Interleukin-6 in Papillary Thyroid Carcinoma. J. Thyroid. Res. 2016, 6178921. [Google Scholar] [CrossRef]
- Bao, Y.; Zhao, T.L.; Zhang, Z.Q.; Liang, X.L.; Wang, Z.X.; Xiong, Y.; Lu, X.; Wang, L.H. High eukaryotic translation elongation factor 1 alpha 1 expression promotes proliferation and predicts poor prognosis in clear cell renal cell carcinoma. Neoplasma 2020, 67, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Lin, J.D.; Chen, J.T.; Chang, C.M.; Weng, H.F.; Hsueh, C.; Chien, H.P.; Yu, J.S. Integrated analysis of fine-needle-aspiration cystic fluid proteome, cancer cell secretome, and public transcriptome datasets for papillary thyroid cancer biomarker discovery. Oncotarget 2018, 9, 12079–12100. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Chen, H.; Lu, P.; Li, X.; Long, M.; Peng, X.; Huang, M.; Huang, K.; Lin, S.; Tan, L.; et al. CHI3L1 overexpression is associated with metastasis and is an indicator of poor prognosis in papillary thyroid carcinoma. Cancer Biomark. 2017, 18, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.P.; Lee, J.J.; Chang, Y.C.; Lin, C.H.; Li, Y.S.; Liu, C.L. Overexpression of chitinase-3-like protein 1 is associated with structural recurrence in patients with differentiated thyroid cancer. J. Pathol. 2020, e5503. [Google Scholar] [CrossRef]
- Oczko-Wojciechowska, M.; Pfeifer, A.; Jarzab, M.; Swierniak, M.; Rusinek, D.; Tyszkiewicz, T.; Kowalska, M.; Chmielik, E.; Zembala-Nozynska, E.; Czarniecka, A.; et al. Impact of the Tumor Microenvironment on the Gene Expression Profile in Papillary Thyroid Cancer. Pathobiology 2020, 87, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Jevnikar, Z.; Rojnik, M.; Jamnik, P.; Doljak, B.; Fonovic, U.P.; Kos, J. Cathepsin H mediates the processing of talin and regulates migration of prostate cancer cells. J. Biol. Chem. 2013, 288, 2201–2209. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Werner, P.; Scemes, E.; Spray, D.C. Alteration of transcriptomic networks in adoptive-transfer experimental autoimmune encephalomyelitis. Front. Integr. Neurosci. 2007, 1. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S. Towards a Personalized Cancer Gene Therapy: A Case of Clear Cell Renal Cell Carcinoma. Cancer Oncol. Res. 2017, 5, 45–52. [Google Scholar] [CrossRef]
- Frigeri, A.; Iacobas, D.A.; Iacobas, S.; Nicchia, G.P.; Desaphy, J.-F.; Camerino, D.C.; Svelto, M.; Spray, D.C. Effect of microagravity on brain gene expression in mice. Exp. Brain Res. 2008, 191, 289–300. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Fan, C.; Iacobas, S.; Spray, D.C.; Haddad, G.G. Transcriptomic changes in developing kidney exposed to chronic hypoxia. Biochem. Biophys. Res. Commun. 2006, 349, 329–338. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Fan, C.; Iacobas, S.; Haddad, G.G. Integrated transcriptomic response to cardiac chronic hypoxia: Translation regulators and response to stress in cell survival. Funct. Integr. Genom. 2008, 8, 265–275. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iacobas, D.A.; Iacobas, S.; Haddad, G.G. Heart rhythm genomic fabric in hypoxia. Biochem. Biophys. Res. Commun. 2010, 391, 1769–1774. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A.; Urban, M.; Iacobas, S.; Scemes, E.; Spray, D.C. Array analysis of gene expression in connexin43 null astrocytes. Physiol. Genom. 2003, 15, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Iacobas, D.A.; Iacobas, S.; Urban-Maldonado, M.; Scemes, E.; Spray, D.C. Similar transcriptomic alterations in Cx43 knock-down and knock-out astrocytes. Cell Commun. Adhes. 2008, 15, 195–206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Surmiak, M.; Hubalewska-Mazgaj, M.; Wawrzycka-Adamczyk, K.; Musiał, J.; Sanak, M. Delayed neutrophil apoptosis in granulomatosis with polyangiitis: Dysregulation of neutrophil gene signature and circulating apoptosis-related proteins. Scand. J. Rheumatol. 2020, 49, 57–67. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.; Zhong, L.; He, M.; Chen, S.; Wang, T.; Ma, S. The combined use of miRNAs and mRNAs as biomarkers for the diagnosis of papillary thyroid carcinoma. Int. J. Mol. Med. 2015, 36, 1097–1103. [Google Scholar] [CrossRef]
- Deligiorgi, M.V.; Mahaira, H.; Eftychiadis, C.; Kafiri, G.; Georgiou, G.; Theodoropoulos, G.; Konstadoulakis, M.M.; Zografos, E.; Zografos, G.C. RANKL, OPG, TRAIL, KRas, and c-Fos expression in relation to central lymph node metastases in papillary thyroid carcinoma. J. BU ON Off. J. Balk. Union Oncol. 2018, 23, 1029–1040. [Google Scholar]
- Kataki, A.; Sotirianakos, S.; Memos, N.; Karayiannis, M.; Messaris, E.; Leandros, E.; Manouras, A.; Androulakis, G. P53 and C-FOS overexpression in patients with thyroid cancer: An immunohistochemical study. Neoplasma 2003, 50, 26–30. [Google Scholar]
- Franceschi, S.; Lessi, F.; Panebianco, F.; Tantillo, E.; La Ferla, M.; Menicagli, M.; Aretini, P.; Apollo, A.; Naccarato, A.G.; Marchetti, I.; et al. Loss of c-KIT expression in thyroid cancer cells. PLoS ONE 2017, 12, e0173913. [Google Scholar] [CrossRef]
- Chu, Y.H.; Sadow, P.M. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP): Diagnostic Updates and Molecular Advances. In Seminars in Diagnostic Pathology; WB Saunders: Philadelphia, PA, USA, 2020. [Google Scholar]
- Lubos, E.; Kelly, N.J.; Oldebeken, S.R.; Leopold, J.A.; Zhang, Y.Y.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 deficiency augments proinflammatory cytokine-induced redox signaling and human endothelial cell activation. J. Biol. Chem. 2011, 286, 35407–35417. [Google Scholar] [CrossRef]
- Cardoso, M.F.S.; Castelletti, C.H.M.; Lima-Filho, J.L.; Martins, D.B.G.; Teixeira, J.A.C. Putative biomarkers for cervical cancer: SNVs, methylation and expression profiles. Mutat. Res. 2017, 773, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Shen, C.; Miao, R.; Wang, J.Z.; Cao, M.D.; Zhang, Y.S.; Shi, L.H.; Zhao, G.H.; Wang, M.H.; Wu, L.S.; et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57Kip2 by an m5C-dependent manner. Cell Death Dis. 2020, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Murakami, M.; Ikeda, Y.; Nakagawa, Y.; Tsuji, A.; Kitagishi, Y. Role of tumor suppressor molecules in genomic perturbations and damaged DNA repair involved in the pathogenesis of cancer and neurodegeneration (Review). Biomed. Rep. 2020, 13, 10. [Google Scholar] [CrossRef]
- Księżakowska-Łakoma, K.; Żyła, M.; Wilczyński, J.R. Mitochondrial dysfunction in cancer. Prz. Menopauzalny 2014, 13, 136–144. [Google Scholar] [CrossRef]
- Huang, Y.P.; Chang, N.W. PPARα modulates gene expression profiles of mitochondrial energy metabolism in oral tumorigenesis. Biomedicine 2016, 6, 3. [Google Scholar] [CrossRef]
- Roversi, F.M.; Olalla Saad, S.T.; Machado-Neto, J.A. Serine peptidase inhibitor Kunitz type 2 (SPINT2) in cancer development and progression. Biomed. Pharmacother. 2018, 101, 278–286. [Google Scholar] [CrossRef]
- Guan, X.; Guan, Z.; Song, C. Expression profile analysis identifies key genes as prognostic markers for metastasis of osteosarcoma. Cancer Cell Int. 2020, 20, 104. [Google Scholar] [CrossRef]
- Graumann, J.; Finkernagel, F.; Reinartz, S.; Stief, T.; Brödje, D.; Renz, H.; Jansen, J.M.; Wagner, U.; Worzfeld, T.; Pogge von Strandmann, E.; et al. Multi-platform affinity proteomics identify proteins linked to metastasis and immune suppression in ovarian cancer plasma. Front. Oncol. 2019, 9, 1150. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Cox, C.D.; Chowdhury, R.; Dovek, L.; Nguyen, H.; Li, T.; Li, S.; Ozer, B.; Chou, A.; Nguyen, N.; et al. SPINT2 is hypermethylated in both IDH1 mutated and wild-type glioblastomas, and exerts tumor suppression via reduction of c-Met activation. J. Neurooncol. 2019, 142, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.S.; Celeiro, S.P.; Costa, Â.M.; Pinto, F.; Popov, S.; de Almeida, G.C.; Amorim, J.; Pires, M.M.; Pinheiro, C.; Lopes, J.M.; et al. Loss of SPINT2 expression frequently occurs in glioma, leading to increased growth and invasion via MMP2. Cell. Oncol. 2020, 43, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Shu, X.; Bao, J.; Guo, X.; Kote-Jarai, Z.; Haiman, C.A.; Eeles, R.A.; Zheng, W.; PRACTICAL, CRUK, BPC3, CAPS, PEGASUS Consortia. Analysis of Over 140,000 European Descendants Identifies Genetically Predicted Blood Protein Biomarkers Associated with Prostate Cancer Risk. Cancer Res. 2019, 79, 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Liu, D.; Li, W.; Di, S.; Zhang, Z.; Zhang, J.; Xu, L.; Guo, K.; Zhu, Y.; Han, J.; et al. STYK1 promotes tumor growth and metastasis by reducing SPINT2/HAI-2 expression in non-small cell lung cancer. Cell Death Dis. 2019, 10, 435. [Google Scholar] [CrossRef] [PubMed]
- Roversi, F.M.; Cury, N.M.; Lopes, M.R.; Ferro, K.P.; Machado-Neto, J.A.; Alvarez, M.C.; Dos Santos, G.P.; Giardini Rosa, R.; Longhini, A.L.; Duarte, A.D.S.S.; et al. Up-regulation of SPINT2/HAI-2 by Azacytidine in bone marrow mesenchymal stromal cells affects leukemic stem cell survival and adhesion. J. Cell. Mol. Med. 2019, 23, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Che, Y.; Yin, F.; Yu, F.; Bi, X.; Wang, Y. Study on the methylation status of SPINT2 gene and its expression in cervical carcinoma. Cancer Biomark. 2018, 22, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, C.F.; Llorca, O. RPAP3 C-Terminal Domain: A Conserved Domain for the Assembly of R2TP Co-Chaperone Complexes. Cells 2020, 9, 1139. [Google Scholar] [CrossRef] [PubMed]
- Kawachi, T.; Tanaka, S.; Fukuda, A.; Sumii, Y.; Setiawan, A.; Kotoku, N.; Kobayashi, M.; Arai, M. Target identification of the marine natural products Dictyoceratin-A and -C as selective growth inhibitors in cancer cells adapted to hypoxic environments. Mar. Drugs 2019, 17, 163. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, H.; Gong, L.; Yao, L.; Li, Y.; Zhang, W. MicroRNA-129-3p functions as a tumor suppressor in serous ovarian cancer by targeting BZW1. Int. J. Clin. Exp. Pathol. 2018, 11, 5901–5908. [Google Scholar]
- Chiou, J.; Chang, Y.C.; Jan, Y.H.; Tsai, H.F.; Yang, C.J.; Huang, M.S.; Yu, Y.L.; Hsiao, M. Overexpression of BZW1 is an independent poor prognosis marker and its down-regulation suppresses lung adenocarcinoma metastasis. Sci. Rep. 2019, 9, 14624. [Google Scholar] [CrossRef]
- Li, S.; Chai, Z.; Li, Y.; Liu, D.; Bai, Z.; Li, Y.; Li, Y.; Situ, Z. BZW1, a novel proliferation regulator that promotes growth of salivary muocepodermoid carcinoma. Cancer Lett. 2009, 284, 86–94. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Spray, D.C. Connexin-dependent transcellular transcriptomic networks in mouse brain. Prog. Biophys. Mol. Biol. 2007, 94, 168–184. [Google Scholar] [CrossRef]
| GENE | DESCRIPTION | NOR | PTC | REG |
|---|---|---|---|---|
| ACTG1 | actin, γ 1 | S | ||
| AKT1 | v-akt murine thymoma viral oncogene homolog 1 | U | ||
| AKT3 | v-akt murine thymoma viral oncogene homolog 3 | S | ||
| ATF4 | activating transcription factor 4 | A | ||
| ATM | ataxia telangiectasia mutated | S | ||
| BAK1 | BCL2-antagonist/killer 1 | S | ||
| BAX | BCL2-associated X protein | U | ||
| BBC3 | BCL2 binding component 3 | S | U | |
| BCL2 | B-cell CLL/lymphoma 2 | I | ||
| BCL2A1 | BCL2-related protein A1 | U | ||
| BCL2L1 | BCL2-like 1 | S | ||
| BCL2L11 | BCL2-like 11 | S | ||
| BID | BH3 interacting domain death agonist | U | ||
| BIRC5 | baculoviral IAP repeat containing 5 | U | ||
| CAPN1 | calpain 1, (mu/I) large subunit | S | ||
| CASP2 | caspase 2, apoptosis-related cysteine peptidase | S | ||
| CASP3 | caspase 3, apoptosis-related cysteine peptidase | U | ||
| CASP6 | caspase 6, apoptosis-related cysteine peptidase | S | ||
| CASP9 | caspase 9, apoptosis-related cysteine peptidase | S | ||
| CFLAR | CASP8 and FADD-like apoptosis regulator | S | ||
| CTSC | cathepsin C | U | ||
| CTSD | cathepsin D | S | ||
| CTSH | cathepsin H | U | ||
| CTSK | cathepsin K | D | ||
| CTSL | cathepsin L | S | ||
| CTSV | cathepsin V | S | ||
| CYCS | cytochrome c, somatic | S | D | |
| DAB2IP | DAB2 interacting protein | S | U | |
| DFFA | DNA fragmentation factor | S | ||
| DIABLO | diablo, IAP-binding mitochondrial protein | S | U | |
| EIF2AK3 | eukaryotic translation initiation factor 2-α kinase 3 | S | ||
| EIF2S1 | eukaryotic translation initiation factor 2, subunit 1 α | S | ||
| FAS | Fas cell surface death receptor | U | ||
| FOS | FBJ murine osteosarcoma viral oncogene homolog | D | ||
| GZMB | granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | S | D | |
| IKBKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase β | I | S | |
| ITPR2 | inositol 1,4,5-trisphosphate receptor, type 2 | U | ||
| JUN | jun proto-oncogene | D | ||
| LMNA | lamin A/C | U | ||
| LMNB2 | lamin B2 | S | ||
| MAP2K1 | mitogen-activated protein kinase kinase 1 | U | ||
| MAP2K2 | mitogen-activated protein kinase kinase 2 | S | ||
| MAP3K9 | mitogen-activated protein kinase kinase kinase 9 | S | ||
| MAPK1 | mitogen-activated protein kinase 1 | S | ||
| MAPK3 | mitogen-activated protein kinase 3 | S | ||
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | S | ||
| NFKBIA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | D | ||
| NRAS | neuroblastoma RAS viral (v-ras) oncogene homolog | S | ||
| PARP1 | poly (ADP-ribose) polymerase 1 | U | ||
| PARP4 | poly (ADP-ribose) polymerase family, member 4 | S | ||
| PDPK1 | 3-phosphoinositide dependent protein kinase 1 | S | ||
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α | S | ||
| PIK3R1 | phosphoinositide-3-kinase, regulatory subunit 1 | D | ||
| PIK3R2 | phosphoinositide-3-kinase, regulatory subunit 2 | S | U | |
| PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 | S | U | |
| RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 | S | ||
| RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | S | ||
| TNFRSF10B | tumor necrosis factor receptor superfamily, member 10b | U | ||
| TNFRSF1A | tumor necrosis factor receptor superfamily, member 1A | S | U | |
| TRAF2 | TNF receptor-associated factor 2 | S | ||
| TUBA1C | tubulin, α 1b | I | ||
| TUBA3D | tubulin, α 3c | I | ||
| TUBA4A | tubulin, α 4a | U | ||
| XIAP | X-linked inhibitor of apoptosis | S |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobas, D.A. Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma. Genes 2020, 11, 1030. https://doi.org/10.3390/genes11091030
Iacobas DA. Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma. Genes. 2020; 11(9):1030. https://doi.org/10.3390/genes11091030
Chicago/Turabian StyleIacobas, Dumitru A. 2020. "Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma" Genes 11, no. 9: 1030. https://doi.org/10.3390/genes11091030
APA StyleIacobas, D. A. (2020). Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma. Genes, 11(9), 1030. https://doi.org/10.3390/genes11091030





