A Highly Polymorphic Panel Consisting of Microhaplotypes and Compound Markers with the NGS and Its Forensic Efficiency Evaluations in Chinese Two Groups
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Novel Microhaplotypes and Compound Markers
2.2. Sample Preparation and DNA Extraction
2.3. Reference Populations
2.4. Libraries Construction and Sequencing Using the NGS
2.5. Statistical Analyses
3. Results
3.1. General Information of the 29 Microhaplotypes and Compound Markers
3.2. Genetic Diversities and Forensic Efficiencies of 29 Loci in Five Continental Populations
3.3. Genetic Divergences and Population Structure Evaluations of Different Continental Populations
3.4. Sequencing Results of the Developed Multiplex System Using the NGS Platform
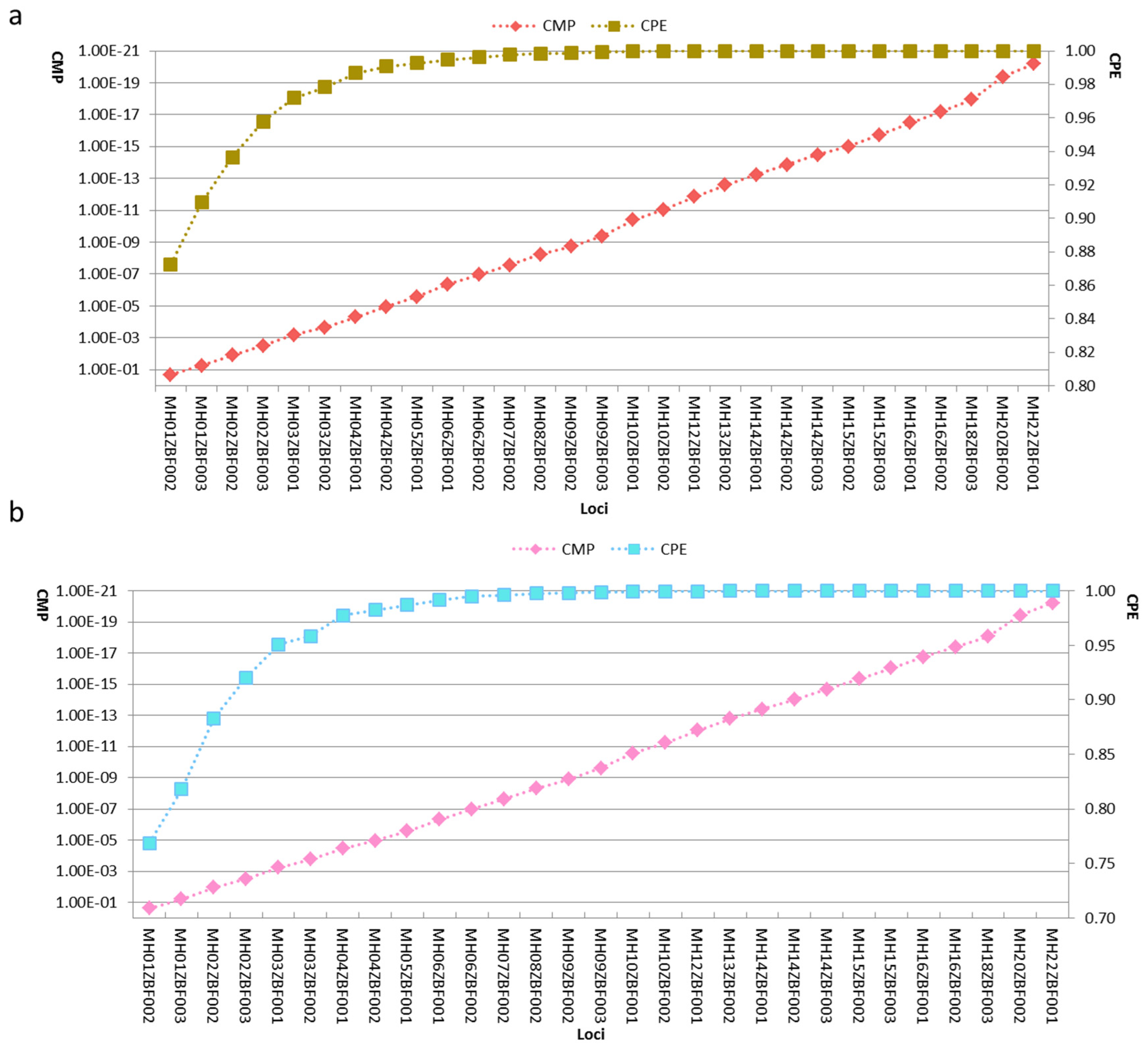
3.5. Genetic Distributions and Forensic Parameters of the 29 Loci in Kazak and Mongolian Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sheng, X.; Wang, Y.; Zhang, J.; Chen, L.; Lin, Y.; Zhao, Z.; Li, C.; Zhang, S. Forensic investigation of 23 autosomal STRs and application in Han and Mongolia ethnic groups. Forensic Sci. Res. 2018, 3, 138–144. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, B.; Jin, X.; Yan, J.; Meng, H.; Zhu, B. Genetic and structural characterization of 20 autosomal short tandem repeats in the Chinese Qinghai Han population and its genetic relationships and interpopulation differentiations with other reference populations. Forensic Sci. Res. 2018, 3, 145–152. [Google Scholar] [CrossRef]
- Butler, J.M.; Shen, Y.; McCord, B.R. The development of reduced size STR amplicons as tools for analysis of degraded DNA. J. Forensic Sci. 2003, 48, 1054–1064. [Google Scholar] [CrossRef]
- Kidd, K.K.; Pakstis, A.J.; Speed, W.C.; Grigorenko, E.L.; Kajuna, S.L.; Karoma, N.J.; Kungulilo, S.; Kim, J.J.; Lu, R.B.; Odunsi, A.; et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci. Int. 2006, 164, 20–32. [Google Scholar] [CrossRef]
- Avent, I.; Kinnane, A.G.; Jones, N.; Petermann, I.; Daniel, R.; Gahan, M.E.; McNevin, D. The QIAGEN 140-locus single-nucleotide polymorphism (SNP) panel for forensic identification using massively parallel sequencing (MPS): An evaluation and a direct-to-PCR trial. Int. J. Leg. Med. 2019, 133, 677–688. [Google Scholar] [CrossRef]
- Borsting, C.; Mogensen, H.S.; Morling, N. Forensic genetic SNP typing of low-template DNA and highly degraded DNA from crime case samples. Forensic Sci. Int. Genet. 2013, 7, 345–352. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, H.; Liu, Y.; Guo, J.; Sun, Y.; Fu, X.; Xiao, D.; Cai, J.; Lan, L.; Xie, P.; et al. Developing a new nonbinary SNP fluorescent multiplex detection system for forensic application in China. Electrophoresis 2017, 38, 1154–1162. [Google Scholar] [CrossRef]
- Pan, X.; Liu, C.; Du, W.; Chen, L.; Han, X.; Yang, X.; Liu, C. Genetic analysis and forensic evaluation of 47 autosomal InDel markers in four different Chinese populations. Int. J. Leg. Med. 2019. [Google Scholar] [CrossRef]
- Li, C.; Zhao, S.; Zhang, S.; Li, L.; Liu, Y.; Chen, J.; Xue, J. Genetic polymorphism of 29 highly informative InDel markers for forensic use in the Chinese Han population. Forensic Sci. Int. Genet. 2011, 5, e27–e30. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Wang, J.; Chen, D.; Liu, Z.; Shi, J.; Li, Z.; Li, W.; Zhang, G.; Du, B. A set of 14 DIP-SNP markers to detect unbalanced DNA mixtures. Biochem. Biophys. Res. Commun. 2018, 497, 591–596. [Google Scholar] [CrossRef]
- Kidd, K.K.; Pakstis, A.J.; Speed, W.C.; Lagace, R.; Chang, J.; Wootton, S.; Haigh, E.; Kidd, J.R. Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci. Int. Genet. 2014, 12, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Oldoni, F.; Kidd, K.K.; Podini, D. Microhaplotypes in forensic genetics. Forensic Sci. Int. Genet. 2019, 38, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Turchi, C.; Melchionda, F.; Pesaresi, M.; Tagliabracci, A. Evaluation of a microhaplotypes panel for forensic genetics using massive parallel sequencing technology. Forensic Sci. Int. Genet. 2019, 41, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Deng, C.; Li, Z.; Pu, Y.; Yang, J.; Yu, Y.; Li, K.; Li, D.; Liang, W.; Zhang, L.; et al. A microhaplotypes panel for massively parallel sequencing analysis of DNA mixtures. Forensic Sci. Int. Genet. 2019, 40, 140–149. [Google Scholar] [CrossRef]
- Pang, J.B.; Rao, M.; Chen, Q.F.; Ji, A.Q.; Zhang, C.; Kang, K.L.; Wu, H.; Ye, J.; Nie, S.J.; Wang, L. A 124-plex Microhaplotype Panel Based on Next-generation Sequencing Developed for Forensic Applications. Sci. Rep. 2020, 10, 1945. [Google Scholar] [CrossRef]
- Cheung, E.Y.; Phillips, C.; Eduardoff, M.; Lareu, M.V.; McNevin, D. Performance of ancestry-informative SNP and microhaplotype markers. Forensic Sci. Int. Genet. 2019, 43, 102141. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, P.; Qu, S.; Wang, Y.; Jian, H.; Cao, S.; Liu, Y.; Zhang, R.; Lv, M.; Liang, W.; et al. Evaluation of the microhaplotype markers in kinship analysis. Electrophoresis 2019, 40, 1091–1095. [Google Scholar] [CrossRef]
- Mo, S.K.; Ren, Z.L.; Yang, Y.R.; Liu, Y.C.; Zhang, J.J.; Wu, H.J.; Li, Z.; Bo, X.C.; Wang, S.Q.; Yan, J.W.; et al. A 472-SNP panel for pairwise kinship testing of second-degree relatives. Forensic Sci. Int. Genet. 2018, 34, 178–185. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Yang, S.; Xia, M.; Yang, Y.; Wang, J.; Lu, D.; Pan, X.; Ma, T.; Jiang, P.; et al. Genome-wide screening for highly discriminative SNPs for personal identification and their assessment in world populations. Forensic Sci. Int. Genet. 2017, 28, 118–127. [Google Scholar] [CrossRef]
- Zhang, S.; Bian, Y.; Chen, A.; Zheng, H.; Gao, Y.; Hou, Y.; Li, C. Developmental validation of a custom panel including 273 SNPs for forensic application using Ion Torrent PGM. Forensic Sci. Int. Genet. 2017, 27, 50–57. [Google Scholar] [CrossRef]
- Zha, L.; Yun, L.; Chen, P.; Luo, H.; Yan, J.; Hou, Y. Exploring of tri-allelic SNPs using pyrosequencing and the SNaPshot methods for forensic application. Electrophoresis 2012, 33, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Westen, A.A.; Matai, A.S.; Laros, J.F.; Meiland, H.C.; Jasper, M.; de Leeuw, W.J.; de Knijff, P.; Sijen, T. Tri-allelic SNP markers enable analysis of mixed and degraded DNA samples. Forensic Sci. Int. Genet. 2009, 3, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Chen, X.; Zhao, Y.; Zhao, X.; Zhang, S.; Yang, Y.; Wang, Y.; Zhang, J. Forensic genetic informativeness of an SNP panel consisting of 19 multi-allelic SNPs. Forensic Sci. Int. Genet. 2018, 34, 49–56. [Google Scholar] [CrossRef]
- Jin, X.Y.; Wei, Y.Y.; Cui, W.; Chen, C.; Guo, Y.X.; Zhang, W.Q.; Zhu, B.F. Development of a novel multiplex polymerase chain reaction system for forensic individual identification using insertion/deletion polymorphisms. Electrophoresis 2019, 40, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Wendt, F.R.; Warshauer, D.H.; Zeng, X.; Churchill, J.D.; Novroski, N.M.M.; Song, B.; King, J.L.; LaRue, B.L.; Budowle, B. Massively parallel sequencing of 68 insertion/deletion markers identifies novel microhaplotypes for utility in human identity testing. Forensic Sci. Int. Genet. 2016, 25, 198–209. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Gouy, A.; Zieger, M. STRAF-A convenient online tool for STR data evaluation in forensic genetics. Forensic Sci. Int. Genet. 2017, 30, 148–151. [Google Scholar] [CrossRef]
- Kidd, K.K.; Speed, W.C. Criteria for selecting microhaplotypes: Mixture detection and deconvolution. Investig. Genet. 2015, 6, 1. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Package Version 1.0.12; R Studio: Boston, MA, USA, 2019. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A.; Li, L.M.; Ward, R.; Pritchard, J.K. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 2003, 73, 1402–1422. [Google Scholar] [CrossRef]
- Jin, X.Y.; Guo, Y.X.; Chen, C.; Cui, W.; Liu, Y.F.; Tai, Y.C.; Zhu, B.F. Ancestry Prediction Comparisons of Different AISNPs for Five Continental Populations and Population Structure Dissection of the Xinjiang Hui Group via a Self-Developed Panel. Genes 2020, 11, 505. [Google Scholar] [CrossRef]
- Kidd, K.K. Proposed nomenclature for microhaplotypes. Hum. Genom. 2016, 10, 16. [Google Scholar] [CrossRef]
- Chen, P.; Yin, C.; Li, Z.; Pu, Y.; Yu, Y.; Zhao, P.; Chen, D.; Liang, W.; Zhang, L.; Chen, F. Evaluation of the Microhaplotypes panel for DNA mixture analyses. Forensic Sci. Int. Genet. 2018, 35, 149–155. [Google Scholar] [CrossRef]
- Liu, J.; Li, W.; Wang, J.; Chen, D.; Liu, Z.; Shi, J.; Cheng, F.; Li, Z.; Ren, J.; Zhang, G.; et al. A new set of DIP-SNP markers for detection of unbalanced and degraded DNA mixtures. Electrophoresis 2019, 40, 1795–1804. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Phillips, C. Forensic genetic analysis of bio-geographical ancestry. Forensic Sci. Int. Genet. 2015, 18, 49–65. [Google Scholar] [CrossRef]
- Kong, T.; Chen, Y.; Guo, Y.; Wei, Y.; Jin, X.; Xie, T.; Mu, Y.; Dong, Q.; Wen, S.; Zhou, B.; et al. Autosomal InDel polymorphisms for population genetic structure and differentiation analysis of Chinese Kazak ethnic group. Oncotarget 2017, 8, 56651–56658. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.M.; Zhang, S.H.; Li, C.T. InDel_typer30: A multiplex PCR system for DNA identification among five Chinese populations. Fa Yi Xue Za Zhi 2010, 26, 343–348, 356. [Google Scholar] [PubMed]
- Zhang, W.; Jin, X.; Wang, Y.; Kong, T.; Cui, W.; Chen, C.; Guo, Y.; Zhu, B. Genetic Polymorphisms and Forensic Efficiencies of a Set of Novel Autosomal InDel Markers in a Chinese Mongolian Group. Biomed. Res. Int. 2020, 2020, 3925189. [Google Scholar] [CrossRef] [PubMed]
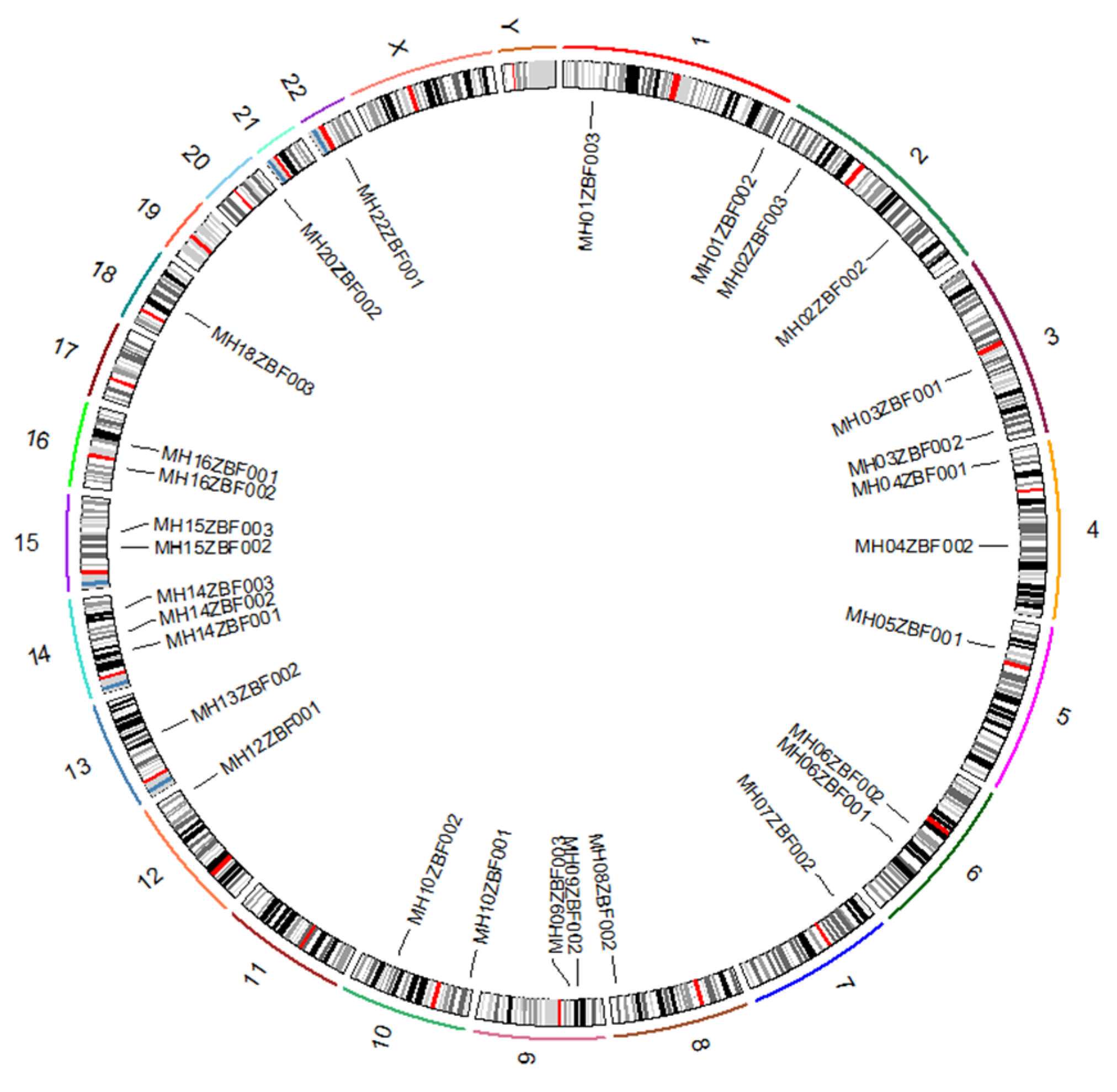
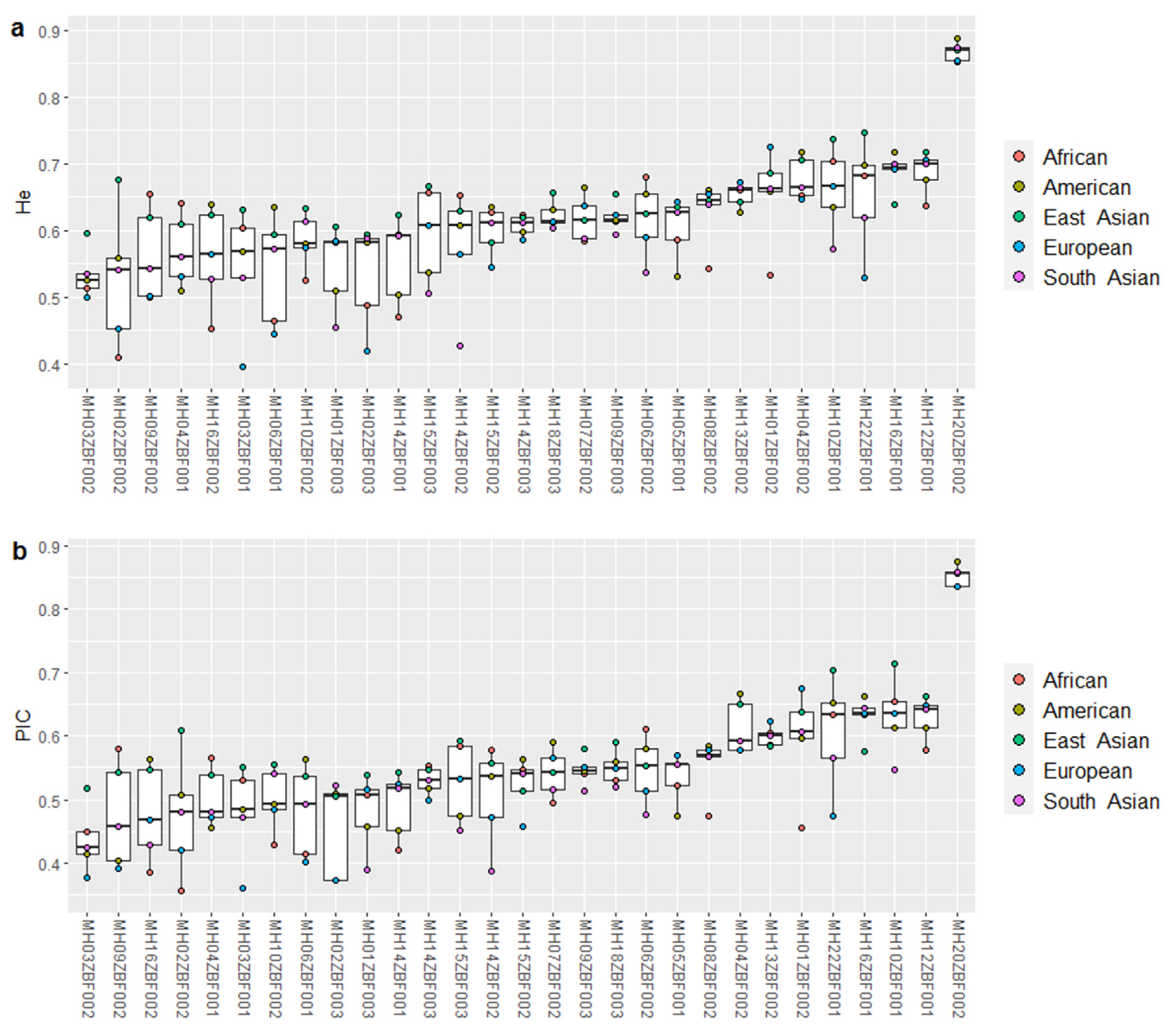

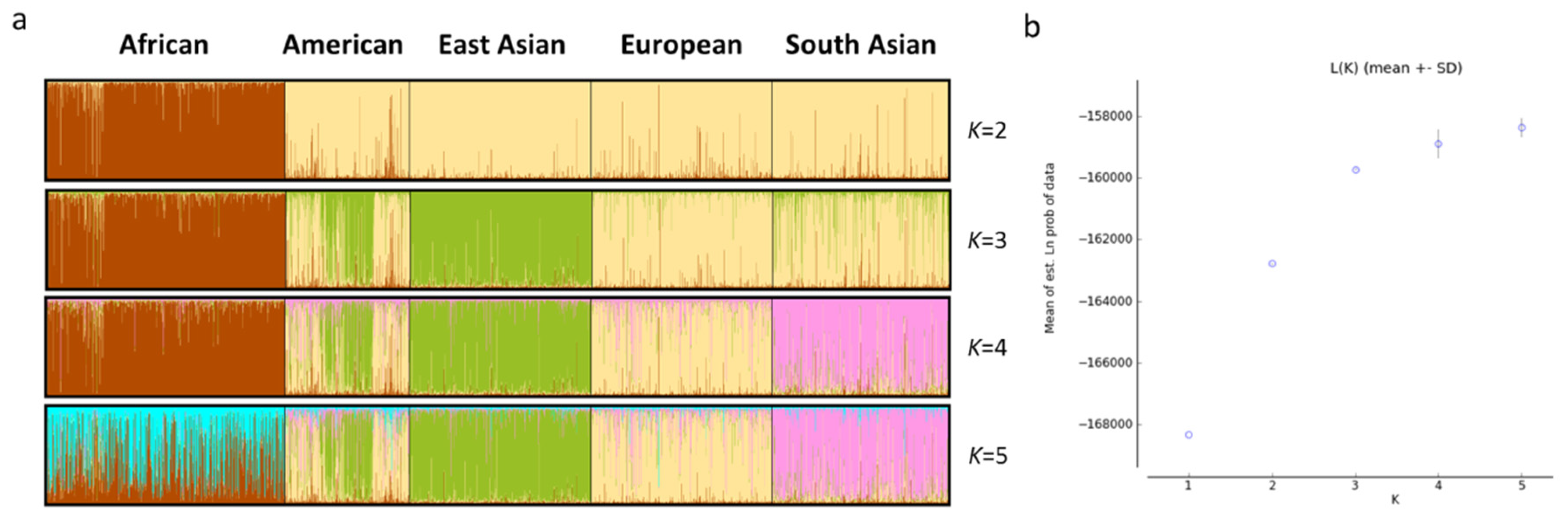
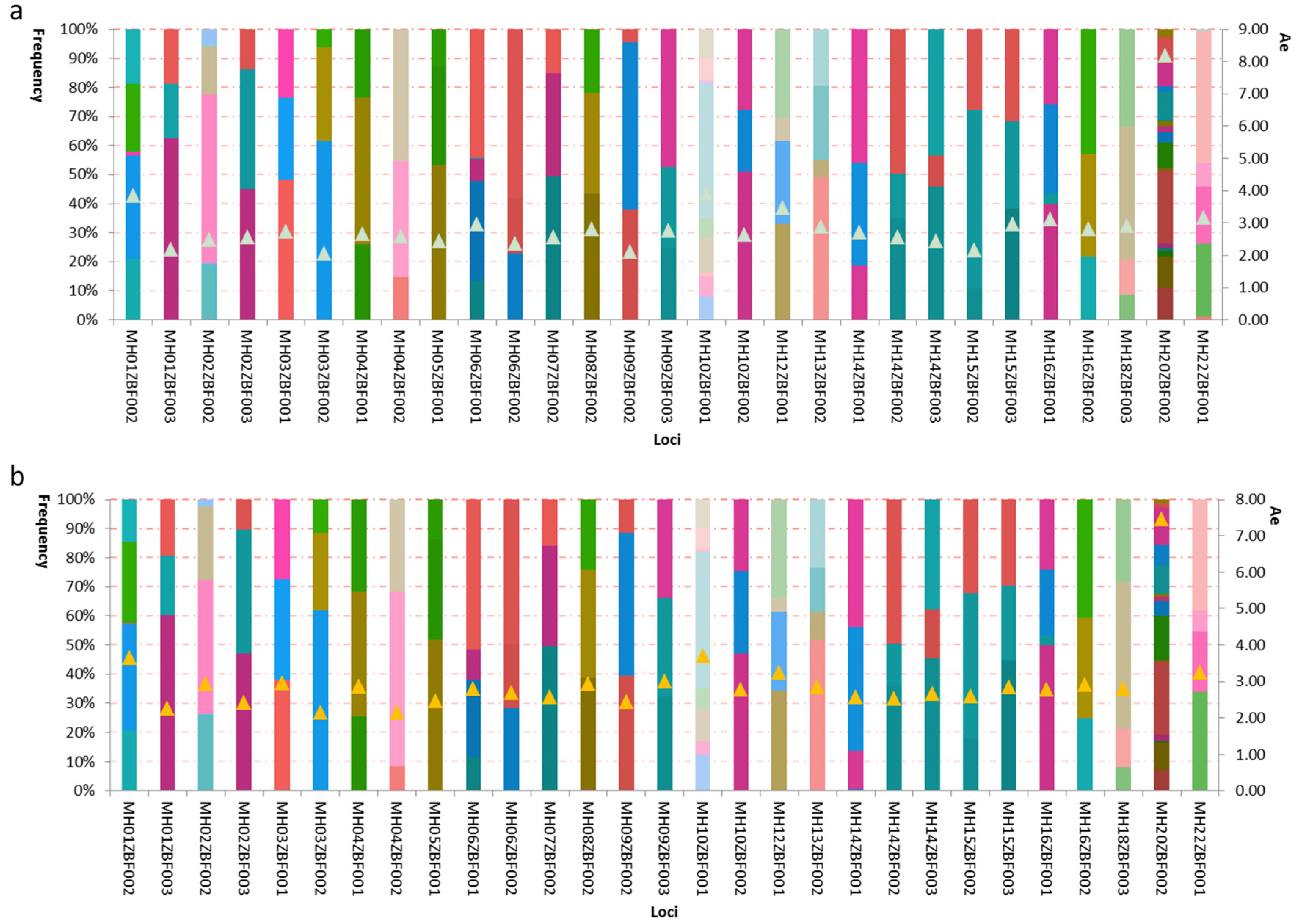
| Continents | CDP | CMP | CPE |
|---|---|---|---|
| African | 0.99999999999999999990425 | 9.5749 × 10−20 | 0.999982 |
| American | 0.9999999999999999999930322 | 6.9679 × 10−21 | 0.999983 |
| European | 0.99999999999999999982977 | 1.7023 × 10−19 | 0.999954 |
| East Asian | 0.99999999999999999999968073 | 3.1928 × 10−22 | 0.999998 |
| South Asian | 0.9999999999999999998964 | 1.036 × 10−19 | 0.999975 |
| Loci | He | Ho | PIC | MP | DP | PE | p |
|---|---|---|---|---|---|---|---|
| MH01ZBF002 | 0.7425 | 0.9375 | 0.6935 | 0.2044 | 0.7956 | 0.8725 | 0.0000 |
| MH01ZBF003 | 0.5415 | 0.5982 | 0.4817 | 0.2761 | 0.7239 | 0.2887 | 0.5120 |
| MH02ZBF002 | 0.5988 | 0.6071 | 0.5486 | 0.2167 | 0.7833 | 0.2995 | 0.6480 |
| MH02ZBF003 | 0.6116 | 0.6339 | 0.5260 | 0.2588 | 0.7412 | 0.3336 | 0.0380 |
| MH03ZBF001 | 0.6353 | 0.6339 | 0.5608 | 0.2068 | 0.7932 | 0.3336 | 1.0000 |
| MH03ZBF002 | 0.5155 | 0.5446 | 0.4310 | 0.3310 | 0.6690 | 0.2296 | 0.4680 |
| MH04ZBF001 | 0.6253 | 0.6786 | 0.5524 | 0.2251 | 0.7749 | 0.3959 | 0.6220 |
| MH04ZBF002 | 0.6158 | 0.5982 | 0.5318 | 0.2250 | 0.7750 | 0.2887 | 0.7210 |
| MH05ZBF001 | 0.5885 | 0.5357 | 0.5076 | 0.2414 | 0.7586 | 0.2207 | 0.3690 |
| MH06ZBF001 | 0.6658 | 0.5804 | 0.6015 | 0.1674 | 0.8326 | 0.2680 | 0.1970 |
| MH06ZBF002 | 0.5771 | 0.6071 | 0.5109 | 0.2481 | 0.7519 | 0.2995 | 0.6970 |
| MH07ZBF002 | 0.6097 | 0.6518 | 0.5289 | 0.2463 | 0.7537 | 0.3577 | 0.8450 |
| MH08ZBF002 | 0.6463 | 0.6161 | 0.5683 | 0.2127 | 0.7873 | 0.3106 | 0.0670 |
| MH09ZBF002 | 0.5247 | 0.5357 | 0.4250 | 0.3276 | 0.6724 | 0.2207 | 0.9010 |
| MH09ZBF003 | 0.6395 | 0.7143 | 0.5647 | 0.2242 | 0.7758 | 0.4507 | 0.3740 |
| MH10ZBF001 | 0.7457 | 0.7589 | 0.7211 | 0.0969 | 0.9031 | 0.5252 | 0.0140 |
| MH10ZBF002 | 0.6212 | 0.6161 | 0.5480 | 0.2229 | 0.7771 | 0.3106 | 0.3440 |
| MH12ZBF001 | 0.7138 | 0.7589 | 0.6540 | 0.1583 | 0.8417 | 0.5252 | 0.0400 |
| MH13ZBF002 | 0.6545 | 0.6250 | 0.5942 | 0.1830 | 0.8170 | 0.3220 | 0.4470 |
| MH14ZBF001 | 0.6318 | 0.6518 | 0.5528 | 0.2160 | 0.7840 | 0.3577 | 0.5690 |
| MH14ZBF002 | 0.6115 | 0.6161 | 0.5313 | 0.2403 | 0.7597 | 0.3106 | 0.6730 |
| MH14ZBF003 | 0.5922 | 0.5446 | 0.5011 | 0.2417 | 0.7583 | 0.2296 | 0.3770 |
| MH15ZBF002 | 0.5348 | 0.5804 | 0.4637 | 0.2953 | 0.7047 | 0.2680 | 0.4940 |
| MH15ZBF003 | 0.6656 | 0.6696 | 0.5887 | 0.1923 | 0.8077 | 0.3829 | 0.6800 |
| MH16ZBF001 | 0.6820 | 0.6964 | 0.6143 | 0.1711 | 0.8289 | 0.4228 | 0.8670 |
| MH16ZBF002 | 0.6470 | 0.6429 | 0.5689 | 0.2038 | 0.7962 | 0.3455 | 0.9240 |
| MH18ZBF003 | 0.6577 | 0.6607 | 0.5931 | 0.1786 | 0.8214 | 0.3701 | 0.7510 |
| MH20ZBF002 | 0.8818 | 0.8839 | 0.8674 | 0.0362 | 0.9638 | 0.7627 | 0.0080 |
| MH22ZBF001 | 0.6858 | 0.6786 | 0.6308 | 0.1518 | 0.8482 | 0.3959 | 0.8050 |
| Loci | He | Ho | PIC | MP | DP | PE | p |
|---|---|---|---|---|---|---|---|
| MH01ZBF002 | 0.7296 | 0.8868 | 0.6769 | 0.2415 | 0.7585 | 0.7685 | 0.0000 |
| MH01ZBF003 | 0.5596 | 0.5283 | 0.4966 | 0.2560 | 0.7440 | 0.2135 | 0.5600 |
| MH02ZBF002 | 0.6610 | 0.6509 | 0.5934 | 0.1808 | 0.8192 | 0.3565 | 0.8810 |
| MH02ZBF003 | 0.5893 | 0.6226 | 0.4976 | 0.2768 | 0.7232 | 0.3189 | 0.5320 |
| MH03ZBF001 | 0.6637 | 0.6698 | 0.5864 | 0.1928 | 0.8072 | 0.3831 | 0.9040 |
| MH03ZBF002 | 0.5356 | 0.4623 | 0.4662 | 0.2789 | 0.7211 | 0.1566 | 0.2500 |
| MH04ZBF001 | 0.6541 | 0.7170 | 0.5773 | 0.2140 | 0.7860 | 0.4550 | 0.5280 |
| MH04ZBF002 | 0.5366 | 0.5377 | 0.4557 | 0.2992 | 0.7008 | 0.2227 | 0.9450 |
| MH05ZBF001 | 0.5963 | 0.5755 | 0.5151 | 0.2391 | 0.7609 | 0.2625 | 0.9170 |
| MH06ZBF001 | 0.6442 | 0.6415 | 0.5875 | 0.1817 | 0.8183 | 0.3437 | 0.4410 |
| MH06ZBF002 | 0.6284 | 0.6792 | 0.5542 | 0.2250 | 0.7750 | 0.3969 | 0.7500 |
| MH07ZBF002 | 0.6133 | 0.5849 | 0.5335 | 0.2225 | 0.7775 | 0.2732 | 0.8860 |
| MH08ZBF002 | 0.6604 | 0.6415 | 0.5837 | 0.2164 | 0.7836 | 0.3437 | 0.0000 |
| MH09ZBF002 | 0.5923 | 0.5660 | 0.5038 | 0.2583 | 0.7417 | 0.2521 | 0.4110 |
| MH09ZBF003 | 0.6696 | 0.6321 | 0.5924 | 0.1857 | 0.8143 | 0.3312 | 0.2430 |
| MH10ZBF001 | 0.7316 | 0.7075 | 0.7043 | 0.1150 | 0.8850 | 0.4400 | 0.0100 |
| MH10ZBF002 | 0.6403 | 0.6698 | 0.5652 | 0.2147 | 0.7853 | 0.3831 | 0.6320 |
| MH12ZBF001 | 0.6948 | 0.6698 | 0.6290 | 0.1563 | 0.8437 | 0.3831 | 0.5500 |
| MH13ZBF002 | 0.6505 | 0.6132 | 0.5967 | 0.1780 | 0.8220 | 0.3070 | 0.3370 |
| MH14ZBF001 | 0.6128 | 0.6604 | 0.5275 | 0.2496 | 0.7504 | 0.3697 | 0.8110 |
| MH14ZBF002 | 0.6077 | 0.5849 | 0.5258 | 0.2433 | 0.7567 | 0.2732 | 0.2290 |
| MH14ZBF003 | 0.6267 | 0.5943 | 0.5453 | 0.2090 | 0.7910 | 0.2841 | 0.5610 |
| MH15ZBF002 | 0.6179 | 0.6226 | 0.5409 | 0.2223 | 0.7777 | 0.3189 | 0.4660 |
| MH15ZBF003 | 0.6491 | 0.5660 | 0.5730 | 0.1924 | 0.8076 | 0.2521 | 0.0560 |
| MH16ZBF001 | 0.6428 | 0.6698 | 0.5785 | 0.2010 | 0.7990 | 0.3831 | 0.5270 |
| MH16ZBF002 | 0.6575 | 0.7358 | 0.5800 | 0.2218 | 0.7782 | 0.4859 | 0.1210 |
| MH18ZBF003 | 0.6443 | 0.7453 | 0.5843 | 0.2205 | 0.7795 | 0.5017 | 0.0370 |
| MH20ZBF002 | 0.8699 | 0.8585 | 0.8527 | 0.0418 | 0.9582 | 0.7117 | 0.0080 |
| MH22ZBF001 | 0.6939 | 0.7264 | 0.6314 | 0.1630 | 0.8370 | 0.4703 | 0.1710 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Zhang, X.; Shen, C.; Liu, Y.; Cui, W.; Chen, C.; Guo, Y.; Zhu, B. A Highly Polymorphic Panel Consisting of Microhaplotypes and Compound Markers with the NGS and Its Forensic Efficiency Evaluations in Chinese Two Groups. Genes 2020, 11, 1027. https://doi.org/10.3390/genes11091027
Jin X, Zhang X, Shen C, Liu Y, Cui W, Chen C, Guo Y, Zhu B. A Highly Polymorphic Panel Consisting of Microhaplotypes and Compound Markers with the NGS and Its Forensic Efficiency Evaluations in Chinese Two Groups. Genes. 2020; 11(9):1027. https://doi.org/10.3390/genes11091027
Chicago/Turabian StyleJin, Xiaoye, Xingru Zhang, Chunmei Shen, Yanfang Liu, Wei Cui, Chong Chen, Yuxin Guo, and Bofeng Zhu. 2020. "A Highly Polymorphic Panel Consisting of Microhaplotypes and Compound Markers with the NGS and Its Forensic Efficiency Evaluations in Chinese Two Groups" Genes 11, no. 9: 1027. https://doi.org/10.3390/genes11091027
APA StyleJin, X., Zhang, X., Shen, C., Liu, Y., Cui, W., Chen, C., Guo, Y., & Zhu, B. (2020). A Highly Polymorphic Panel Consisting of Microhaplotypes and Compound Markers with the NGS and Its Forensic Efficiency Evaluations in Chinese Two Groups. Genes, 11(9), 1027. https://doi.org/10.3390/genes11091027





