Distinct Alteration of Gene Expression Programs in the Small Intestine of Male and Female Mice in Response to Ablation of Intestinal Fabp Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Preparation of RNA and Hybridization to DNA Microarray
2.3. Analysis of DNA Microarray Data
2.3.1. Processing of Raw DNA Microarray Data
2.3.2. Gene Annotation and Functional Enrichment Analysis
2.3.3. Protein–Protein Interaction Network Analysis
2.3.4. Availability of DNA Microarray Data
3. Results
3.1. Gene Expression Patterns in Male and Female Murine Small Intestine Are Distinct
3.2. Genomic Responses to Ablation of Specific Fabp Genes Are Sexually Dimorphic
3.3. Predicted Nutrient Metabolism Processes in the Small Intestine of Fabp Gene Ablated Mice Are Sex Biased
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Storch, J.; Córsico, B. The Emerging Functions and Mechanisms of Mammalian Fatty Acid–Binding Proteins. Annu. Rev. Nutr. 2008, 28, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.B.; Boguski, M.S.; Sweetser, D.A.; Elshourbagy, N.; Taylor, J.M.; Gordon, J.I. Human liver fatty acid binding protein. Isolation of a full length cDNA and comparative sequence analyses of orthologous and paralogous proteins. J. Biol. Chem. 1985, 260, 3413–3417. [Google Scholar] [PubMed]
- Ockner, R.K.; A Manning, J.; Kane, J.P. Fatty acid binding protein. Isolation from rat liver, characterization, and immunochemical quantification. J. Biol. Chem. 1982, 257, 7872–7878. [Google Scholar] [PubMed]
- Sweetser, D.A.; Birkenmeier, E.H.; Klisak, I.J.; Zollman, S.; Sparkes, R.S.; Mohandas, T.; Lusis, A.J.; I Gordon, J. The human and rodent intestinal fatty acid binding protein genes. A comparative analysis of their structure, expression, and linkage relationships. J. Biol. Chem. 1987, 262, 16060–16071. [Google Scholar]
- Walz, D.A.; Wider, M.D.; Snow, J.W.; Dass, C.; Desiderio, D.M. The complete amino acid sequence of porcine gastrotropin, an ileal protein which stimulates gastric acid and pepsinogen secretion. J. Biol. Chem. 1988, 263, 14189–14195. [Google Scholar]
- Gong, Y.Z.; Everett, E.T.; Schwartz, D.A.; Norris, J.S.; Wilson, F.A. Molecular cloning, tissue distribution, and expression of a 14-kDa bile acid-binding protein from rat ileal cytosol. Proc. Natl. Acad. Sci. USA 1994, 91, 4741–4745. [Google Scholar] [CrossRef]
- Agellon, L.B.; Toth, M.J.; Thomson, A.B. Intracellular lipid binding proteins of the small intestine. Mol. Cell. Biochem. 2002, 239, 79–82. [Google Scholar] [CrossRef]
- Zimmerman, A.W.; Van Moerkerk, H.T.; Veerkamp, J.H. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 2001, 33, 865–876. [Google Scholar] [CrossRef]
- Richieri, G.V.; Ogata, R.T.; Zimmerman, A.W.; Veerkamp, J.H.; Kleinfeld, A.M. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 2000, 39, 7197–7204. [Google Scholar] [CrossRef]
- Schroeder, F.; Petrescu, A.D.; Huang, H.; Atshaves, B.P.; McIntosh, A.L.; Martin, G.G.; Hostetler, H.A.; Vespa, A.; Landrock, D.; Landrock, K.K.; et al. Role of Fatty Acid Binding Proteins and Long Chain Fatty Acids in Modulating Nuclear Receptors and Gene Transcription. Lipids 2007, 43, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, C.; Borrmann, C.M.; Börchers, T.; Spener, F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors α- and γ-mediated gene expression via liver fatty acid binding protein: A signaling path to the nucleus. Proc. Natl. Acad. Sci. USA 2001, 98, 2323–2328. [Google Scholar] [CrossRef] [PubMed]
- Newberry, E.P.; Xie, Y.; Kennedy, S.M.; Han, X.; Buhman, K.K.; Luo, J.; Gross, R.W.; Davidson, N.O. Decreased Hepatic Triglyceride Accumulation and Altered Fatty Acid Uptake in Mice with Deletion of the Liver Fatty Acid-binding Protein Gene. J. Biol. Chem. 2003, 278, 51664–51672. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.G.; Danneberg, H.; Kumar, L.S.; Atshaves, B.P.; Erol, E.; Bader, M.; Schroeder, F.; Binas, B. Decreased Liver Fatty Acid Binding Capacity and Altered Liver Lipid Distribution in Mice Lacking the Liver Fatty Acid-binding Protein Gene. J. Biol. Chem. 2003, 278, 21429–21438. [Google Scholar] [CrossRef]
- Xie, Y.; Newberry, E.P.; Kennedy, S.M.; Luo, J.; Davidson, N.O. Increased susceptibility to diet-induced gallstones in liver fatty acid binding protein knockout mice. J. Lipid Res. 2009, 50, 977–987. [Google Scholar] [CrossRef]
- Weickert, M.O.; Loeffelholz, C.V.; Roden, M.; Chandramouli, V.; Brehm, A.; Nowotny, P.; Osterhoff, M.A.; Isken, F.; Spranger, J.; Landau, B.R.; et al. A Thr94Ala mutation in human liver fatty acid-binding protein contributes to reduced hepatic glycogenolysis and blunted elevation of plasma glucose levels in lipid-exposed subjects. Am. J. Physiol. Metab. 2007, 293, E1078–E1084. [Google Scholar] [CrossRef]
- Brouillette, C.; Bossé, Y.; Pérusse, L.; Gaudet, D.; Vohl, M.-C. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J. Hum. Genet. 2004, 49, 424–432. [Google Scholar] [CrossRef]
- Fisher, E.; Weikert, C.; Klapper, M.; Lindner, I.; Möhlig, M.; Spranger, J.; Boeing, H.; Schrezenmeir, J.; Döring, F. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol. Genet. Metab. 2007, 91, 278–284. [Google Scholar] [CrossRef]
- Hsu, K.T.; Storch, J. Fatty Acid Transfer from Liver and Intestinal Fatty Acid-binding Proteins to Membranes Occurs by Different Mechanisms. J. Biol. Chem. 1996, 271, 13317–13323. [Google Scholar] [CrossRef]
- Agellon, L.B.; Li, L.; Luong, L.; Uwiera, R.R.E. Adaptations to the loss of intestinal fatty acid binding protein in mice. Mol. Cell. Biochem. 2006, 284, 159–166. [Google Scholar] [CrossRef]
- Vassileva, G.; Huwyler, L.; Poirier, K.; Agellon, L.B.; Toth, M.J. The intestinal fatty acid binding protein is not essential for dietary fat absorption in mice. FASEB J. 2000, 14, 2040–2046. [Google Scholar] [CrossRef] [PubMed]
- Hasnain, S. The fatty acid binding protein 2 (FABP2) polymorphism Ala54Thr and obesity in Pakistan: A population based study and a systematic meta-analysis. Gene 2015, 574, 106–111. [Google Scholar] [CrossRef]
- Tavridou, A.; Arvanitidis, K.I.; Tiptiri-Kourpeti, A.; Petridis, I.; Ragia, G.; Kyroglou, S.; Christakidis, D.; Manolopoulos, V.G. Thr54 allele of fatty-acid binding protein 2 gene is associated with obesity but not type 2 diabetes mellitus in a Caucasian population. Diabetes Res. Clin. Pract. 2009, 84, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, K.K.; Khan, I.A.; Bazzi, M.D.; Al-Daghri, N.M.; Hasan, T.N.; Al-Nbaheen, M.S.; Alharbi, F.K.; Al-Sheikh, Y.A.; Syed, R.; Aboul-Soud, M.A. A54T polymorphism in the fatty acid binding protein 2 studies in a Saudi population with type 2 diabetes mellitus. Lipids Health Dis. 2014, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Damcott, C.M.; Feingold, E.; Moffett, S.P.; Barmada, M.M.; Marshall, J.A.; Hamman, R.F.; Ferrell, R.E. Variation in the FABP2 promoter alters transcriptional activity and is associated with body composition and plasma lipid levels. Hum. Genet. 2003, 112, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Labonté, E.D.; Li, Q.; Kay, C.M.; Agellon, L.B. The relative ligand binding preference of the murine ileal lipid binding protein. Protein Expr. Purif. 2003, 28, 25–33. [Google Scholar] [CrossRef]
- Praslickova, D.; Torchia, E.C.; Sugiyama, M.G.; Magrane, E.J.; Zwicker, B.L.; Kolodzieyski, L.; Agellon, L.B. The Ileal Lipid Binding Protein Is Required for Efficient Absorption and Transport of Bile Acids in the Distal Portion of the Murine Small Intestine. PLoS ONE 2012, 7, e50810. [Google Scholar] [CrossRef]
- Duggavathi, R.; Siddappa, D.; Schuermann, Y.; Pansera, M.; Menard, I.J.; Praslickova, D.; Agellon, L.B. The fatty acid binding protein 6 gene (Fabp6) is expressed in murine granulosa cells and is involved in ovulatory response to superstimulation. J. Reprod. Dev. 2015, 61, 237–240. [Google Scholar] [CrossRef]
- Nakahara, M.; Furuya, N.; Takagaki, K.; Sugaya, T.; Hirota, K.; Fukamizu, A.; Kanda, T.; Fujii, H.; Sato, R. Ileal Bile Acid-binding Protein, Functionally Associated with the Farnesoid X Receptor or the Ileal Bile Acid Transporter, Regulates Bile Acid Activity in the Small Intestine. J. Biol. Chem. 2005, 280, 42283–42289. [Google Scholar] [CrossRef]
- Landrier, J.F.; Grober, J.; Zaghini, I.; Besnard, P. Regulation of the ileal bile acid-binding protein gene: An approach to determine its physiological function(s). Mol. Cell. Biochem. 2002, 239, 149–155. [Google Scholar] [CrossRef]
- Fisher, E.; Grallert, H.; Klapper, M.; Pfäfflin, A.; Schrezenmeir, J.; Illig, T.; Boeing, H.; Döring, F. Evidence for the Thr79Met polymorphism of the ileal fatty acid binding protein (FABP6) to be associated with type 2 diabetes in obese individuals. Mol. Genet. Metab. 2009, 98, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.G.; Atshaves, B.P.; McIntosh, A.L.; Mackie, J.T.; Kier, A.B.; Schroeder, F. Liver fatty-acid-binding protein (L-FABP) gene ablation alters liver bile acid metabolism in male mice. Biochem. J. 2005, 391, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Bourgon, R.; Gentleman, R.; Huber, W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. USA 2010, 107, 9546–9551. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 31 July 2020).
- Hulsen, T.; De Vlieg, J.; Alkema, W. BioVenn–A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Smith, C.L.; Blake, J.A.; Kadin, J.A.; Richardson, J.E.; Bult, C.J. Mouse Genome Database Group Mouse Genome Database (MGD)-2018: Knowledgebase for the laboratory mouse. Nucleic Acids Res. 2018, 46, D836–D842. [Google Scholar] [CrossRef]
- Xia, J.; Benner, M.J.; Hancock, R.E.W. NetworkAnalyst-integrative approaches for protein—protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef]
- Li, B.; Qing, T.; Zhu, J.; Wen, Z.; Yu, Y.; Fukumura, R.; Zheng, Y.; Gondo, Y.; Shi, L. A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Sci. Rep. 2017, 7, 4200. [Google Scholar] [CrossRef] [PubMed]
- Steegenga, W.T.; Mischke, M.; Lute, C.; Boekschoten, M.V.; Pruis, M.G.; Lendvai, A.; Verkade, H.J.; Boekhorst, J.; Timmerman, H.M.; Plosch, T.; et al. Sexually dimorphic characteristics of the small intestine and colon of prepubescent C57BL/6 mice. Biol. Sex Differ. 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Soldin, O.P.; Mattison, D.W. Sex differences in pharmacokinetics and pharmacodynamics. Clin. Pharmacokinet. 2009, 48, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.G.; Agellon, L.B. Sex differences in lipid metabolism and metabolic disease risk. Biochem. Cell Biol. 2012, 90, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.G.; Hobson, L.; Agellon, A.B.; Agellon, L.B. Visualization of Sex-Dimorphic Changes in the Intestinal Transcriptome of Fabp2 Gene-Ablated Mice. Lifestyle Genom. 2012, 5, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Burton-Freeman, B.; Davis, P.A.; Schneeman, B.O. Interaction of fat availability and sex on postprandial satiety and cholecystokinin after mixed-food meals. Am. J. Clin. Nutr. 2004, 80, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Knuth, N.D.; Horowitz, J.F. The Elevation of Ingested Lipids within Plasma Chylomicrons Is Prolonged in Men Compared with Women. J. Nutr. 2006, 136, 1498–1503. [Google Scholar] [CrossRef]
- Klaassen, C.D.; Aleksunes, L.M. Xenobiotic, bile acid, and cholesterol transporters: Function and regulation. Pharmacol. Rev. 2010, 62, 1–96. [Google Scholar] [CrossRef]
- Pédrono, F.; Blanchard, H.; Kloareg, M.; D’Andréa, S.; Daval, S.; Rioux, V.; Legrand, P. The fatty acid desaturase 3 gene encodes for different FADS3 protein isoforms in mammalian tissues. J. Lipid Res. 2009, 51, 472–479. [Google Scholar] [CrossRef]
- France, M.; Skorich, E.; Kadrofske, M.; Swain, G.M.; Galligan, J.J. Sex-related differences in small intestinal transit and serotonin dynamics in high-fat-diet-induced obesity in mice. Exp. Physiol. 2015, 101, 81–99. [Google Scholar] [CrossRef]
- Priego, T.; Sanchez, J.; Picó, C.; Palou, A. Sex-differential Expression of Metabolism-related Genes in Response to a High-fat Diet. Obesity 2008, 16, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Pelsers, M.M.; Namiot, Z.; Kisielewski, W.; Namiot, A.; Januszkiewicz, M.; Hermens, W.T.; Glatz, J.F.C. Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue distribution and clinical utility. Clin. Biochem. 2003, 36, 529–535. [Google Scholar] [CrossRef]
- Agellon, L.B.; Drozdowski, L.; Li, L.; Iordache, C.; Luong, L.; Clandinin, M.T.; Uwiera, R.R.; Toth, M.J.; Thomson, A.B. Loss of intestinal fatty acid binding protein increases the susceptibility of male mice to high fat diet-induced fatty liver. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2007, 1771, 1283–1288. [Google Scholar] [CrossRef]
- Martin, G.G.; Landrock, D.; Chung, S.; Dangott, L.J.; Seeger, D.R.; Murphy, E.J.; Golovko, M.Y.; Kier, A.B.; Schroeder, F. Fabp1gene ablation inhibits high-fat diet-induced increase in brain endocannabinoids. J. Neurochem. 2016, 140, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Melé, M.; Ferreira, P.G.; Reverter, F.; DeLuca, D.S.; Monlong, J.; Sammeth, M.; Young, T.R.; Goldmann, J.M.; Pervouchine, D.D.; Sullivan, T.J.; et al. The human transcriptome across tissues and individuals. Science 2015, 348, 660–665. [Google Scholar] [CrossRef]
- Gershoni, M.; Pietrokovski, S. The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol. 2017, 15, 7. [Google Scholar] [CrossRef]
- Yang, X.; Schadt, E.E.; Wang, S.; Wang, H.; Arnold, A.P.; Ingram-Drake, L.; Drake, T.A.; Lusis, A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006, 16, 995–1004. [Google Scholar] [CrossRef]
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef]
- Grath, S.; Parsch, J. Sex-Biased Gene Expression. Annu. Rev. Genet. 2016, 50, 29–44. [Google Scholar] [CrossRef]
- Rinn, J.L.; Snyder, M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005, 21, 298–305. [Google Scholar] [CrossRef]
- Bermejo-Álvarez, P.; Rizos, D.; Lonergan, P.; Gutiérrez-Adán, A. Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction 2011, 141, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Jalouli, M.; Carlsson, L.; Améen, C.; Lindén, D.; Ljungberg, A.; Michalik, L.; Edén, S.; Wahli, W.; Oscarsson, J. Sex Difference in Hepatic Peroxisome Proliferator-Activated Receptor α Expression: Influence of Pituitary and Gonadal Hormones. Endocrinology 2003, 144, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Selwyn, F.P.; Csanaky, I.L.; Zhang, Y.; Klaassen, C.D. Importance of Large Intestine in Regulating Bile Acids and Glucagon-Like Peptide-1 in Germ-Free Mice. Drug Metab. Dispos. 2015, 43, 1544–1556. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Jena, P.K.; Liu, H.-X.; Kalanetra, K.M.; Gonzalez, F.J.; French, S.W.; Krishnan, V.V.; Mills, D.A.; Wan, Y.-J.Y. Gender Differences in Bile Acids and Microbiota in Relationship with Gender Dissimilarity in Steatosis Induced by Diet and FXR Inactivation. Sci. Rep. 2017, 7, 1748. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Michalak, M.; Agellon, L.B. Importance of Nutrients and Nutrient Metabolism on Human Health. Yale J. Biol. Med. 2018, 91, 95–103. [Google Scholar] [PubMed]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef]
- Altmann, S.W.; Davis, H.R.; Zhu, L.J.; Yao, X.; Hoos, L.M.; Tetzloff, G.; Iyer, S.P.N.; Maguire, M.; Golovko, A.; Zeng, M.; et al. Niemann-Pick C1 Like 1 Protein Is Critical for Intestinal Cholesterol Absorption. Science 2004, 303, 1201–1204. [Google Scholar] [CrossRef]

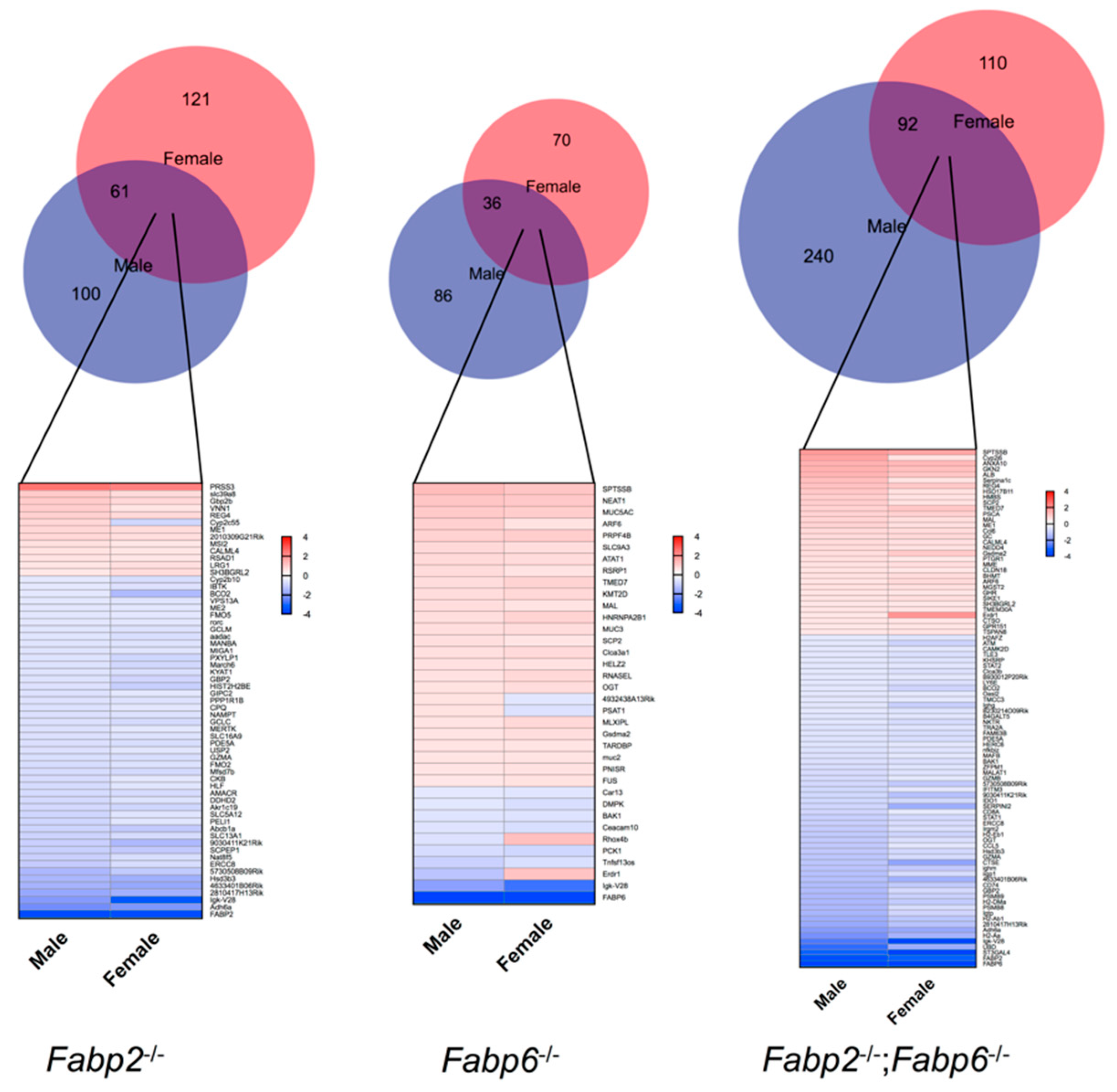
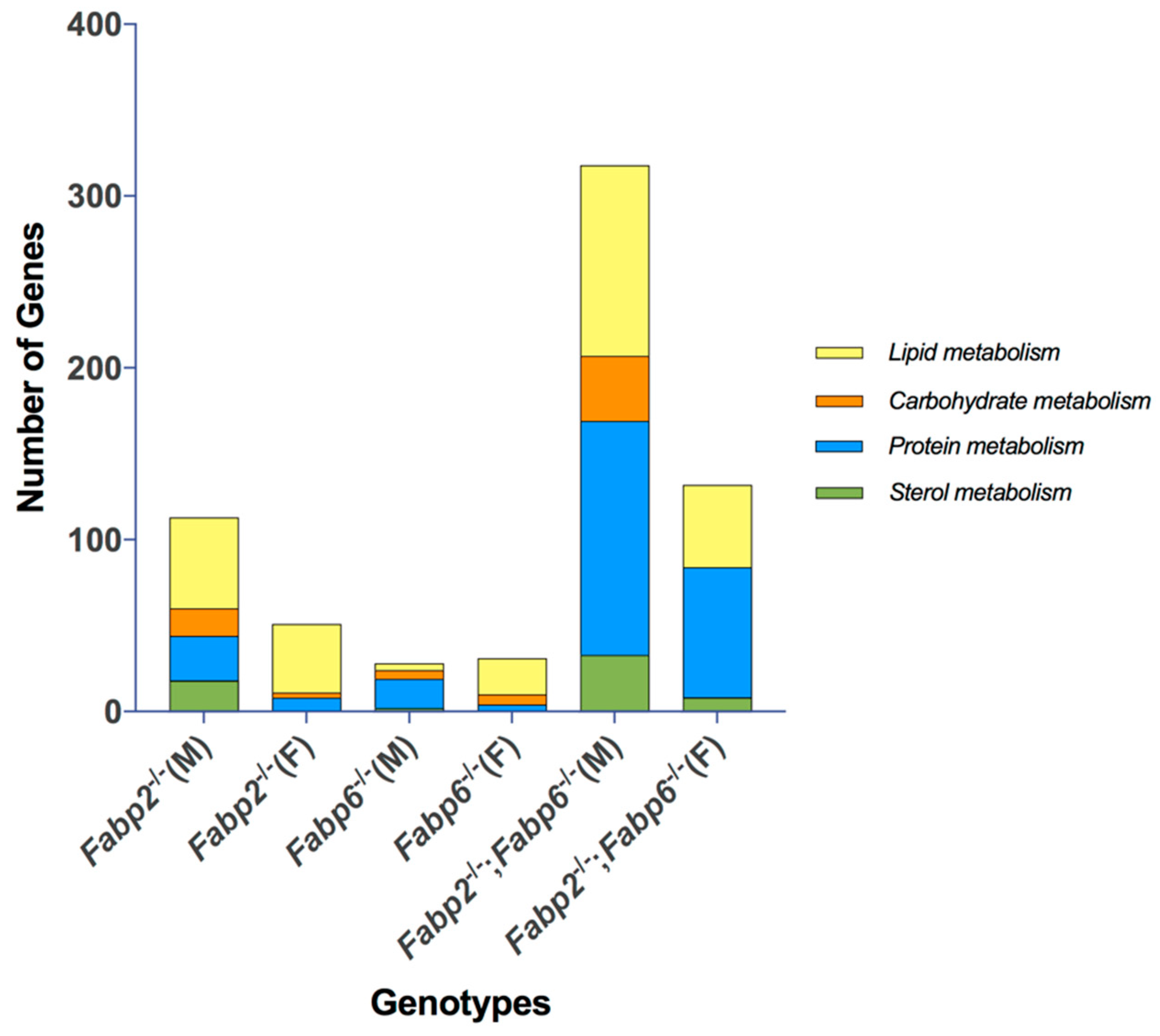
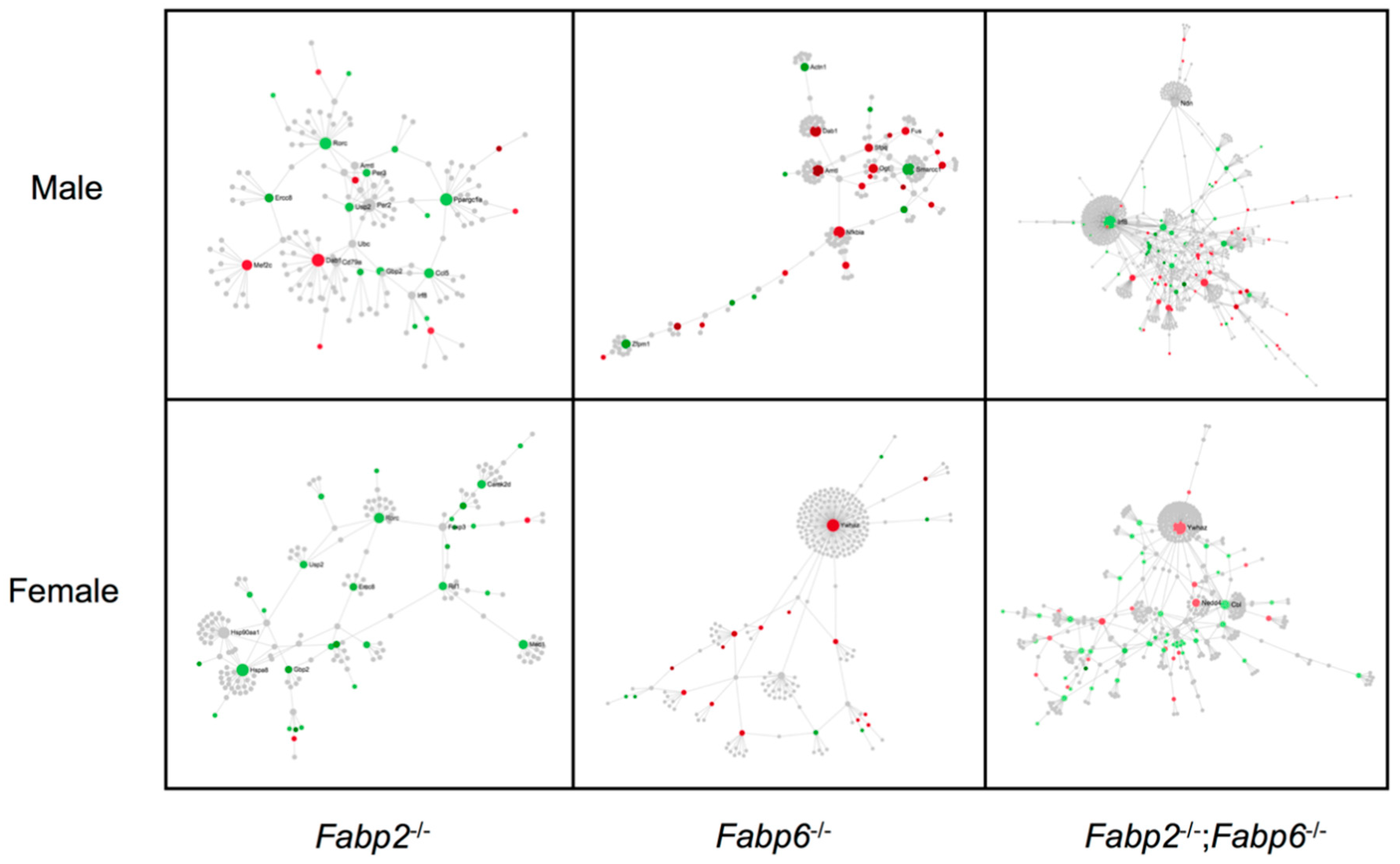
| Fabp2–/– (M) | Count | p-Value | Fabp2–/– (F) | Count | p-Value |
|---|---|---|---|---|---|
| GO:0055114~oxidation-reduction process | 22 | 6.42 × 10−8 | GO:0006749~glutathione metabolic process | 5 | 6.03 × 10−4 |
| GO:0006629~lipid metabolic process | 17 | 6.02 × 10−7 | GO:0006805~xenobiotic metabolic process | 4 | 8.68 × 10−4 |
| GO:0032922~circadian regulation of gene expression | 7 | 7.89 × 10−6 | GO:0008152~metabolic process | 12 | 0.001148 |
| GO:0008202~steroid metabolic process | 7 | 4.98 × 10−5 | GO:0042130~negative regulation of T cell proliferation | 4 | 0.003863 |
| GO:0048511~rhythmic process | 8 | 6.51 × 10−5 | GO:0035458~cellular response to interferon-β | 4 | 0.004144 |
| GO:0006631~fatty acid metabolic process | 8 | 2.24 × 10−4 | GO:0035729~cellular response to hepatocyte growth factor stimulus | 3 | 0.00693 |
| GO:0006805~xenobiotic metabolic process | 4 | 8.33 × 10−4 | GO:0055085~transmembrane transport | 9 | 0.00846 |
| GO:0006694~steroid biosynthetic process | 5 | 0.001392 | GO:0017144~drug metabolic process | 3 | 0.008746 |
| GO:0007623~circadian rhythm | 6 | 0.001573 | GO:0032922~circadian regulation of gene expression | 4 | 0.012478 |
| GO:0006641~triglyceride metabolic process | 4 | 0.003454 | GO:0006807~nitrogen compound metabolic process | 3 | 0.012935 |
| Fabp6–/– (M) | Fabp6–/– (F) | ||||
| GO:0045944~positive regulation of transcription from RNA polymerase II promoter | 14 | 0.002734 | GO:0006915~apoptotic process | 10 | 0.001953 |
| GO:0006814~sodium ion transport | 5 | 0.004634 | GO:0045779~negative regulation of bone resorption | 3 | 0.002748 |
| GO:0048511~rhythmic process | 5 | 0.0051837 | GO:0006397~mRNA processing | 7 | 0.005133 |
| GO:0006351~transcription, DNA-templated | 20 | 0.005575 | GO:0008652~cellular amino acid biosynthetic process | 3 | 0.006677 |
| GO:0033137~negative regulation of peptidyl-serine phosphorylation | 3 | 0.008042 | GO:0033137~negative regulation of peptidyl-serine phosphorylation | 3 | 0.006677 |
| GO:0008652~cellular amino acid biosynthetic process | 3 | 0.008042 | GO:0006094~gluconeogenesis | 3 | 0.00721 |
| GO:0051726~regulation of cell cycle | 4 | 0.023003 | GO:0061430~bone trabecula morphogenesis | 2 | 0.014694 |
| GO:0035020~regulation of Rac protein signal transduction | 2 | 0.032085 | GO:0051726~regulation of cell cycle | 4 | 0.017872 |
| GO:0006915~apoptotic process | 8 | 0.03525 | GO:0006564~L-serine biosynthetic process | 2 | 0.019545 |
| GO:0006810~transport | 17 | 0.035583 | GO:0045944~positive regulation of transcription from RNA polymerase II promoter | 11 | 0.024183 |
| Fabp2–/–;Fabp6–/– (M) | Fabp2–/–;Fabp6–/– (F) | ||||
| GO:0002376~immune system process | 26 | 5.43 × 10−9 | GO:0019886~antigen processing and presentation of exogenous peptide antigen via MHC class II | 6 | 9.87 × 10−8 |
| GO:0034341~response to interferon-γ | 8 | 1.55 × 10−7 | GO:0002376~immune system process | 17 | 3.32 × 10−7 |
| GO:0019882~antigen processing and presentation | 10 | 2.21 × 10−7 | GO:0019882~antigen processing and presentation | 8 | 4.85 × 10−7 |
| GO:0055114~oxidation-reduction process | 32 | 2.85 × 10−7 | GO:0034341~response to interferon-γ | 6 | 2.97 × 10−6 |
| GO:0035458~cellular response to interferon-β | 9 | 2.95 × 10−7 | GO:0035458~cellular response to interferon-β | 6 | 3.04 × 10−5 |
| GO:0042572~retinol metabolic process | 7 | 2.00 × 10−6 | GO:0006955~immune response | 12 | 3.31 × 10−5 |
| GO:0019886~antigen processing and presentation of exogenous peptide antigen via MHC class II | 6 | 2.08 × 10−6 | GO:0002504~antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 4 | 5.60 × 10−5 |
| GO:0006955~immune response | 17 | 1.23 × 10−5 | GO:0042130~negative regulation of T cell proliferation | 5 | 4.31 × 10−4 |
| GO:0007584~response to nutrient | 9 | 2.63 × 10−5 | GO:0060337~type I interferon signaling pathway | 3 | 0.001154 |
| GO:0006629~lipid metabolic process | 22 | 2.64 × 10−5 | GO:0031175~neuron projection development | 7 | 0.00154 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Agellon, L.B. Distinct Alteration of Gene Expression Programs in the Small Intestine of Male and Female Mice in Response to Ablation of Intestinal Fabp Genes. Genes 2020, 11, 943. https://doi.org/10.3390/genes11080943
Chen Y, Agellon LB. Distinct Alteration of Gene Expression Programs in the Small Intestine of Male and Female Mice in Response to Ablation of Intestinal Fabp Genes. Genes. 2020; 11(8):943. https://doi.org/10.3390/genes11080943
Chicago/Turabian StyleChen, Yiheng, and Luis B. Agellon. 2020. "Distinct Alteration of Gene Expression Programs in the Small Intestine of Male and Female Mice in Response to Ablation of Intestinal Fabp Genes" Genes 11, no. 8: 943. https://doi.org/10.3390/genes11080943
APA StyleChen, Y., & Agellon, L. B. (2020). Distinct Alteration of Gene Expression Programs in the Small Intestine of Male and Female Mice in Response to Ablation of Intestinal Fabp Genes. Genes, 11(8), 943. https://doi.org/10.3390/genes11080943





