Abstract
Wolbachia (Anaplasmataceae) is an endosymbiont of arthropods and nematodes that resides within host cells and is well known for manipulating host biology to facilitate transmission via the female germline. The effects Wolbachia has on host physiology, combined with reproductive manipulations, make this bacterium a promising candidate for use in biological- and vector-control. While it is becoming increasingly clear that Wolbachia’s effects on host biology are numerous and vary according to the host and the environment, we know very little about the molecular mechanisms behind Wolbachia’s interactions with its host. Here, I analyze 29 Wolbachia genomes for the presence of systems that are likely central to the ability of Wolbachia to respond to and interface with its host, including proteins for sensing, signaling, gene regulation, and secretion. Second, I review conditions under which Wolbachia alters gene expression in response to changes in its environment and discuss other instances where we might hypothesize Wolbachia to regulate gene expression. Findings will direct mechanistic investigations into gene regulation and host-interaction that will deepen our understanding of intracellular infections and enhance applied management efforts that leverage Wolbachia.
1. Introduction to Wolbachia
Wolbachia pipientis is an alpha-proteobacterium within the Anaplasmataceae, a family of anciently intracellular bacteria belonging to the Rickettsiales [1]. The Anaplasmataceae family includes well known intracellular pathogens such as Rickettsia, Anaplasma, Erlichia, and Orientia, as well as Midichloria, which reside within the mitochondria of certain ticks [2,3]. Wolbachia are somewhat unique within the Anaplasmataceae: they are not transmitted to vertebrate hosts via an arthropod vector like many of the pathogenic species. Instead, Wolbachia colonize many species of arthropods and nematodes and are transmitted from mother to offspring via the maternal germline [4,5]. Additionally, Wolbachia’s interactions with hosts are highly variable, and include pathogenic and mutualistic effects, even within the lifespan of a single Wolbachia-host association [6,7,8].
Wolbachia were initially categorized as “reproductive parasites” due to their colonization of insect germlines, inclusion bodies reminiscent of other intracellular infections (i.e., viruses), relationship to the pathogenic Rickettsia, and the reproductive phenotypes induced in hosts, including skewing sex ratios and creating sperm-egg incompatibilities [9,10,11,12]. However, it is becoming clear that manipulating the reproductive biology of hosts is not a feature of all Wolbachia strains, nor is it essential for the spread of Wolbachia through a population of hosts [13,14]. Despite the absence of reproductive manipulations in many Wolbachia-host associations, Wolbachia are effective at spreading throughout a population of hosts [14,15]. Indeed, many populations of arthropods and nematodes are fixed for Wolbachia infection, and an estimated 40% of arthropod species have Wolbachia infections at some level [16,17]. Proposed mechanisms for the advantage Wolbachia provides that would result in spread through a population include metabolic provisioning [18], higher fecundity relative to Wolbachia-free hosts [8], and protection against viruses [19]. Additionally, while Wolbachia are primarily transmitted from mother to offspring, they are generally capable of host-switching and establishing within a new, phylogenetically distant host species, as is evident by highly incongruent host and Wolbachia phylogenies [11,20].
Again, in contrast to the pathogenic members of the Anaplasmataceae, we know comparatively little about the mechanisms Wolbachia uses to interact with hosts and manipulate their biology. Wolbachia studies across the past few decades have been overwhelmingly focused on evolutionary biology and genomics [21,22,23,24], anti-viral protection [19,25,26,27,28,29,30], or molecular biology of host factors [31,32,33], rather than bacteriology of Wolbachia. This is in part due to the combination of research groups that have studied the Wolbachia-host symbiosis over the years, and the availability of genetic tools for host insects (e.g., Drosophila) and not for Wolbachia. However, within the last couple of years, several key studies have been published that provide some insight into the mechanisms Wolbachia uses to interface with host biology. Namely, the first link between Wolbachia genotype and phenotype was identified [34], the Wolbachia loci that induce and rescue sperm-egg incompatibilities (known as “cytoplasmic incompatibility” or CI) have been identified and enzymatically characterized [35,36,37], a suite of putatively secreted effector proteins have been identified and made available as vector constructs [38,39,40], and significant attention has been given to the potential functions encoded within Wolbachia’s prophage, “prophage WO”, beyond CI [41,42]. While these studies are particularly exciting and will be beneficial for directing applied efforts that leverage Wolbachia for insect management, a vast majority of Wolbachia genes are uncharacterized. Additionally, very little is known about what pathways and loci are critical for colonizing hosts and manipulating their biology. Furthermore, how Wolbachia regulates these interactions across host life spans or upon encountering novel hosts is almost completely unknown.
While obligately intracellular bacteria in general have more stable environments (i.e., the host cell) than free-living bacteria, even bacteria that are the most reduced in complexity and are obligate for their host have maintained some level of regulatory capacity [43]. For example, in Buchnera, the highly reduced nutritional symbiont of aphids, there is differential protein expression that appears to be regulated by small RNAs [44]. While Wolbachia have relatively reduced genomes (typically between 0.9 and 1.5 Mbp [45,46]) and they are obligately intracellular, Wolbachia are not obligate for the host in most circumstances, and they are not as strictly maternally transmitted as some obligate symbionts such as Buchnera. Thus, the ability to respond to their host environment is likely to be even more critical for establishment and transmission.
2. How do Other Bacteria Interface with Their Environment?
In facultatively intracellular bacteria that transition between environmental and host-associated states (e.g., pathogens such as Shigella [47], Brucella [48], Francisella [49], Mycobacterium [50], Salmonella [51], and Legionella [52]) and symbionts that regularly alternate between intracellular and extracellular states within a host (e.g., Spiroplasma [53]), bacteria must be able to sense whether or not they are within a host cell and mount an environment-appropriate response (e.g., virulence, growth and division, changes to metabolism, persistence, etc). Obligately intracellular bacteria (including Wolbachia and other Rickettsiales, Coxiella [54], and Chlamydia [55]) have a much more stable environment than free-living or facultatively intracellular bacteria, but must still be able to respond to changes in their surroundings. Regulation in response to host signals is important for (1) the bacterium to modify its own physiology, and/or (2) for the bacterium to exert reciprocal effects back on host physiology. Ultimately, this might allow a bacterium to avoid immune responses, regulate their metabolism and virulence, and alter host physiology in other ways that enhances their own fitness. To accomplish this, bacteria must be able to (1) detect important intra- and extracellular signals (e.g., changes in the bacterium’s own nucleotide pool, or the presence of an antimicrobial peptide), (2) relay that signal appropriately, (3) modulate gene and protein expression, and (4) properly traffic and/or export gene products and metabolites.
In bacteria, including those that live intracellularly, signals might include mechanical contact, pH, temperature, osmotic pressure, or the presence of metabolites, toxins, and proteins, all of which can be sensed by a variety of receptors [50,56,57,58]. These receptors either work alone (as a one-component signaling system) or they may be members of two- or three- component signaling systems that involve other proteins to relay the signal and initiate changes in metabolism or on gene expression [59,60]. One-component signaling systems typically involve an intracellular protein that responds to a stimulus and causes a response, with the input and output functionalities encoded in different domains [59]. Two-component systems (TCS) are the canonical pathways that bacteria use to link the environment to gene regulatory responses [61]. TCS employ a transmembrane histidine kinase that autophosphorylates upon receiving a signal, and a cognate response regulator that typically has enzymatic or transcription factor activity [62]. Finally, three component systems work similarly to TCS, except that signal reception is encoded on a separate transmembrane protein which then activates an intracellular histidine kinase [59,60]. Bacteria regularly encode for dozens of these systems and are highly specific for a given input and the downstream response [63]. For example, in Escherichia coli, there are dozens of pairs of histidine kinases and cognate response regulators that are critical for sensing and mounting responses to stimuli such as nitrite, phosphate, cell envelope structure, and osmolarity [59]. In host-associated bacteria, TCS are essential for regulating the secretion of toxins and effector proteins, internalization into host cells, trafficking within a host cell, modifying host metabolism, and egress [47,48,64,65,66]. Because TCS sensor histidine kinases are embedded in the inner membrane, factors such as outer membrane permeability affect the ability of TCS to receive and transduce signals [67].
After receiving and transducing a signal, the cell will initiate a response. The response could be at the protein level (e.g., levels of a metabolite are high enough, and negative regulation shuts off the enzyme making said metabolite), or at the level of gene expression. In one-component systems, the response is enacted by the same protein that received the signal [59]. In some two- and three-component systems, the response regulator has transcription factor activity. In some intracellular bacteria, groups of pathogenicity genes are regulated by a single factor (that may or not be a member of a TCS). For example, in Francisella, MglA regulates the expression of a genomic island that is required for intracellular growth [49]. In Legionella, amino acid starvation seems to induce virulence via an increase in RpoS, the sigma factor that guides RNA polymerase to initiate transcription at specific sites [52].
Downstream effects on virulence and pathogenicity are often mediated through the regulation of secretion systems. Secretion systems are membrane-embedded protein complexes that transport substrates across the membrane into a host, the environment, or into a neighboring bacterium [65,68]. In pathogenic bacteria, secretion systems are key to the success of the infection: the pathogen secretes effector proteins into the host that affect numerous physiological processes, ultimately enhancing the pathogen’s fitness. These secretion systems are tightly regulated by two-component systems and other transcription factors [69,70,71,72,73]. Bacteria regularly encode for multiple secretion systems, with each system specialized to transport substrates. For example, Coxiella encodes for a Type I Secretion System (T1SS), a Type II Secretion System (T2SS), and a Type IV Secretion System (T4SS) [54]. Francisella encodes for both a T2SS and a T4SS [49]. Many secretion systems are homologous to other structures evolved to span membranes. Type III Secretion Systems (T3SS) were initially discovered in Yersinia pestis, are found across many highly pathogenic bacteria, and are related to the basal body of flagella [74]. Several kinds of T4SS are related to bacterial conjugation machinery and some are capable of secreting DNA in addition to effector proteins [75,76,77]. Type VI Secretion Systems (T6SS), discovered in Vibrio cholerae and Pseudomonas aeruginosa, are highly similar to the tail spike of T4 bacteriophage [78,79,80].
These secretion systems use many different mechanisms to move substrates across membranes. The T1SS is a relatively simple system, encoded by only three proteins: an ABC transporter, a membrane fusion protein (MFP), and an outer membrane protein (OMP) (Figure 1) [81]. These proteins form a tunnel when the ABC transporter in the inner membrane interacts with the substrate, allowing the substrate to pass across the periplasm and out of the cell [81]. Like T1SS, the T3SS, T4SS, and T6SS also transport substrates from the cytoplasm to the outside of the cell (or into a target cell), and span both membranes and the periplasm. However, these secretion systems are made up of many more protein subunits. T3SS, related to flagella, move proteins through a needle-like “injectisome” after chaperones deliver substrates to a portal on the cytoplasmic side of the inner membrane [82]. T4SS are typically composed of 12 protein subunits present in variable copies that form a pilus, a translocation channel scaffold, and ATP-ases that drive a substrate through the channel (Figure 1) [75]. One of the ATP-ases, known as VirD4 in many T4SS, is the “coupling protein” that is responsible for substrate recruitment [75]. T6SS, which functions as an inverted phage tail, exports substrates via polymerization, which pushes the substrate out of the cell [80]. Finally, the T2SS and Type V Secretion Systems (T5SS) span the outer membrane and move substrates from the periplasmic space to the exterior of the cell. In T2SS this is thought to be mediated by a pseudopilus that functions as a piston, pushing a substrate out of the cell [83]. T5SS are autotransporters: they have a β-barrel domain that embeds in the outer membrane and facilitates movement of the passenger domain(s) across the membrane [84].
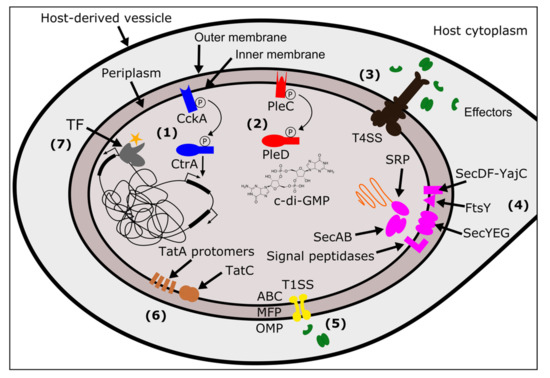
Figure 1.
Wolbachia’s systems for sensing, responding to, and modifying the host. Wolbachia cells are surrounded by three membranes: two bacterial-derived membranes (inner and outer), and one host-derived membrane of golgi- or endoplasmic reticular-origin [31,85]. The host-derived membrane may totally enclose Wolbachia, or, it may connect to the rest of the host endomembrane system. Between Wolbachia’s inner and outer membranes is the periplasmic space. The TCS sensor histidine kinases detect changes in the periplasmic space and initiate phosphorelays. (1) The CckA/CtrA TCS, (blue), results in changes to gene expression of target genes. (2) The PleC/PleD TCS (red) regulates levels of c-di-GMP, a ubiquitous bacterial second messenger. (3) The T4SS (black) secretes effector proteins (green) across both Wolbachia-derived membranes. (4) The Sec system (pink) translocates proteins across or into the inner membrane. Unfolded proteins are brought to the SecYEG channel by either SecA/B or SRP/FtsY. SecDF-YajC facilitate movement through the channel and signal peptidases cleave the signal sequence within the periplasm. (5) The T1SS (yellow) is another secretion system that spans both inner and outer membranes for the secretion of proteins. An ABC transporter/ATPase (ABC) is embedded in the inner membrane, which connects to a membrane fusion protein (MFP) that crosses the periplasmic space. Finally, the outer membrane protein (OMP) spans the outer membrane to facilitate the last step of transfer out of the bacterial cell. (6) The Tat system (brown) translocates folded proteins to the periplasm using a complex of TatC and TatA proteins. (7) TFs (grey) modulate gene expression in response to a range of signals (orange star) including reactive oxygen species, heavy metals, the binding of other proteins, and various metabolites.
In addition to secretion systems that span the outer membrane of the bacteria, there are additional systems that translocate proteins from the cytoplasm into the inner membrane, or through the inner membrane into the periplasm [86]. The general secretion system (Sec) directs unfolded proteins through a channel in the inner membrane, composed of Sec proteins Y, E, ang G. Proteins are directed to the SecYEG complex via either (1) a SecA ATPase motor protein that leverages SecB as a chaperone, or, (2) a signal recognition particle ribonucleoprotein complex (SRP) in combination with YidC (an insertase) and a membrane receptor FtsY [86,87]. Additional proteins, SecD, SecF, and YajC, enhance the efficiency of translocation through SecYEG. Finally, signal peptidases embedded in the inner membrane will cleave signal domains off the polypeptide, releasing the mature protein [88]. A separate system, twin-arginine translocation or “Tat” moves folded proteins across the inner membrane [86]. The number of Tat proteins that a given bacterium encodes for and requires for functional Tat secretion is variable [89]. The most minimal Tat systems contain two proteins: TatA and TatC [90]. Other systems use a third protein, TatB, and some bacteria encode for a paralog of TatA, named TatE [90]. All Tat components are embedded within the inner membrane. Folded proteins with the Tat signal sequence are bound by TatC (or a TatBC complex) which then recruits TatA protomers to form a translocation site in the membrane [90]. The aforementioned T2SS and T5SS only span the outer membrane of the bacterium, and thus rely on Tat and Sec systems for the initial translocation of effectors from the cytoplasm to the periplasm [83,84,88]. In addition to secretion outside of the bacterial cell, the Sec and Tat systems are important for embedding proteins in membranes. This includes proteins destined to the inner membrane (e.g., histidine kinases of a TCS), and proteins destined for the outer membrane that are localized to the periplasm via Sec/Tat and then located to the outer membrane by other pathways. These outer membrane proteins may directly interact with host factors [88].
Across the Rickettsiales, many of these bacterial systems have been identified and play roles in establishment, pathogenicity, and transmission. Genes encoding TCS have been identified across all genera in the Rickettsiales (Rickettsia, Anaplasma, Ehrlichia, Orientia, Neorickettsia, Wolbachia, Midichloria, and the newly described Aquarickettsia), and in some cases have been shown to be differentially expressed across development [91,92,93,94,95,96,97,98]. Several transcription factors have been identified in Rickettsiales that are differentially regulated according to host context (e.g., in the vertebrate host vs the arthropod vector) [92], and there are clear transcriptional responses of Rickettsiales species to stress, temperature, and the host [99,100]. Finally, all members of the Rickettsiales encode for a T4SS which was likely acquired by the ancestor of the clade, and facilitated the evolution of intracellularity [101].
As compared to the other Rickettsiales, relatively few studies have looked at Wolbachia’s ability to sense and respond to the environment. However, it seems the field of Wolbachia research is seeing a shift as more and more studies are focusing on molecular and cell biological aspects of the Wolbachia-host relationship, and of Wolbachia bacteriology [102]. This is particularly exciting given the rich understanding of the evolution and natural history of Wolbachia across arthropods and nematodes, which we can now begin to link to a mechanistic understanding of the host-Wolbachia interface. To facilitate a better understanding of the mechanistic basis of host interaction in Wolbachia, I characterize the presence of two-component systems, transcriptional regulators, and secretion systems across 29 Wolbachia genomes, and discuss what is currently known about how Wolbachia interfaces with its host.
3. Materials and Methods
Twenty-nine Wolbachia strains with complete or near-complete genomes were selected for analyses. These strains belong to Supergroups A, B, C, D, E, F, and L. Wolbachia genomes and their annotations (all annotated with NCBI’s PGAP [103]) were downloaded from RefSeq [104] on 02/10/2020 (for accession numbers, see Supplementary Materials Table S1). I queried annotations for proteins involved in sensing and signaling, transcriptional regulation, and secretion using a combination of annotation mining, orthology, BLAST, and manual curation. First, annotations were searched for terms related to the processes (for code, see Supplementary Materials Table S2). Then, previously published clusters of orthologous proteins across Wolbachia [38,105] were used to annotate proteins and ensure that proteins were not missed due to any differences in annotation. Manual curation further aided in grouping proteins based on domain structure available from NCBI. Previously published data on genes of interest (TCS [95], T4SS [106]) were cross referenced to ensure I recovered expected proteins. Pseudogenization and partial annotations are derived from the PGAP/RefSeq annotations.
Proteins annotated as T6SS components were manually inspected and found to be mis-annotated. They were all located within predicted T4SS operons, had more recent NCBI annotations that re-named these proteins as T4SS components, were homologous to T4SS proteins, and/or had conserved domain structure of either the VirB3 or VirB4 proteins of the T4SS. Given the presence of T1SS proteins HlyD and a T1SS ATPase, I specifically searched for the presence of an outer membrane protein that would complete a T1SS and indicate the potential for it to be functional. A protein annotated as TolC was identified in single copy across the majority of Wolbachia strains. Strains missing a protein annotated as TolC had a single putative TolC (labeled as “membrane protein”) that was homologous to the other TolC Wolbachia proteins, as inferred by previously published ortholog clustering [38,105].
Plotting was performed in R [107] version 3.5.0 (23-04-2018) “Joy in Playing” with the heatmap.2() function from the gplots package [108]. Figure polishing and schematics were created in Inkscape (https://inkscape.org/).
4. Sensing and Signaling
I identified two sets of TCS across the 29 Wolbachia genomes: PleC/PleD and CckA/CtrA (Figure 1). These are relatively highly conserved in their presence across Wolbachia, with just a handful of losses or pseudogenizations. These findings are in agreement with previously published data on the evolution of TCS across 12 strains of Wolbachia [95]. Here, with the addition of many more genomes, a few patterns begin to emerge.
Most losses are in the PleC/PleD TCS, where PleC is the sensor histidine kinase and PleD is the response regulator. PleC typically has enzymatic activity that regulates levels of cyclic diguanylate (c-di-GMP) in the cell, a ubiquitous bacterial second messenger that is involved in the regulation of many complex processes [109,110]. While the signal that PleC detects is unknown, the PleC/PleD system is present in many bacteria and is a key component of cycle regulation [111,112]. Here, I identify six losses or pseudogenizations in the PleC/PleD system, all of which are restricted to the monophyletic clade composed of the C and F Supergroups (represented by the strains wOo and wOv from Supergroup C, and wCle from Supergroup F). Both pleC and pleD are pseudogenized in wOo and wCle, and in wOv, pleD is annotated as pseudogenized while no pleC was recovered (Supplementary Materials Tables S3 and S4).
The second TCS present in Wolbachia is the CckA/CtrA system, where CckA is the sensor histidine kinase, and CtrA is the response regulator. Again, the signal for activation of CckA is unknown, but the downstream response regulator CtrA is a well described master regulator with transcriptional factor activity that is present across alpha-proteobacteria [112,113,114,115]. I identified two instances of pseudogenization of cckA: in wMel and in wAlbB (Supplementary Materials Tables S3 and S4). The wNfe strain had two copies of cckA, one predicted to be complete and functional, and the other partially present which may indicate an assembly or annotation issue (Supplementary Materials Tables S3 and S4).
The PleC/PleD and CckA/CtrA TCS are present across other Rickettisales, many of which encode for a third TCS not found in Wolbachia: NtrY/NtrX, which is involved in the regulation of nitrogen fixation [116]. In Anaplasma and Erlichia, the PleC/PleD and CckA/CtrA systems are differentially expressed, are enzymatically active (histidine kinases are capable of phosphorylation and PleD can make c-di-GMP), and they have been implicated in regulating intracellular infection [116,117,118]. The role of these TCS in Wolbachia biology remains to be determined, but their strong conservation across Wolbachia strains indicates these TCS are likely critical for Wolbachia’s fitness.
5. Transcriptional Regulation
Transcription in bacteria is a highly regulated process and includes three major steps: (1) initiation, (2) elongation, and (3) termination. Initiation involves RNA polymerase (bacteria have one RNA polymerase, whereas eukaryotes have three), sigma factors (“σ factor” or “specificity factor”) which are unique to bacteria, transcription factors (TF), specific DNA sequences where proteins bind, and signals that interact with the proteins (e.g., small molecules or other activating or repressing proteins). Transcriptional regulation in bacteria is more nuanced than described here, but a brief introduction aids to contextualize the genes that Wolbachia encode for that regulate these processes.
In bacteria, RNA polymerase requires a σ factor for proper and specific binding to a promotor. Bacteria typically encode for a “housekeeping” or “primary” σ factor (in E. coli this is σ70—RpoD), as well as additional “alternative” σ factors which are specialized for responses to particular environmental conditions (e.g., heat, stress) [119]. Wolbachia all encode for one copy of a RpoD-like protein (in wPpe the copy is pseudogenized, but we cannot rule out assembly or annotation artifacts) (Figure 2). Additionally, all strains have one copy of a σ32-like protein, RpoH, which in other bacteria is the heat shock σ factor (in the wVitB assembly this gene is only partially present, which may again be an assembly-related problem). Beyond that, some Wolbachia strains encode for up to four additional σ factors, all in the size range of 158–181 amino acids long. While the target genes for Wolbachia’s σ factors remains to be determined, the presence of multiple σ factors in varying copy number indicates that Wolbachia likely controls its transcriptional response to environmental conditions.
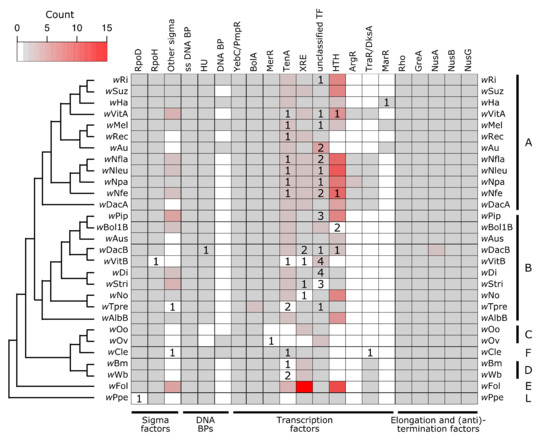
Figure 2.
Wolbachia encode for a suite of proteins that are involved in transcriptional regulation. Cladogram was drawn based on previously published Wolbachia phylogenies to show relationships between strains [132,133,134]. Colors on the heatmap indicate how many putatively functional copies are present within a Wolbachia strain for a given gene, with white indicating no copies, and the grey to red color scale indicating between one and 15 copies. Numbers inside of the heatmap indicate how many additional copies of the gene are present but annotated as pseudogenized or partial. Supergroup membership (monophyletic clades of Wolbachia historically used for designation) of the strains is indicated with the capital letters on the right side of the heatmap. Abbreviations: ss = Single-Stranded; BP = Binding Protein; TF = Transcription Factor; HTH = Helix-Turn-Helix.
TF’s play a role in gene expression via regulating RNA polymerase/σ factor complex binding. TFs typically occlude the promotor region, preventing RNA polymerase and the σ factor from binding, or they bind upstream of the promotor to aid in RNA polymerase/σ factor complex recruitment [120]. TFs typically have two domains: one that binds signals, and one that binds to the DNA at a Transcription Factor Binding Site (TFBS) [120]. TFs may work in concert with each other, some have the ability to bind to multiple sites, and many TFBS can each be bound by multiple TFs [121,122]. Variation in the affinity for any given TFBS, the TF concentration, gene expression of the TF itself, and stoichiometry of the pool of TFs are important aspects of downstream transcriptional regulation for target genes [123].
For TFs and RNA polymerase/σ factor complexes to bind DNA, the DNA must be open and not occluded by other proteins. Wolbachia encode for three additional proteins which are predicted to bind DNA and putatively structure chromatin: a single-stranded DNA binding protein (known to stabilize single-stranded DNA in E. coli [124]), a few instances of an unidentified DNA binding protein, and HU (Heat Unstable), a histone-like protein found in bacteria that can also have effects on gene regulation [125] (Figure 2). Wolbachia also encode for a suite of TFs (Figure 2, Supplementary Materials Tables S5 and S6), several of which are present across all or nearly all Wolbachia strains surveyed, and others which are highly variable across the Wolbachia phylogeny. There are two putative TFs which are present in at least single copy across all strains: a YebC/PmpR-like TF and BolA. A YebC/PmpR homolog was previously identified as a Type III Secretion System (T3SS) regulator in the pathogen Edwardsiella (Enterobacteriaceae) [126] and as a pathogenicity regulator in Pseudomonas aeruginosa [127]. In Escherichia coli, BolA has roles in cell morphology, growth, motility, and stress responses, regulating a large set of genes [128,129,130,131].
The HU DNA binding protein and a MerR TF were present in all Wolbachia except for wOo and wOv, indicating a Supergroup C-specific loss of these two proteins. There were seven other classes of TFs that were highly variable in their presence across Wolbachia strains. Many strains have pseudogenized versions of these TFs, which might be indicative of recent selective pressures and genome reduction. The largest class of TFs were Helix-Turn-Helix type TFs. While XRE and MerR are subclasses of HTH proteins present in a subset of the Wolbachia strains, there were additional HTH-domain containing proteins present in the majority of Wolbachia. Relatively few of these Helix-Turn-Helix TFs were annotated as pseudogenized or partial. Additionally, there is a large amount of variability in the size of these predicted HTH proteins (92–328 amino acids), likely indicating a range of functions.
After transcription is initiated, proteins involved in elongation and termination can regulate gene expression through a variety of mechanisms that interact with both -cis and -trans factors. Elongation ensures that the RNA polymerase complex continues to transcribe the mRNA molecule, and termination stops transcription. Anti-termination factors result in the RNA polymerase complex ignoring a termination signal that is present in the middle of a transcript, thus preventing premature termination. In prokaryotes, Gre proteins aid in elongation [135]. All Wolbachia strains encoded for one copy of GreA, which is involved in resolving paused RNA polymerase during transcription [135,136,137] (Figure 2). Wolbachia’s GreA protein has been previously explored as an anti-filarial target [138], as clearing Wolbachia infections has been a successful strategy for treating filariasis due to the obligate nature of Wolbachia-nematode symbioses [139]. In addition to GreA, all Wolbachia encoded for one copy each of the proteins Rho, NusA, NusB, and NusG, except for wDacB which appears to encode for two copies of NusA (Figure 2, Supplementary Materials Tables S5 and S6). Rho is the canonical terminator protein in prokaryotes that initiates Rho-dependent transcriptional termination [140]. However, there are other -cis and -trans factors that can also result in termination, which are Rho-independent (for example, example hairpin structures or riboswitches) [141,142,143].
NusA is multifunctional: it can stimulate pausing of the RNA polymerase complex leading to termination (for example, at hairpins), or NusA can result in antitermination when in complex with NusB and NusG [135,144]. The bimodal functionality of NusA allows for the switching of transcription to either include or exclude 3’ sequence, which is often involved in regulating the expression of genes in an operon or of overlapping ORFs. For example, the two genes that induce and rescue CI in Wolbachia (cifA and cifB [145]) are, at least in the wMel strain, separated by a Rho-independent transcription terminator that is predicted to form a GC-rich hairpin [105]. NusA might initiate termination at the hairpin (and thus only transcribe cifA), but when in complex with NusB and NusG, the hairpin might be ignored or resolved, leading to antitermination and thus transcription of a monocystronic cifA/cifB mRNA [105]. Indeed, the expression of cifA is always equal to or higher than the level of cifB expression [105]. The stoichiometry of cifA and cifB expression significantly varies due to host age and sex [105], and the bimodal functionality of NusA is one hypothesis for how Wolbachia might regulate these dynamics. The ratio and concentration of CifA and CifB could have downstream effects on the strength of CI induction in sperm and the ability to effectively rescue CI in eggs.
6. Secretion Systems
Wolbachia encode for several systems used to transport proteins. I find evidence for four putatively functional secretion systems across Wolbachia: Sec, Tat, T1SS, T4SS. Additionally many Wolbachia strains encoded for a protein annotated as (or homologous to) a T2SS protein (Figure 3, Supplementary Materials Tables S7–S9). However, there is some evidence that this T2SS protein is related to T4 pilus proteins, so it may in fact be part of the T4SS, which is derived from bacterial conjugation machinery [146]. Because of this discrepancy, this protein was left out of Figure 3, but the annotations are available in Supplementary Materials Tables S7–S9.
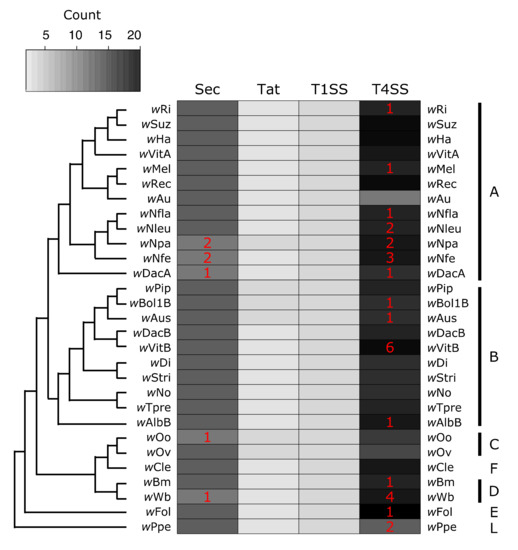
Figure 3.
Presence of secretion systems across the Wolbachia phylogeny. Cladogram was drawn based off previously published Wolbachia phylogenies to show relationships between strains [132,133,134]. The grey to black scale indicates the number of putatively functional protein copies present within a Wolbachia strain for a given secretion system. Red numbers inside of the heatmap indicate how many additional gene copies are present but annotated as pseudogenized or partial (for example, wPpe encodes for 12 proteins that make up a T4SS, plus an additional two pseudogenized ORFs also corresponding to T4SS proteins). Supergroup membership (monophyletic clades of Wolbachia historically used for designation) of the strains is indicated with the capital letters on the right side of the heatmap.
The first two systems, Sec and Tat, move proteins from the cytoplasm to the periplasm (or into the inner membrane). The Sec system is composed of Sec proteins (SecABDEFGY), YajC, SRP, FtsY, and signal peptidases [88] (Figure 1). All Wolbachia strains had one copy of each protein, with just a few examples of pseudogenizations or split open reading frames (ORFs) (Figure 3, Supplementary Materials Tables S7–S9). In wNpa and wNfe, there were two partial copies of FtsY (the SRP-docking protein), but this may be an assembly error resulting in split ORFs. The other partially present Sec system proteins were SRP in wDacA, and signal peptidase I in wWb. In the wOo strain, SecY was pseudogenized. It is likely that the Sec system is functional across most Wolbachia, and additional work is needed to verify the loss of proteins in in the handful of strains with partial or pseudogeized Sec components. The second secretion system that translocates proteins into the periplasm is the Tat system. All Wolbachia encoded for two or three Tat proteins, always with at least one copy each of TatA and TatC, which are required for Tat function [90]. Strain wNpa coded for two copies of TatC, and there were two sets of monophyletic clades that encoded for two copies of the TatA protein (the group composed of wPip, wBol1B, and wAus, as well as the C Supergroup strains wOo and wOv) (Figure 3, Supplementary Materials Tables S7–S9).
The second group of secretion systems are those that translocate proteins across the outer membrane. All Wolbachia encoded for three proteins constituting a complete T1SS: a T1SS permease/ATPase that putatively spans the inner membrane, HlyD (the membrane fusing protein that spans the periplasm, and TolC (the component that spans the outer membrane) (Figure 1 and Figure 3, Supplementary Materials Tables S7–S9) [81]. Very little is known about T1SS function or substrates in Wolbachia, but the T1SS has been implicated in virulence in other pathogens including Legionella [147,148], Rickettsia [149], Orientia [150], and Anaplasma [151]. In the Rickettsiales, several of the secreted T1SS effectors are ankyrin repeat containing proteins, predicted to be involved in protein-protein interactions [149,150,151].
Wolbachia’s T4SS is perhaps the most well understood of all the systems discussed here. Previous studies have described the evolution of T4SS proteins and genomic organization [101,106,152], identified putative T4SS substrates [38,153] (including the first described Wolbachia effector protein [39]), and analyzed T4SS expression patterns in the host [154]. Several microbiology resources are available to study Wolbachia’s T4SS including a candidate effector library [40] and a heterologous expression system in E. coli that uses a chimeric VirD4 (the T4SS coupling protein) to determine whether or not a substrate is secreted [155]. Many functional studies focus on the wMel strain that infects the model insect Drosophila melanogaster. Here, I identify putatively functional T4SS across 29 Wolbachia strains and show that pseudogenization is relatively common. However, T4SS proteins in Rickettsiales are often encoded redundantly [101], and Wolbachia undergo high levels of genomic re-arrangements [156], which together might explain the dynamics of T4SS gene gain and loss across Wolbachia. For example, approximately a third of all the pseudogenized or partial T4SS proteins are virB2 (which codes for the pilin subunit), which is highly duplicated and present in up to six putatively functional copies in some strains (Supplementary Materials Table S7).
In wBm, T4SS operon promotors are bound by two XRE-like TFs that share homology with EcxR, a Ehrlichia chaffeensis T4SS regulator [70,71]. XRE-like TFs were identified across most of the Wolbachia strains (Figure 2, Supplementary Materials Tables S5 and S6), so it is plausible they are a relatively conserved feature of T4SS regulation in Wolbachia. It remains to be determined whether additional TFs also contribute to T4SS regulation, how regulation varies across strains, and what the TFs signals or ligands are.
7. When do Wolbachia Respond to Their Environment?
While we have limited functional data on the systems and pathways described above, there are several studies that clearly indicate Wolbachia modifies gene or protein expression in response to changes in the host environment. Transcription and translation are metabolically expensive processes, so limiting un-necessary gene expression is important. Additionally, many proteins can be toxic or have off-target effects and would be harmful to Wolbachia and/or the host if expressed under the wrong conditions [157]. A strain of Wolbachia that is native to Drosophila melanogaster, wMelPop, regulates gene expression during doxycycline-induced stress in a transinfected mosquito cell line [158]. In the Drosophila melanogaster host, there are significant changes in Wolbachia gene expression patterns that correlate with fly age, life stage, and sex [159]. Interestingly, genes showing sex-biased expression patterns were also differentially expressed according to host age, perhaps indicating they are coregulated. The two genes which induce and rescue CI also show clear patterns of expression dependent on host age and sex [105]. The dynamics of this are likely critical for the proper expression of CI in the host and avoiding any toxic effects of these proteins [36]. Genes with the most significant changes in expression level are those previously predicted to be involved in stress-responses and host-interaction. These include molecular chaperones, components of secretion systems, and Ankyrin repeat domain containing proteins [159]. Additionally, genes with these dynamic transcriptional patterns were also more likely to be evolutionarily conserved across Wolbachia, perhaps highlighting their importance in host colonization and manipulation across the genus [159].
In nematode-infecting Wolbachia, which are more reduced in genome size, coding content, and the number of sensing systems and transcriptional regulators, we still see changes in Wolbachia physiology that correlate with some aspect of host biology. For example, the wBm Wolbachia symbiont of the filarial nematode Brugia malayi expresses proteins in a host-stage and host-sex dependent manner [160]. Similar patterns in protein expression were identified in the wOv symbiont of Onchocerca volvulus [161]. Additionally, wOv also had different gene expression patterns in male worm somas, versus in female worm gonads, further indicating gene regulation in accordance with host context [162]. Lastly, the filarial nematode Dirofilaria immitis and its Wolbachia symbiont both display dynamic and coordinated changes in gene expression across the life of the worm [163]. Interestingly, most of the clade specific losses were in the Supergroup C strains wOo and wOv (Figure 2, Supplementary Materials Tables S3 and S4), which, as mentioned, still regulate transcription and protein expression in response to the host context.
There are numerous other scenarios in which we might imagine advantages for Wolbachia to regulate gene expression patterns in response to the host environment. Many of the studies find that host age and sex are commonly correlated with changes in Wolbachia gene expression. Wolbachia are obligately associated with the female germline, so it is quite likely that Wolbachia respond to host cues involved in or present during gonad maturation and oogenesis. Not only is this important for successful transmission to the next generation, but also the timing and dosage of expression for reproductive manipulation genes is critical for recapitulating phenotypes (such as CI) in a transgenic model [164,165]. Additionally, while Wolbachia are obligately associated with the female germline, they also infect a range of other tissue and cell types in the host in non-random patterns that depend on the host and Wolbachia strain combination [166]. In holometabolous insects (i.e., those with complete metamorphosis including a pupal stage), not only are organ structures and cell physiology different across development, but the host insect often feeds on very different diets as a larva and as an adult, resulting in drastically different metabolite availabilities across the host lifespan. Furthermore, many insects overwinter: a physiological state that Wolbachia too must be able to navigate. It would be advantageous for Wolbachia to respond to specific changes in physiology across all these conditions to limit fitness costs for the host (upon which Wolbachia relies for transmission) while simultaneously enhancing colonization and transmission for Wolbachia. Finally, while Wolbachia may respond to host-specific changes in their environment, there are many other external stressors that Wolbachia might respond to, including temperature and antibiotics.
In addition to physiological and physical changes associated with host age, tissue, and diet, Wolbachia are in many cases capable of horizontal transfer to new hosts, especially across insects [167]. These horizontal transfers present a drastic change in environment (including the potential for time outside of a host, as well as the new host itself), and likely come with many challenges for Wolbachia. New challenges might include differences in host immune responses with which Wolbachia is not co-adapted, the presence of different metabolites, and differences in host structure. For example, ovaries across insect evolution have major differences in yolk, the interfollicular tissue, and the organization of connections between nurse cells and oocytes [168]. Finally, there is also the potential for other intracellular symbionts or pathogens present in a host (including other Wolbachia strains) competing for similar niches as a newly arrived Wolbachia.
8. Future Directions
Across Wolbachia strains we see a relatively high level of conservation in systems that would aid in sensing the environment, regulating gene expression, and exporting proteins to the host. These include the PleC/PleD and CckA/CrtA TCS, at least two σ factors, a diversity of TFs, a set of termination, anti-termination, and elongation factors, and four secretion systems. The high level of evolutionary conservation is highly indicative of their importance across Wolbachia and strong selective pressures to maintain these systems. Indeed, even in strains with high levels of pseudogenization and gene loss, these systems have been maintained [46]. There are a few clear paths for future studies that would significantly advance our understanding of Wolbachia biology and host-microbe interactions.
Firstly, all the structures and functions discussed here are highly reliant on bioinformatic predictions. Pseudogenizations and losses need to be verified, especially for strains with draft genome assemblies that have not been closed into a circle. More in depth bioinformatic analyses will be important for detecting the presence of other proteins involved in gene regulation. Wolbachia genomes have many unannotated or broadly annotated proteins [46] which will require more in depth analysis (including domain predictions, homology searches, and Markov models), along with functional validation. Verifying the expression of these genes and proteins in vivo, using heterologous expression systems, and in vitro analyses to study the biochemistry of these proteins are essential for understanding the role of these proteins in Wolbachia biology. While there are no genetic tools currently available for transforming Wolbachia, complementation experiments can be performed in other systems (e.g., Agrobacterium [169]). For example, does Wolbachia CtrA rescue a CrtA knockout in Agrobacterium, indicating similar functionality? Bacterial two-hybrid systems are useful tools for determining if proteins directly interact with each other [170], which can help identify regulatory partners and other protein-protein interactions.
Many signaling pathways (e.g., PleC/PleD) make or respond to second messengers, such as c-di-GMP. These signaling molecules can be detected and quantified in Wolbachia exposed to different environments to determine if there is a second messenger response [171]. Again, complementation experiments combined with c-di-GMP detection would be useful for determining if proteins such as Wolbachia’s PleD can make and degrade c-di-GMP. Very few studies have studied Wolbachia physiology, and metabolomic approaches will likely be fruitful in understanding the interplay between Wolbachia and host. Additionally, dual RNA-seq of host and Wolbachia transcriptomes, either alone or coupled to metabolomics, is also likely to be a fruitful direction for understanding the symbiosis [172].
Regardless of the level or type of environmental response Wolbachia is capable of mounting, Wolbachia’s regulatory networks, and how they vary across strains, are almost completely unknown. Bioinformatic approaches can be used to predict TFBS [173], and antibodies can be raised against Wolbachia’s TFs to determine promotor sites and putative regulons [174]. Additional experiments will be necessary to identify which signals TFs are responding to, when alternative σ factors are employed, and how elongation and termination are regulated to fine tune expression. Which environmental signals funnel into which responses, and which intermediate proteins and signaling molecules are required?
Finally, there are several computational approaches that will be useful for understating the evolution of Wolbachia more broadly. Here, I identify the conservation of systems as indicated by presence or absence. However, more fine-scale analyses of protein evolution and selection will give clues as to which regions of the protein are particularly important and which points in the Wolbachia phylogeny have undergone rapid evolution or dynamic changes in gene gain or loss. For example, the T4SS pilus (encoded by virB2) is an external structure, and thus detectable by the host. Is VirB2 under strong selection pressure? Are there clade- or host-specific selection pressures on these Wolbachia proteins? Wolbachia are known to undergo high levels of rearrangements and horizontal transfers of genes [156]. Do the phylogenies of these systems all track the strain phylogeny? Or, are there genes that have been transferred between Wolbachia strains? The T4SS is particularly dynamic regarding gains and losses (Figure 3). Wolbachia’s phage WO has been proposed as one method by which horizontal transfer might be achieved [42,105]. Identifying biases in genomic context might give clues to if and how genes are horizontally transferred. For example, the prophage WO region in wMel includes several transcription factors [42] that are recovered here in variable copy number across strains, including some Helix-Turn-Helix domain containing proteins, and the TF MarR (Figure 2).
Even though Wolbachia have relatively reduced genomes and minimal numbers of TCS and secretion systems compared to other intracellular bacteria, they can alter the physiology of diverse hosts in many ways. Expanding our knowledge of the regulatory networks Wolbachia employs to initiate infections and manipulate host biology will facilitate the use of Wolbachia in insect management programs and our understanding of the evolution of intracellular infections.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/7/813/s1, Table S1: Genomes used in analyses with accession numbers, Table S2: Query code, Table S3: Sensing and signaling genes, Table S4: Count data for sensing and signaling genes, Table S5: Transcription genes, Table S6: Count data for transcription genes, Table S7: Secretion system genes, Table S8: Count data for secretion system genes, Table S9: Manual curation of annotations corresponding to each secretion system.
Author Contributions
All analysis, writing, and figure production was carried out by A.R.I.L. Author has read and agreed to the published version of the manuscript.
Funding
ARIL was supported in part by the National Institutes of Health (R01AI144430 to Irene Newton), and the APC was funded by a generous startup grant from the Agricultural Research, Education and Extension Tech Transfer program (AGREETT) to ARIL at the University of Minnesota.
Acknowledgments
Thank you to Irene Newton, Delaney Miller, Audrey Parish, Sylvie Martin-Eberhardt, MaryAnn Martin, and Eric Smith for feedback on an earlier draft of the manuscript. Also thank you to Danny Rice who shared data generated in Rice et al 2017 (GBE) which was helpful for verifying the presence and absence of select genes.
Conflicts of Interest
The author declares no conflict of interest.
References
- Dumler, J.S.; Barbet, A.F.; Bekker, C.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and’HGE agent’as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [PubMed]
- Muñoz-Gómez, S.A.; Hess, S.; Burger, G.; Lang, B.F.; Susko, E.; Slamovits, C.H.; Roger, A.J. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. Elife 2019, 8, e42535. [Google Scholar] [CrossRef] [PubMed]
- Epis, S.; Mandrioli, M.; Genchi, M.; Montagna, M.; Sacchi, L.; Pistone, D.; Sassera, D. Localization of the bacterial symbiont Candidatus Midichloria mitochondrii within the hard tick Ixodes ricinus by whole-mount FISH staining. Ticks and Tick-Borne Diseases 2013, 4, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Micro. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Stouthamer, R.; Breeuwer, J.A.J.; Hurst, G.D.D. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 1999, 53, 71–102. [Google Scholar] [CrossRef]
- Min, K.-T.; Benzer, S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 1997, 94, 10792–10796. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biological Reviews 2015, 90, 89–111. [Google Scholar] [CrossRef]
- Weeks, A.R.; Turelli, M.; Harcombe, W.R.; Reynolds, K.T.; Hoffmann, A.A. From parasite to mutualist: Rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 2007, 5, e114. [Google Scholar] [CrossRef]
- Hertig, M. The Rickettsia, Wolbachia pipientis (gen. et sp.n.) and Associated Inclusions of the Mosquito, Culex pipiens. Parasitology 1936, 28, 453–486. [Google Scholar] [CrossRef]
- Hertig, M.; Wolbach, S.B. Studies on rickettsia-like micro-organisms in insects. The J. Med. Res. 1924, 44, 329. [Google Scholar]
- Werren, J.H.; Zhang, W.; Guo, L.R. Evolution and phylogeny of Wolbachia—reproductive parasites of arthropods. Proc. R. Soc. Lond. B 1995, 261, 55–63. [Google Scholar]
- Bensaadi–Merchermek, N.; Salvado, J.-C.; Cagnon, C.; Karama, S.; Mouchès, C. Characterization of the unlinked 16S rDNA and 23S-5S rRNA operon of Wolbachia pipientis, a prokaryotic parasite of insect gonads. Gene 1995, 165, 81–86. [Google Scholar] [CrossRef]
- Hamm, C.A.; Begun, D.J.; Vo, A.; Smith, C.C.; Saelao, P.; Shaver, A.O.; Jaenike, J.; Turelli, M. Wolbachia do not live by reproductive manipulation alone: Infection polymorphism in Drosophila suzukii and D. subpulchrella. Mol. Ecol. 2014, 23, 4871–4885. [Google Scholar] [CrossRef] [PubMed]
- Kriesner, P.; Hoffmann, A.A. Rapid spread of a Wolbachia infection that does not affect host reproduction in Drosophila simulans cage populations. Evolution 2018, 72, 1475–1487. [Google Scholar] [CrossRef]
- Turelli, M.; Cooper, B.S.; Richardson, K.M.; Ginsberg, P.S.; Peckenpaugh, B.; Antelope, C.X.; Kim, K.J.; May, M.R.; Abrieux, A.; Wilson, D.A. Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr. Biol. 2018, 28, 963–971.e968. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Weinert, L.A.; Araujo-Jnr, E.V.; Ahmed, M.Z.; Welch, J.J. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. R. Soc. Lond. B 2015, 282, 20150249. [Google Scholar] [CrossRef]
- Newton, I.L.; Rice, D.W. The Jekyll and Hyde symbiont: Could Wolbachia be a nutritional mutualist? J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e1000002. [Google Scholar] [CrossRef]
- Raychoudhury, R.; Baldo, L.; Oliveira, D.C.S.G.; Werren, J.H. Modes of acquisition of Wolbachia: Horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 2009, 63, 165–183. [Google Scholar] [CrossRef]
- Conner, W.R.; Blaxter, M.L.; Anfora, G.; Ometto, L.; Rota-Stabelli, O.; Turelli, M. Genome comparisons indicate recent transfer of wRi-like Wolbachia between sister species Drosophila suzukii and D. subpulchrella. Ecol. Evol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lindsey, A.R.I.; Chatterjee, P.; Werren, J.H.; Stouthamer, R.; Yi, S.V. Distinct epigenomic and transcriptomic modifications associated with Wolbachia-mediated asexuality. PLoS Path. 2020, 16, e1008397. [Google Scholar] [CrossRef] [PubMed]
- Bailly-Bechet, M.; Martins-Simões, P.; Szöllősi, G.J.; Mialdea, G.; Sagot, M.-F.; Charlat, S. How long does Wolbachia remain on board? Mol. Biol. Evol. 2017, 34, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Sun, L.V.; Vamathevan, J.; Riegler, M.; Deboy, R.; Brownlie, J.C.; McGraw, E.A.; Martin, W.; Esser, C.; Ahmadinejad, N.; et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004, 2, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Ross, P.A.; Rasic, G. Wolbachia strains for disease control: Ecological and evolutionary considerations. Ecol. Evol. 2015, 8, 751–768. [Google Scholar]
- Schmidt, T.L.; Barton, N.H.; Rašić, G.; Turley, A.P.; Montgomery, B.L.; Iturbe-Ormaetxe, I.; Cook, P.E.; Ryan, P.A.; Ritchie, S.A.; Hoffmann, A.A. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 2017, 15, e2001894. [Google Scholar] [CrossRef]
- Dutra, H.L.C.; Rocha, M.N.; Dias, F.B.S.; Mansur, S.B.; Caragata, E.P.; Moreira, L.A. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host & Microbe 2016, 19, 771–774. [Google Scholar]
- Lindsey, A.R.I.; Bhattacharya, T.; Newton, I.L.G.; Hardy, R.W. Conflict in the intracellular lives of endosymbionts and viruses: A mechanistic look at Wolbachia—mediated pathogen-blocking. Viruses 2018, 10, 141. [Google Scholar] [CrossRef]
- Hedges, L.M.; Brownlie, J.C.; O’neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322, 702. [Google Scholar] [CrossRef]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef]
- White, P.M.; Serbus, L.R.; Debec, A.; Codina, A.; Bray, W.; Guichet, A.; Lokey, R.S.; Sullivan, W. Reliance of Wolbachia on high rates of host proteolysis revealed by a genome-wide RNAi screen of Drosophila cells. Genetics 2017, 205, 1473–1488. [Google Scholar] [CrossRef]
- Grobler, Y.; Yun, C.Y.; Kahler, D.J.; Bergman, C.M.; Lee, H.; Oliver, B.; Lehmann, R. Whole genome screen reveals a novel relationship between Wolbachia levels and Drosophila host translation. PLoS Path. 2018, 14, e1007445. [Google Scholar] [CrossRef] [PubMed]
- Newton, I.L.; Savytskyy, O.; Sheehan, K.B. Wolbachia utilize host actin for efficient maternal transmission in Drosophila melanogaster. PLoS Path. 2015, 11, e1004798. [Google Scholar] [CrossRef] [PubMed]
- Chrostek, E.; Teixeira, L. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol. 2015, 13, e1002065. [Google Scholar] [CrossRef] [PubMed]
- LePage, D.P.; Metcalf, J.A.; Bordenstein, S.R.; On, J.; Perlmutter, J.I.; Shropshire, J.D.; Layton, E.M.; Funkhouser-Jones, L.J.; Beckmann, J.F.; Bordenstein, S.R. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 2017, 543, 243–247. [Google Scholar] [CrossRef]
- Beckmann, J.F.; Ronau, J.A.; Hochstrasser, M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Micro. 2017, 2, 17007. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ronau, J.A.; Beckmann, J.F.; Hochstrasser, M. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl. Acad. Sci. USA 2019, 116, 22314–22321. [Google Scholar] [CrossRef]
- Rice, D.W.; Sheehan, K.B.; Newton, I.L. Large–scale identification of Wolbachia pipientis effectors. Genome Biol. Evol. 2017, 9, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, K.B.; Martin, M.; Lesser, C.F.; Isberg, R.R.; Newton, I.L. Identification and characterization of a candidate Wolbachia pipientis type IV effector that interacts with the actin cytoskeleton. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Newton, I.L.; Sheehan, K.B. Gateway entry vector library of Wolbachia pipientis candidate effectors from strain wMel. Microbiol. Resour. Announc. 2018, 7, e00806–00818. [Google Scholar] [CrossRef]
- Perlmutter, J.I.; Bordenstein, S.R.; Unckless, R.L.; LePage, D.P.; Metcalf, J.A.; Hill, T.; Martinez, J.; Jiggins, F.M.; Bordenstein, S.R. The phage gene wmk is a candidate for male killing by a bacterial endosymbiont. PLoS Path. 2019, 15. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Bordenstein, S.R. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Miravet-Verde, S.; Lloréns-Rico, V.; Serrano, L. Alternative transcriptional regulation in genome-reduced bacteria. Curr. Opin. Microbiol. 2017, 39, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.K.; Degnan, P.H. Widespread expression of conserved small RNAs in small symbiont genomes. ISME J. 2014, 8, 2490–2502. [Google Scholar] [CrossRef] [PubMed]
- Fenn, K.; Blaxter, M. Wolbachia genomes: Revealing the biology of parasitism and mutualism. Trends Parasitol. 2006, 22, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, A.R.I.; Werren, J.H.; Richards, S.; Stouthamer, R. Comparative genomics of a parthenogenesis–inducing Wolbachia symbiont. G3: Genes|Genomes|Genetics 2016, 6, 2113–2123. [Google Scholar] [CrossRef]
- Dorman, C.J.; McKenna, S.; Beloin, C. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int. J. Med. Microbiol. 2001, 291, 89–96. [Google Scholar] [CrossRef]
- Gorvel, J.P.; Moreno, E. Brucella intracellular life: From invasion to intracellular replication. Vet. Microbiol. 2002, 90, 281–297. [Google Scholar] [CrossRef]
- Sjöstedt, A. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microb. Infect. 2006, 8, 561–567. [Google Scholar] [CrossRef]
- Rohde, K.H.; Abramovitch, R.B.; Russell, D.G. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host & Microbe 2007, 2, 352–364. [Google Scholar]
- Ibarra, J.A.; Steele-Mortimer, O. Salmonella-the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 2009, 11, 1579–1586. [Google Scholar] [PubMed]
- Molmeret, M.; Bitar, D.M.; Han, L.; Kwaik, Y.A. Cell biology of the intracellular infection by Legionella pneumophila. Microb. Infect. 2004, 6, 129–139. [Google Scholar] [CrossRef]
- Haselkorn, T.S. The Spiroplasma heritable bacterial endosymbiont of Drosophila. Fly 2010, 4, 80–87. [Google Scholar] [CrossRef]
- Van Schaik, E.J.; Chen, C.; Mertens, K.; Weber, M.M.; Samuel, J.E. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Micro. 2013, 11, 561–573. [Google Scholar] [CrossRef]
- Wyrick, P.B. Intracellular survival by Chlamydia: Microreview. Cell. Microbiol. 2000, 2, 275–282. [Google Scholar] [CrossRef]
- Persat, A.; Nadell, C.D.; Kim, M.K.; Ingremeau, F.; Siryaporn, A.; Drescher, K.; Wingreen, N.S.; Bassler, B.L.; Gitai, Z.; Stone, H.A. The mechanical world of bacteria. Cell 2015, 161, 988–997. [Google Scholar] [CrossRef]
- Kotte, O.; Zaugg, J.B.; Heinemann, M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol. Syst. Biol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiol. 2005, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Marijuán, P.C.; Navarro, J.; del Moral, R. How prokaryotes ‘encode’their environment: Systemic tools for organizing the information flow. BioSyst. 2018, 164, 26–38. [Google Scholar] [CrossRef]
- Marijuán, P.C.; Navarro, J.; del Moral, R. On prokaryotic intelligence: Strategies for sensing the environment. BioSyst. 2010, 99, 94–103. [Google Scholar] [CrossRef]
- Gao, R.; Mack, T.R.; Stock, A.M. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007, 32, 225–234. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Podgornaia, A.I.; Laub, M.T. Determinants of specificity in two-component signal transduction. Curr. Opin. Microbiol. 2013, 16, 156–162. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Lewenza, S.; Hancock, R.E. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Beier, D.; Gross, R. Regulation of bacterial virulence by two–component systems. Curr. Opin. Microbiol. 2006, 9, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.B.; Dubnau, E.; Kolesnikova, I.; Laval, F.; Daffe, M.; Smith, I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006, 60, 312–330. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Batut, J.; Andersson, S.G.; O’Callaghan, D. The evolution of chronic infection strategies in the α-proteobacteria. Nat. Rev. Micro. 2004, 2, 933–945. [Google Scholar] [CrossRef]
- Martínez-Núñez, C.; Altamirano-Silva, P.; Alvarado-Guillén, F.; Moreno, E.; Guzmán-Verri, C.; Chaves-Olarte, E. The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol. 2010, 192, 5603–5608. [Google Scholar] [CrossRef]
- Cheng, Z.; Wang, X.; Rikihisa, Y. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J. Bacteriol. 2008, 190, 2096–2105. [Google Scholar] [CrossRef]
- Li, Z.; Carlow, C.K. Characterization of transcription factors that regulate the type IV secretion system and riboflavin biosynthesis in Wolbachia of Brugia malayi. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Altman, E.; Segal, G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 2008, 190, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Zusman, T.; Aloni, G.; Halperin, E.; Kotzer, H.; Degtyar, E.; Feldman, M.; Segal, G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 2007, 63, 1508–1523. [Google Scholar] [CrossRef] [PubMed]
- Büttner, D. Protein export according to schedule: Architecture, assembly, and regulation of type III secretion systems from plant-and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 262–310. [Google Scholar] [CrossRef]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef]
- Cascales, E.; Christie, P.J. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 2004, 304, 1170–1173. [Google Scholar] [CrossRef]
- Alvarez-Martinez, C.E.; Christie, P.J. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009, 73, 775–808. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef]
- Kanonenberg, K.; Spitz, O.; Erenburg, I.N.; Beer, T.; Schmitt, L. Type I secretion system—it takes three and a substrate. FEMS Microbiol. Lett. 2018, 365, fny094. [Google Scholar] [CrossRef] [PubMed]
- Radics, J.; Königsmaier, L.; Marlovits, T.C. Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol. 2014, 21, 82–87. [Google Scholar] [CrossRef]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Micro. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Leo, J.C.; Grin, I.; Linke, D. Type V secretion: Mechanism (s) of autotransport through the bacterial outer membrane. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-O.; Kim, G.-W.; Lee, O.-K. Wolbachia bacteria reside in host Golgi–related vesicles whose position is regulated by polarity proteins. PLoS ONE 2011, 6, e22703. [Google Scholar] [CrossRef]
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Müller, M.; Koch, H.-G. Protein translocation across the inner membrane of Gram-negative bacteria: The Sec and Tat dependent protein transport pathways. Res. Microbiol. 2013, 164, 505–534. [Google Scholar] [CrossRef] [PubMed]
- Angelini, S.; Deitermann, S.; Koch, H.G. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Reports 2005, 6, 476–481. [Google Scholar] [CrossRef]
- Tsirigotaki, A.; De Geyter, J.; Šoštaric, N.; Economou, A.; Karamanou, S. Protein export through the bacterial Sec pathway. Nat. Rev. Micro. 2017, 15, 21. [Google Scholar] [CrossRef]
- Palmer, T.; Berks, B.C. The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Micro. 2012, 10, 483–496. [Google Scholar] [CrossRef]
- Berks, B.C. The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 2015, 84, 843–864. [Google Scholar] [CrossRef]
- Zhang, J.z.; Popov, V.L.; Gao, S.; Walker, D.H.; Yu, X.j. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 2007, 9, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Rikihisa, Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: Subversive manipulators of host cells. Nat. Rev. Micro. 2010, 8, 328–339. [Google Scholar] [CrossRef]
- McLeod, M.P.; Qin, X.; Karpathy, S.E.; Gioia, J.; Highlander, S.K.; Fox, G.E.; McNeill, T.Z.; Jiang, H.; Muzny, D.; Jacob, L.S. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 2004, 186, 5842–5855. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Yamashita, A.; Kurokawa, K.; Morimoto, T.; Ogawa, M.; Fukuhara, M.; Urakami, H.; Ohnishi, M.; Uchiyama, I.; Ogura, Y. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008, 15, 185–199. [Google Scholar] [CrossRef]
- Christensen, S.; Serbus, L.R. Comparative analysis of Wolbachia genomes reveals streamlining and divergence of minimalist two-component systems. G3: Genes|Genomes|Genetics 2015, 5, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Zhang, C.; Gibson, K.; Rikihisa, Y. Analysis of complete genome sequence of Neorickettsia risticii: Causative agent of Potomac horse fever. Nucleic Acids Res. 2009, 37, 6076–6091. [Google Scholar] [CrossRef] [PubMed]
- Klinges, J.G.; Rosales, S.M.; McMinds, R.; Shaver, E.C.; Shantz, A.A.; Peters, E.C.; Eitel, M.; Wörheide, G.; Sharp, K.H.; Burkepile, D.E. Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. The ISME journal 2019, 13, 2938–2953. [Google Scholar] [CrossRef] [PubMed]
- Sassera, D.; Lo, N.; Epis, S.; D’Auria, G.; Montagna, M.; Comandatore, F.; Horner, D.; Peretó, J.; Luciano, A.M.; Franciosi, F. Phylogenomic evidence for the presence of a flagellum and cbb 3 oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 2011, 28, 3285–3296. [Google Scholar] [CrossRef]
- Rovery, C.; Renesto, P.; Crapoulet, N.; Matsumoto, K.; Parola, P.; Ogata, H.; Raoult, D. Transcriptional response of Rickettsia conorii exposed to temperature variation and stress starvation. Res. Microbiol. 2005, 156, 211–218. [Google Scholar] [CrossRef]
- Nelson, C.M.; Herron, M.J.; Felsheim, R.F.; Schloeder, B.R.; Grindle, S.M.; Chavez, A.O.; Kurtti, T.J.; Munderloh, U.G. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 2008, 9, 364. [Google Scholar] [CrossRef]
- Gillespie, J.J.; Brayton, K.A.; Williams, K.P.; Diaz, M.A.Q.; Brown, W.C.; Azad, A.F.; Sobral, B.W. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 2010, 78, 1809–1823. [Google Scholar] [CrossRef]
- Newton, I.L.; Slatko, B.E. Symbiosis comes of age at the 10th biennial meeting of Wolbachia researchers. Am Soc. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Lindsey, A.R.I.; Rice, D.W.; Bordenstein, S.R.; Brooks, A.W.; Bordenstein, S.R.; Newton, I.L.G. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol. Evol. 2018, 10, 434–451. [Google Scholar] [CrossRef]
- Bhattacharya, T.; Newton, I.L. Mi Casa es Su Casa: How an intracellular symbiont manipulates host biology. Environ. Microbiol. 2017, 21, 3188–3196. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: URL http://www.R-project.org/ (accessed on 23 April 2010).
- Warnes, M.G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W. gplots: Various R Programming Tools for Plotting Data; ScienceOpen, Inc.: Middlesex Turnpike Burlington, MA, USA, 2016. [Google Scholar]
- Römling, U.; Gomelsky, M.; Galperin, M.Y. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005, 57, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, R.; Pratt, J.T.; Camilli, A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007, 61, 131–148. [Google Scholar] [CrossRef]
- Del Medico, L.; Cerletti, D.; Schächle, P.; Christen, M.; Christen, B. The type IV pilin PilA couples surface attachment and cell–cycle initiation in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2020, 117, 9546–9553. [Google Scholar] [CrossRef] [PubMed]
- Brilli, M.; Fondi, M.; Fani, R.; Mengoni, A.; Ferri, L.; Bazzicalupo, M.; Biondi, E.G. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: A comparative genomic analysis. BMC Systems Biology 2010, 4, 52. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Kumar, L.; Radhakrishnan, S.K. Sensory domain of the cell cycle kinase CckA regulates the differential DNA binding of the master regulator CtrA in Caulobacter crescentus. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2018, 1861, 952–961. [Google Scholar] [CrossRef]
- Leung, M.M.; Brimacombe, C.A.; Beatty, J.T. Transcriptional regulation of the Rhodobacter capsulatus response regulator CtrA. Microbiology 2013, 159, 96. [Google Scholar] [CrossRef][Green Version]
- Hallez, R.; Bellefontaine, A.-F.; Letesson, J.-J.; De Bolle, X. Morphological and functional asymmetry in α-proteobacteria. Trends Microbiol. 2004, 12, 361–365. [Google Scholar] [CrossRef]
- Cheng, Z.; Kumagai, Y.; Lin, M.; Zhang, C.; Rikihisa, Y. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell. Microbiol. 2006, 8, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.-H.; Kumagai, Y.; Hyodo, M.; Hayakawa, Y.; Rikihisa, Y. The Anaplasma phagocytophilum PleC histidine kinase and PleD diguanylate cyclase two-component system and role of cyclic Di-GMP in host cell infection. J. Bacteriol. 2009, 191, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, Y.; Cheng, Z.; Lin, M.; Rikihisa, Y. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 2006, 74, 5014–5022. [Google Scholar] [CrossRef]
- Paget, M.S. Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 2015, 5, 1245–1265. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J. The regulation of bacterial transcription initiation. Nat. Rev. Micro. 2004, 2, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Lopez-Bojorquez, L.N.; Martínez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chávez, I.; Balderas-Martínez, Y.I.; Encarnación, S.; Collado-Vides, J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2008, 33, 133–151. [Google Scholar] [CrossRef]
- Browning, D.F.; Butala, M.; Busby, S.J. Bacterial Transcription Factors: Regulation by Pick ‘N’Mix. J. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Martınez-Antonio, A.; Collado-Vides, J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003, 6, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.R.; Laine, P.S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Mol. Biol. Rev. 1990, 54, 342–380. [Google Scholar] [CrossRef]
- Dorman, C.J.; Deighan, P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003, 13, 179–184. [Google Scholar] [CrossRef]
- Wei, L.; Wu, Y.; Qiao, H.; Xu, W.; Zhang, Y.; Liu, X.; Wang, Q. YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol. Lett. 2018, 365, fny137. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Li, L.; Dong, Z.; Surette, M.G.; Duan, K. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 2008, 190, 6217–6227. [Google Scholar] [CrossRef]
- Dressaire, C.; Moreira, R.N.; Barahona, S.; de Matos, A.P.A.; Arraiano, C.M. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Freire, P.; Vicente, M.; Arraiano, C.M. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 1999, 32, 789–798. [Google Scholar] [CrossRef]
- Aldea, M.; Garrido, T.; Hernández-Chico, C.; Vicente, M.; Kushner, S. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989, 8, 3923–3931. [Google Scholar] [CrossRef]
- Freire, P.; Moreira, R.N.; Arraiano, C.M. BolA inhibits cell elongation and regulates MreB expression levels. J. Mol. Biol. 2009, 385, 1345–1351. [Google Scholar] [CrossRef]
- Brown, A.M.; Wasala, S.K.; Howe, D.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci. Rep. 2016, 6, 34955. [Google Scholar] [CrossRef]
- Gerth, M.; Bleidorn, C. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat. Micro. 2016, 2, 16241. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.M.; Baxter, S.W. Draft genome assembly of a Wolbachia endosymbiont of Plutella australiana. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Borukhov, S.; Lee, J.; Laptenko, O. Bacterial transcription elongation factors: New insights into molecular mechanism of action. Mol. Microbiol. 2005, 55, 1315–1324. [Google Scholar] [CrossRef]
- Borukhov, S.; Polyakov, A.; Nikiforov, V.; Goldfarb, A. GreA protein: A transcription elongation factor from Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 8899–8902. [Google Scholar] [CrossRef]
- Kusuya, Y.; Kurokawa, K.; Ishikawa, S.; Ogasawara, N.; Oshima, T. Transcription factor GreA contributes to resolving promoter-proximal pausing of RNA polymerase in Bacillus subtilis cells. J. Bacteriol. 2011, 193, 3090–3099. [Google Scholar] [CrossRef] [PubMed]
- Nag, J.K.; Shrivastava, N.; Chahar, D.; Gupta, C.L.; Bajpai, P.; Misra-Bhattacharya, S. Wolbachia Transcription Elongation Factor “Wol GreA” Interacts with α2ββ′ σ Subunits of RNA Polymerase through Its Dimeric C-Terminal Domain. PLoS Negl. Trop. Dis. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Slatko, B.E.; Taylor, M.J.; Foster, J.M. The Wolbachia endosymbiont as an anti-filarial nematode target. Symbiosis 2010, 51, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Dombroski, A.J.; Platt, T. Transcription termination factor rho is an RNA-DNA helicase. Cell 1987, 48, 945–952. [Google Scholar] [CrossRef]
- Farnham, P.J.; Platt, T. Rho-independent termination: Dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981, 9, 563–577. [Google Scholar] [CrossRef]
- Lesnik, E.A.; Sampath, R.; Levene, H.B.; Henderson, T.J.; McNeil, J.A.; Ecker, D.J. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001, 29, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Gottesman, M.E. Transcription termination and anti-termination in E. coli. Genes Cells 2002, 7, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.F.; Bonneau, M.; Chen, H.; Hochstrasser, M.; Poinsot, D.; Merçot, H.; Weill, M.; Sicard, M.; Charlat, S. The toxin-antidote model of cytoplasmic incompatibility: Genetics and evolutionary implications. Trends Genet. 2019. [Google Scholar] [CrossRef]
- Peabody, C.R.; Chung, Y.J.; Yen, M.-R.; Vidal-Ingigliardi, D.; Pugsley, A.P.; Saier Jr, M.H. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 2003, 149, 3051–3072. [Google Scholar] [CrossRef] [PubMed]
- Fuche, F.; Vianney, A.; Andrea, C.; Doublet, P.; Gilbert, C. Functional type 1 secretion system involved in Legionella pneumophila virulence. J. Bacteriol. 2015, 197, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.L.; Garner, E.; Jospin, G.; Coil, D.A.; Schwake, D.O.; Eisen, J.A.; Mukhopadhyay, B.; Pruden, A.J. Whole genome sequence analysis reveals the broad distribution of the RtxA type 1 secretion system and four novel putative type 1 secretion systems throughout the Legionella genus. PLoS One 2020, 15, e0223033. [Google Scholar] [CrossRef]
- Kaur, S.J.; Rahman, M.S.; Ammerman, N.C.; Beier-Sexton, M.; Ceraul, S.M.; Gillespie, J.J.; Azad, A.F. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J. Bacteriol. 2012, 194, 4920–4932. [Google Scholar] [CrossRef]
- VieBrock, L.; Evans, S.M.; Beyer, A.R.; Larson, C.L.; Beare, P.A.; Ge, H.; Singh, S.; Rodino, K.G.; Heinzen, R.A.; Richards, A.L. Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Frontiers in Cellular and Infection Microbiology 2015, 4, 186. [Google Scholar] [CrossRef]
- Wakeel, A.; den Dulk-Ras, A.; Hooykaas, P.J.; McBride, J.W. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front. Cell. Infect. Microbiol. 2011, 1, 22. [Google Scholar] [CrossRef]
- Pichon, S.; Bouchon, D.; Cordaux, R.; Chen, L.; Garrett, R.A.; Grève, P. Conservation of the Type IV secretion system throughout Wolbachia evolution. Biochem. Biophys. Res. Commun. 2009, 385, 557–562. [Google Scholar] [CrossRef]
- Carpinone, E.M.; Li, Z.; Mills, M.K.; Foltz, C.; Brannon, E.R.; Carlow, C.K.; Starai, V.J. Identification of putative effectors of the Type IV secretion system from the Wolbachia endosymbiont of Brugia malayi. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Pichon, S.; Bouchon, D.; Liu, C.; Chen, L.; Garrett, R.; Greve, P. The expression of one ankyrin pk2 allele of the WO prophage is correlated with the Wolbachia feminizing effect in isopods. BMC Microbiol. 2012, 12, 55. [Google Scholar] [CrossRef]
- Whitaker, N.; Berry, T.M.; Rosenthal, N.; Gordon, J.E.; Gonzalez-Rivera, C.; Sheehan, K.B.; Truchan, H.K.; VieBrock, L.; Newton, I.L.; Carlyon, J.A. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J. Bacteriol. 2016, 198, 2701–2718. [Google Scholar] [CrossRef]
- Ellegaard, K.M.; Klasson, L.; Naslund, K.; Bourtzis, K.; Andersson, S.G.E. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 2013, 9, e1003381. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Darby, A.C.; Gill, A.C.; Armstrong, S.D.; Hartley, C.S.; Xia, D.; Wastling, J.M.; Makepeace, B.L. Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME J. 2014, 8, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Gutzwiller, F.; Carmo, C.R.; Miller, D.E.; Rice, D.W.; Newton, I.L.; Hawley, R.S.; Teixeira, L.; Bergman, C.M. Dynamics of Wolbachia pipientis gene expression across the Drosophila melanogaster life cycle. G3: Genes Genomes Genet. 2015, 5, 2843–2856. [Google Scholar] [CrossRef]
- Bennuru, S.; Meng, Z.; Ribeiro, J.M.; Semnani, R.T.; Ghedin, E.; Chan, K.; Lucas, D.A.; Veenstra, T.D.; Nutman, T.B. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc. Natl. Acad. Sci. USA 2011, 108, 9649–9654. [Google Scholar] [CrossRef] [PubMed]
- Bennuru, S.; Cotton, J.A.; Ribeiro, J.M.; Grote, A.; Harsha, B.; Holroyd, N.; Mhashilkar, A.; Molina, D.M.; Randall, A.Z.; Shandling, A.D.; et al. Stage-specific transcriptome and proteome analyses of the filarial parasite Onchocerca volvulus and its Wolbachia endosymbiont. mBio 2016, 7. [Google Scholar] [CrossRef]
- Darby, A.C.; Armstrong, S.D.; Bah, G.S.; Kaur, G.; Hughes, M.A.; Kay, S.M.; Koldkjær, P.; Rainbow, L.; Radford, A.D.; Blaxter, M.L. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 2012, 22, 2467–2477. [Google Scholar] [CrossRef] [PubMed]
- Luck, A.N.; Evans, C.C.; Riggs, M.D.; Foster, J.M.; Moorhead, A.R.; Slatko, B.E.; Michalski, M.L. Concurrent transcriptional profiling of Dirofilaria immitis and its Wolbachia endosymbiont throughout the nematode life cycle reveals coordinated gene expression. BMC Genomics 2014, 15, 1041. [Google Scholar] [CrossRef]
- Shropshire, J.D.; Bordenstein, S.R. Two-By-One model of cytoplasmic incompatibility: Synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef]
- Shropshire, J.D.; On, J.; Layton, E.M.; Zhou, H.; Bordenstein, S.R. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2018, 115, 4987–4991. [Google Scholar] [CrossRef] [PubMed]
- Pietri, J.E.; DeBruhl, H.; Sullivan, W. The rich somatic life of Wolbachia. Microbiol. Open 2016. [Google Scholar] [CrossRef] [PubMed]
- Vavre, F.; Fleury, F.; Lepetit, D.; Fouillet, P.; Bouletreau, M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol. Biol. Evol. 1999, 16, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Bonhag, P.F. Ovarian structure and vitellogenesis in insects. Annu. Rev. Entomol. 1958, 3, 137–160. [Google Scholar] [CrossRef]
- Berger, B.R.; Christie, P.J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 1994, 176, 3646–3660. [Google Scholar] [CrossRef]
- Karna, S.R.; Zogaj, X.; Barker, J.R.; Seshu, J.; Dove, S.L.; Klose, K.E. A bacterial two-hybrid system that utilizes Gateway cloning for rapid screening of protein-protein interactions. BioTechniques 2010, 49, 831–833. [Google Scholar] [CrossRef]
- Massie, J.P.; Reynolds, E.L.; Koestler, B.J.; Cong, J.-P.; Agostoni, M.; Waters, C.M. Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 12746–12751. [Google Scholar] [CrossRef]
- Lindsey, A.R.; Bhattacharya, T.; Hardy, R.W.; Newton, I. Wolbachia and virus alter the host transcriptome at the interface of nucleotide metabolism pathways. bioRxiv 2020. [Google Scholar]
- Jayaram, N.; Usvyat, D.; Martin, A.C. Evaluating tools for transcription factor binding site prediction. BMC Bioinform. 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Mundade, R.; Ozer, H.G.; Wei, H.; Prabhu, L.; Lu, T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 2014, 13, 2847–2852. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).