Heat Diffusion Kernel Algorithm-Based Interpretation of the Disease Intervention Mechanism for DHA
Abstract
1. Introduction
2. Data Sources and Methods
2.1. cMap Data Preprocessing
2.2. Gene Regulatory Network Calculation
2.3. Biological Function Enrichment
2.4. Similarity Calculation of Gene Regulatory Networks
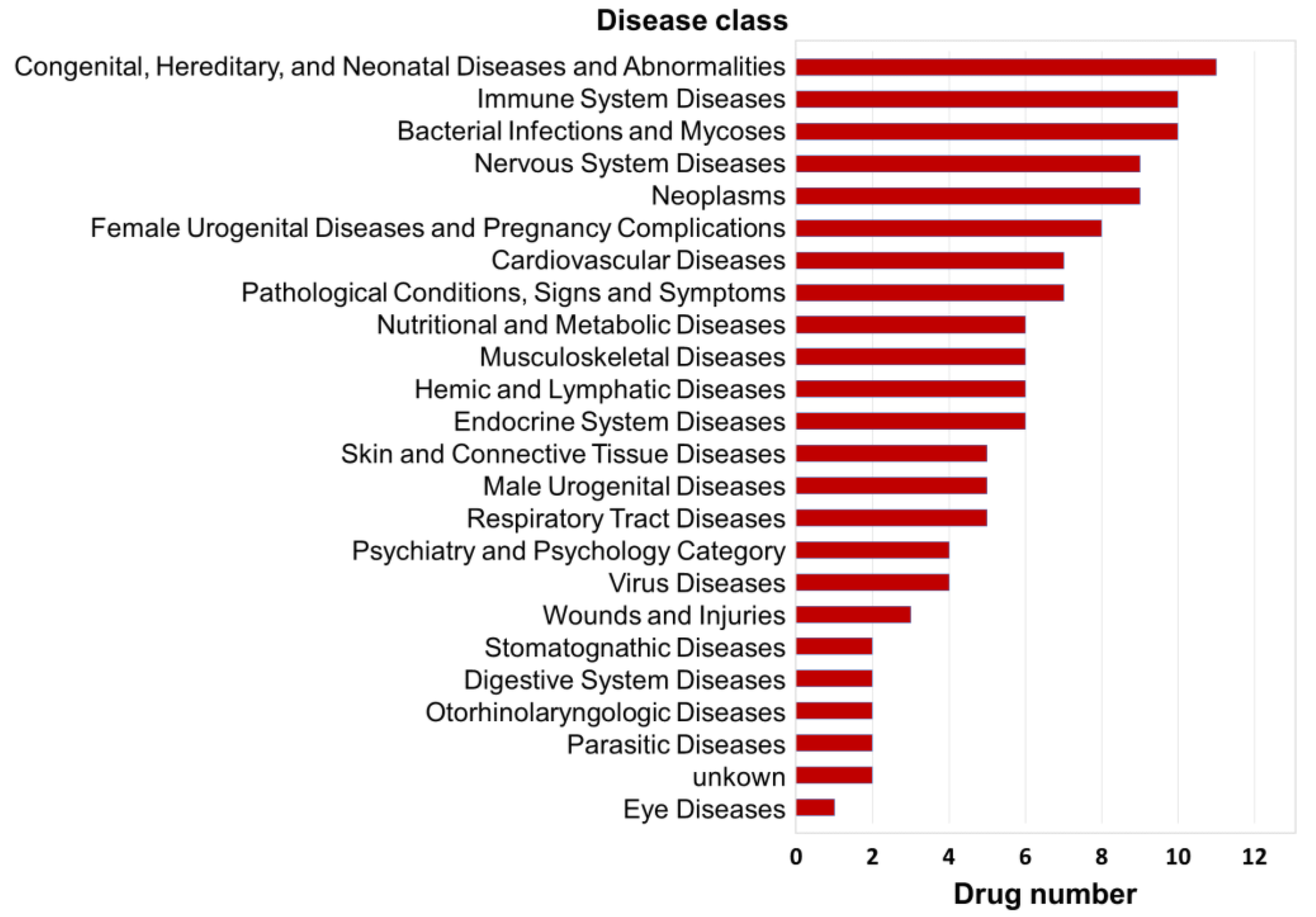
3. Results
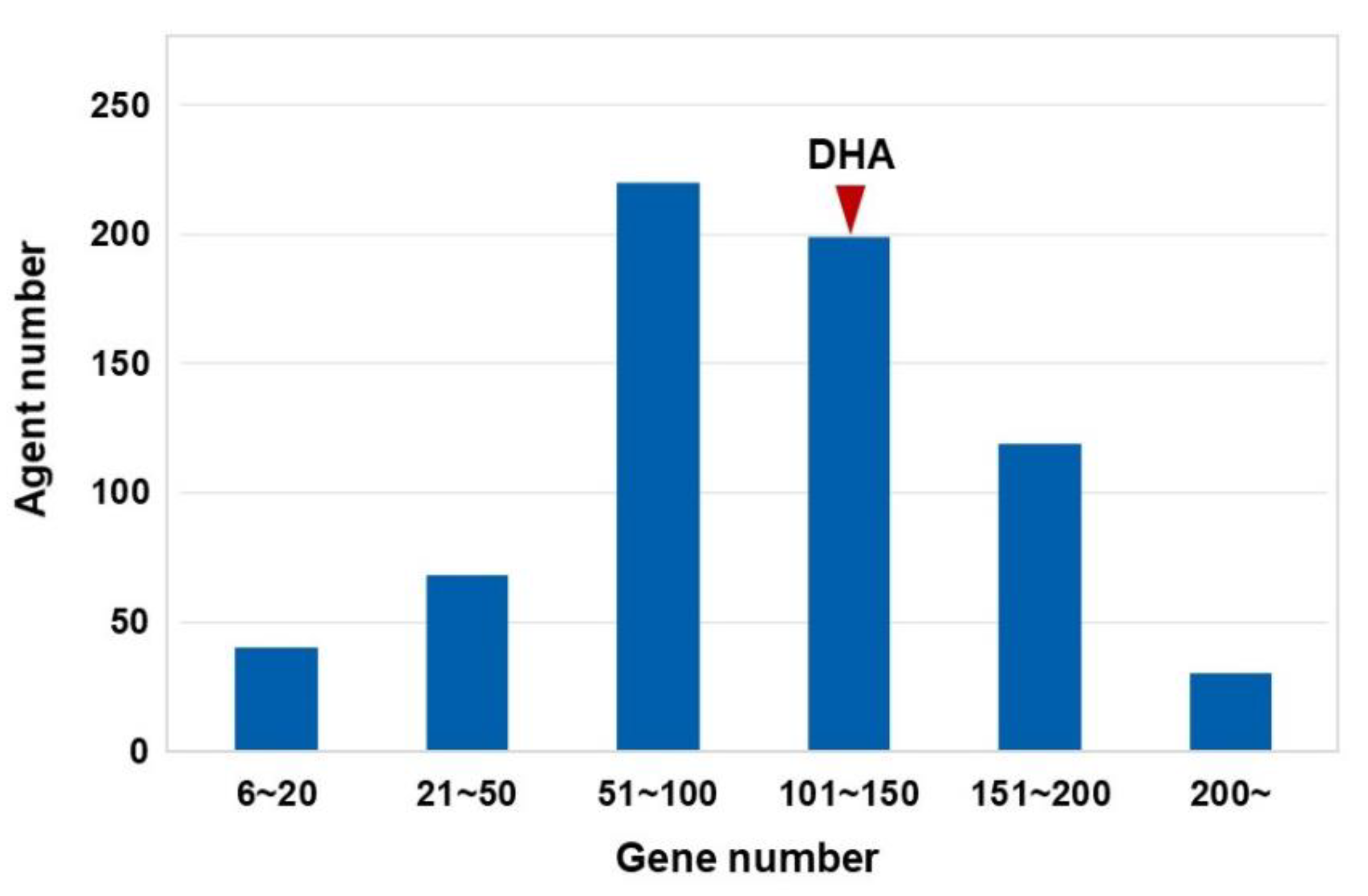
3.1. Gene Regulatory Networks of cMap Agents
3.2. Interpretation of Disease Intervention Mechanisms Based on Biological Functions
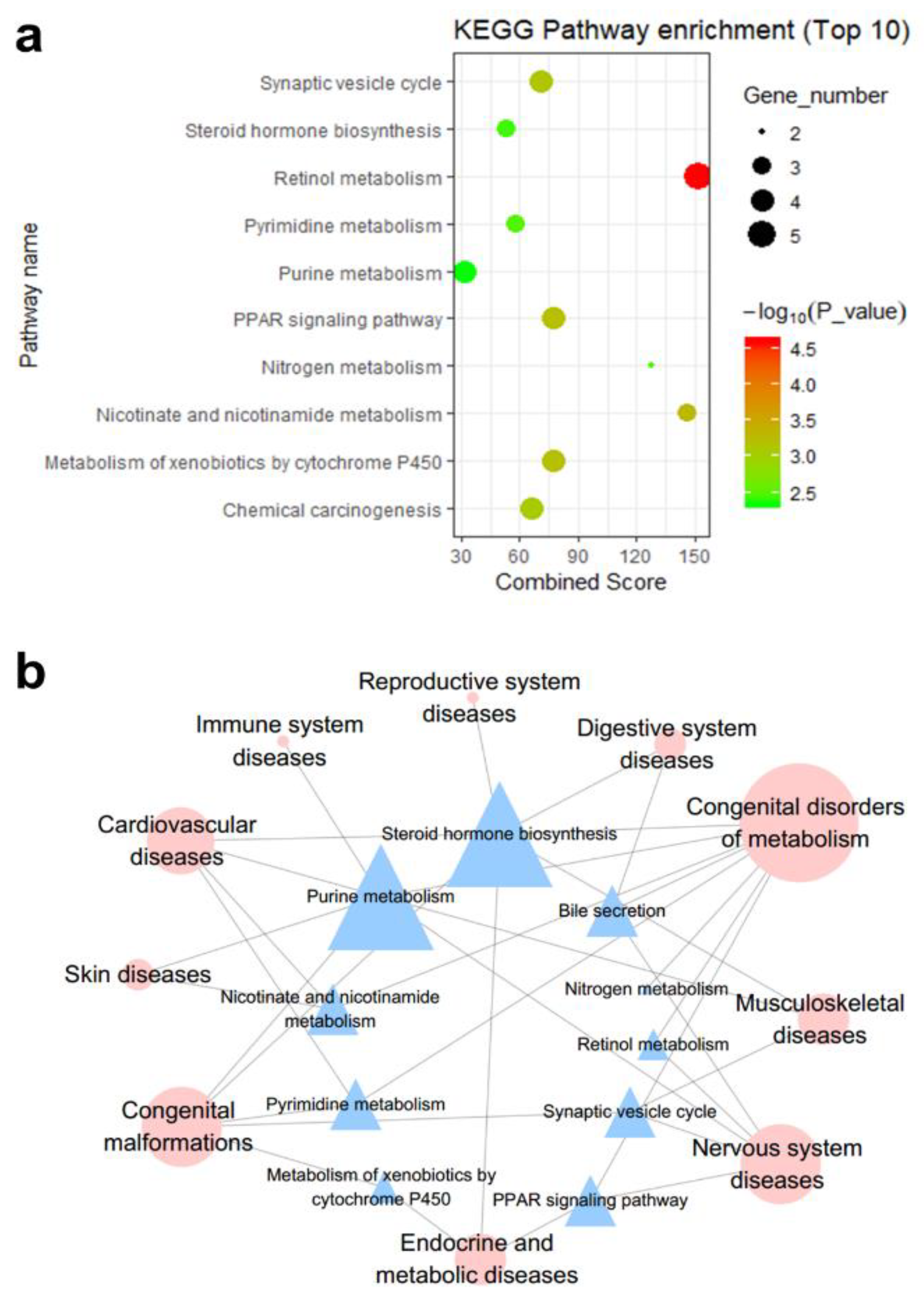
3.2.1. Interpretation by KEGG Pathway Enrichment
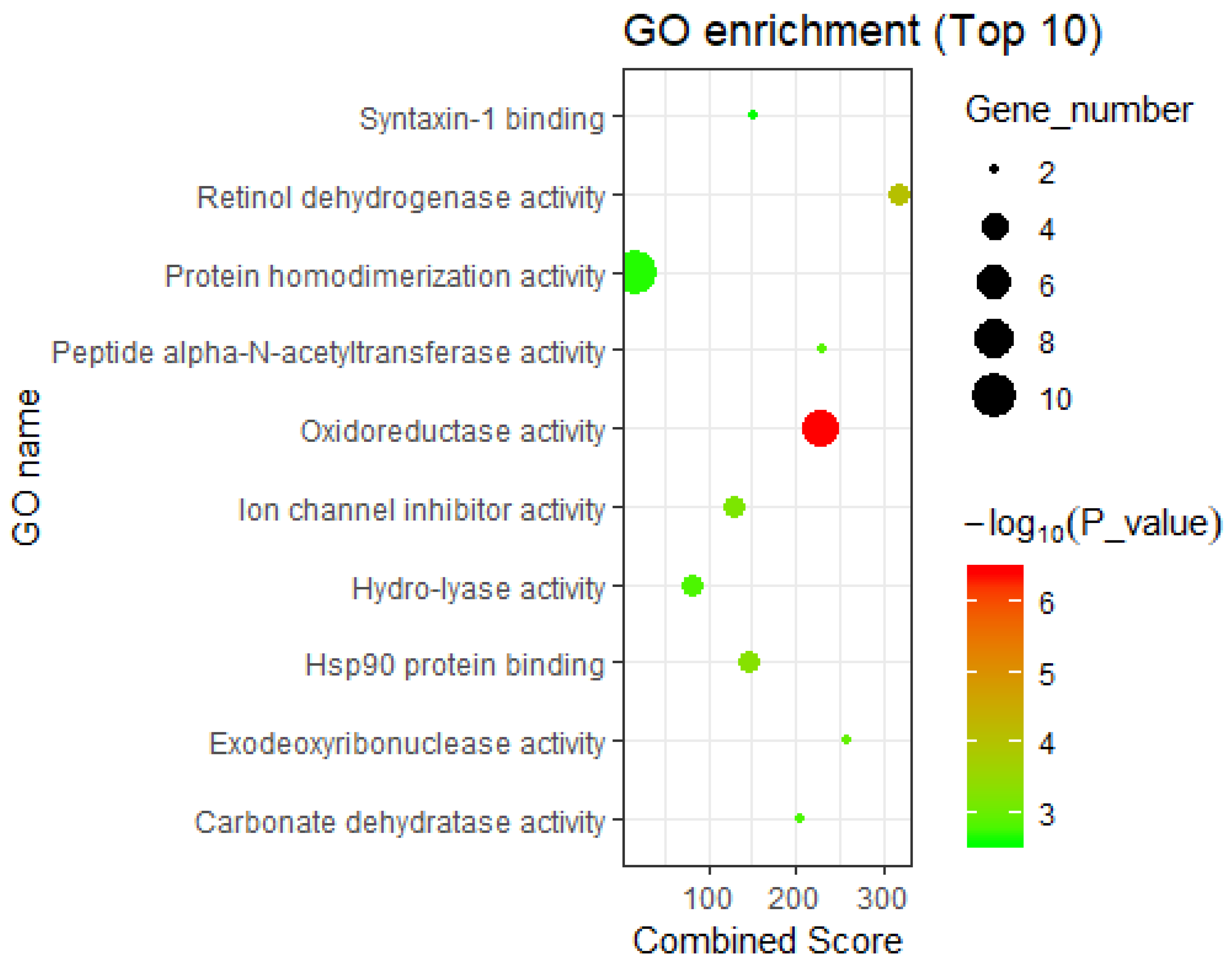
3.2.2. Interpretation by GO Molecular Functional Enrichment
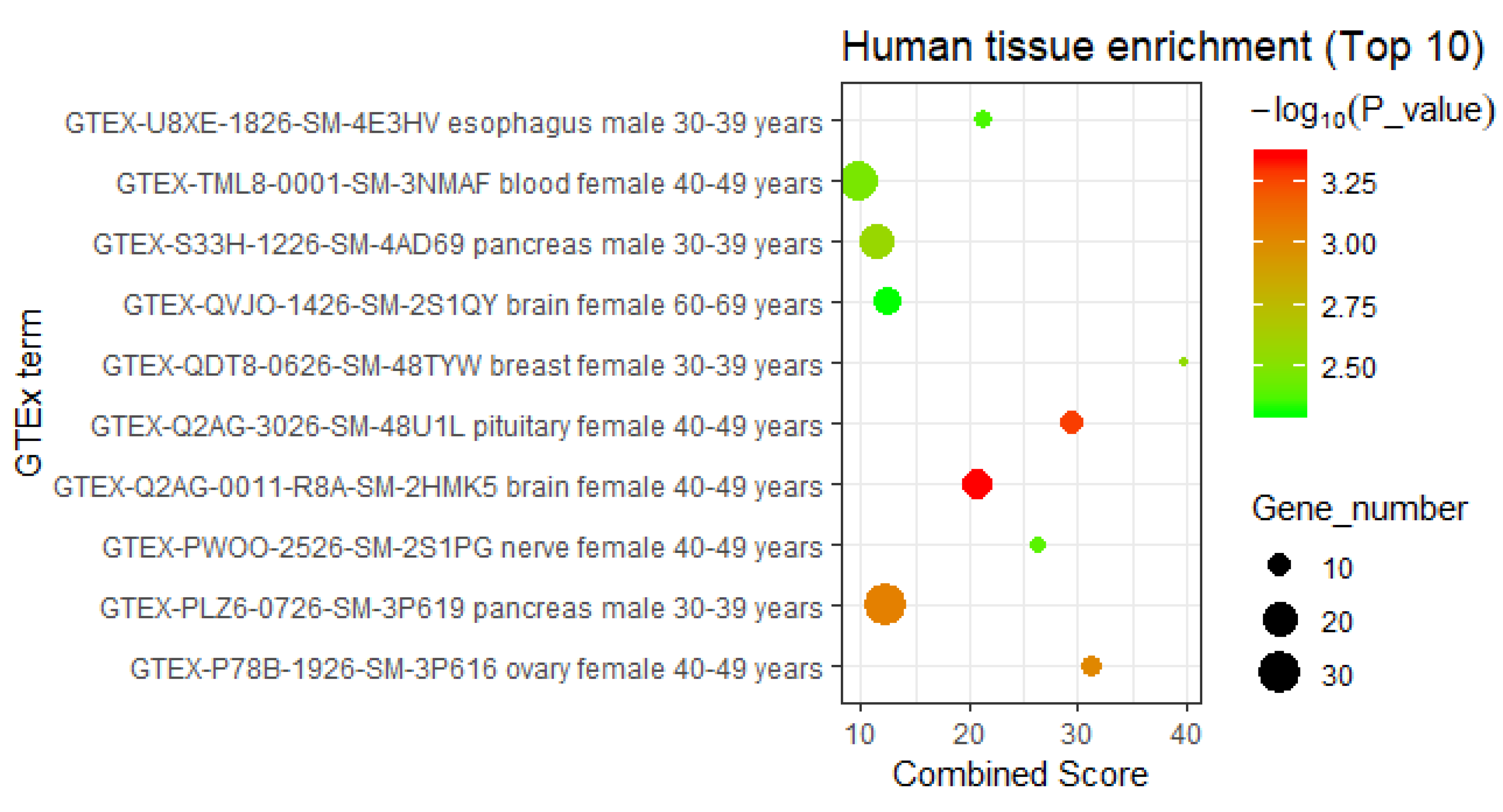
3.2.3. Interpretation by Tissue-Specific Expression Analysis
3.3. Interpretation of the Disease Intervention Mechanism Based on Network Similarity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Calder, P.C. Mechanisms of action of (n-3) fatty acids. J. Nutr. 2012, 142, 592–599. [Google Scholar] [CrossRef]
- Cederholm, T.; Salem, N.; Palmblad, J. ω-3 fatty acids in the prevention of cognitive decline in humans. Adv. Nutr. 2013, 4, 672–676. [Google Scholar] [CrossRef]
- SanGiovanni, J.P.; Chew, E.Y. The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog. Retin Eye Res. 2005, 24, 87–138. [Google Scholar] [CrossRef]
- Pereira, H.; Barreira, L.; Figueiredo, F.; Custódio, L.; Vizetto-Duarte, C.; Polo, C.; Varela, J. Polyunsaturated fatty acids of marine macroalgae: Potential for nutritional and pharmaceutical applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.M.; Burdge, G. Long-chain n-3 PUFA: Plant v. marine sources. Proc. Nutr. Soc. 2006, 65, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Keum, Y.S. Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance—A review. Life Sci. 2018, 203, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, F.S.; Wang, F.; Sinclair, A.J.; Elliott, G.; Turchini, G.M. How does high DHA fish oil affect health? A systematic review of evidence. Crit. Rev. Food Sci. Nutr. 2019, 59, 1684–1727. [Google Scholar] [CrossRef]
- Abdelhamid, A.S.; Brown, T.J.; Brainard, J.S.; Biswas, P.; Thorpe, G.C.; Moore, H.J. Omega-3 fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Database Syst. Rev. 2018, 11, CD003177. [Google Scholar]
- Aung, T.; Halsey, J.; Kromhout, D.; Gerstein, H.C.; Marchioli, R.; Tavazzi, L.; Clarke, R. Associations of omega-3 fatty acid supplement use with cardiovascular disease risks: Meta-analysis of 10 trials involving 77 917 individuals. JAMA Cardiol. 2018, 3, 225–234. [Google Scholar] [CrossRef]
- Kwak, S.M.; Myung, S.K.; Lee, Y.J.; Seo, H.G. Efficacy of omega-3 fatty acid supplements (eicosapentaenoic acid and docosahexaenoic acid) in the secondary prevention of cardiovascular disease: A meta-analysis of randomized, double-blind, placebo-controlled trials. Arch. Intern. Med. 2012, 172, 686–694. [Google Scholar]
- Billman, G.E. The effects of omega-3 polyunsaturated fatty acids on cardiac rhythm: A critical reassessment. Pharmacol. Ther. 2013, 140, 53–80. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, S.; Jun, M.; Sullivan, D.R.; Perkovic, V.; Neal, B. Omega 3 fatty acids and cardiovascular outcomes: Systematic review and meta-analysis. Circ.-Cardiovasc. Qual. Outcomes 2012, 5, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Mazereeuw, G.; Lanctôt, K.L.; Chau, S.A.; Swardfager, W.; Herrmann, N. Effects of ω-3 fatty acids on cognitive performance: A meta-analysis. Neurobiol. Aging 2012, 33, 1482.e17–1482.e29. [Google Scholar] [CrossRef]
- Chew, E.Y.; Clemons, T.E.; Agron, E.; Launer, L.J.; Grodstein, F.; Bernstein, P.S. Effect of omega-3 fatty acids, lutein/zeaxanthin, or other nutrient supplementation on cognitive function: The AREDS2 randomized clinical trial. JAMA 2015, 314, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.C.; Holroyd-Leduc, J.M.; Poulin, M.J.; Hogan, D.B. Effect of nutrients, dietary supplements and vitamins on cognition: A systematic review and meta-analysis of randomized controlled trials. Can. Geriatr. J. 2015, 18, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Daviglus, M.L.; Bell, C.C.; Berrettini, W.; Bowen, P.E.; Conolly, E.S.; Cox, N.J.; Trevisan, M. National institutes of health state-of-the-science conference statement: Preventing Alzheimer disease and cognitive decline. Ann. Intern. Med. 2010, 153, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef]
- Leiserson, M.D.; Vandin, F.; Wu, H.T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Raphael, B.J. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Golub, T.R. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Xiong, M.; Li, B.; Zhu, Q.; Wang, Y.X.; Zhang, H.Y. Identification of transcription factors for drug-associated gene modules and biomedical implications. Bioinformatics 2013, 30, 305–309. [Google Scholar] [CrossRef]
- Huang, D.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.Q. KEGG pathway database. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Maayan, A. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Manley, B.J.; Makrides, M.; Collins, C.T.; Mcphee, A.J.; Gibson, R.A.; Ryan, P.; Davis, P.G. High-dose docosahexaenoic acid supplementation of preterm infants: Respiratory and allergy outcomes. Pediatrics 2011, 128, e71–e77. [Google Scholar] [CrossRef] [PubMed]
- Helland, I.B.; Smith, L.; Saarem, K.; Saugstad, O.D.; Drevon, C.A. Maternal supplementation with very-long-chain n-3 fatty acids during pregnancy and lactation augments children’s IQ at 4 years of age. Pediatrics 2003, 111, e39–e44. [Google Scholar] [CrossRef]
- Lauritzen, L.; Jorgensen, M.H.; Olsen, S.F.; Straarup, E.M.; Michaelsen, K.F. Maternal fish oil supplementation in lactation: Effect on developmental outcome in breast-fed infants. Reprod. Nutr. Dev. 2005, 45, 535–547. [Google Scholar] [CrossRef]
- Dunstan, J.; Simmer, K.; Dixon, G.; Prescott, S.L. Cognitive assessment of children at age 2(1/2) years after maternal fish oil supplementation in pregnancy: A randomised controlled trial. Arch. Dis. Child.-Fetal Neonatal Ed. 2008, 93, F45–F50. [Google Scholar] [CrossRef]
- Lee, L.K.; Shahar, S.; Chin, A.V.; Yusoff, N.A.M. Docosahexaenoic acid-concentrated fish oil supplementation in subjects with mild cognitive impairment (MCI): A 12-month randomised, double-blind, placebo-controlled trial. Psychopharmacology 2013, 225, 605–612. [Google Scholar] [CrossRef]
- Herassandoval, D.; Pedrazachaverri, J.; Perezrojas, J.M. Role of docosahexaenoic acid in the modulation of glial cells in Alzheimer’s disease. J. Neuroinflamm. 2016, 13, 61. [Google Scholar] [CrossRef]
- Stonehouse, W.; Conlon, C.A.; Podd, J.; Hill, S.; Minihane, A.M.; Haskell, C.F.; Kennedy, D.O. DHA supplementation improved both memory and reaction time in healthy young adults: A randomized controlled trial. Am. J. Clin. Nutr. 2013, 97, 1134–1143. [Google Scholar] [CrossRef]
- Levant, B.; Ozias, M.K.; Davis, P.F.; Winter, M.; Russell, K.L.; Carlson, S.E.; Reed, G.A.; McCarson, K.E. Decreased brain docosahexaenoic acid content produces neurobiological effects associated with depression: Interactions with reproductive status in female rats. Psychoneuroendocrinology 2008, 33, 1279–1292. [Google Scholar] [CrossRef]
- Jiang, L.H.; Liang, Q.Y.; Shi, Y. Pure docosahexaenoic acid can improve depression behaviors and affect hpa axis in mice. Eur. Rev. Med. Pharmacol. Sci. 2012, 16, 1765–1773. [Google Scholar]
- Mcmanus, S.A.; Tejera, N.; Awwad, K.; Vauzour, D.; Rigby, N.M.; Fleming, I.; Minihane, A.M. Differential effects of EPA versus DHA on postprandial vascular function and the plasma oxylipin profile in men. J. Lipid Res. 2016, 57, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K. Docosahexaenoic acid regulates vascular endothelial cell function and prevents cardiovascular disease. Lipids Health Dis. 2017, 16, 118. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Kim, H.; Chiba, H.; Matsumoto, A. Anti-obesity effect of fish oil and fish oil-fenofibrate combination in female KK mice. J. Atheroscler. Thromb. 2009, 16, 674–683. [Google Scholar] [CrossRef]
- Newell, M.; Baker, K.; Postovit, L.M.; Field, C.J. A critical review on the effect of docosahexaenoic acid (DHA) on cancer cell cycle progression. Int. J. Mol. Sci. 2017, 18, 1784. [Google Scholar] [CrossRef]
- Ma, Y.; Belyaeva, O.V.; Brown, P.M.; Fujita, K.; Valles, K.; Karki, S.; Rotman, Y. 17-β hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology 2019, 69, 1504–1519. [Google Scholar] [CrossRef]
- Li, X.T.; Li, Y.S.; Shi, Z.Y.; Guo, X.L. New insights into molecular chaperone TRAP1 as a feasible target for future cancer treatments. Life Sci. 2020, 254, 117737. [Google Scholar] [CrossRef]
- Quan, Y.; Luo, Z.H.; Yang, Q.Y.; Li, J.; Zhu, Q.; Liu, Y.M.; Zhang, H.Y. Systems chemical genetics-based drug discovery: Prioritizing agents targeting multiple/reliable disease-associated genes as drug candidates. Front. Genet. 2019, 10, 474. [Google Scholar] [CrossRef]

| Agent | p-Value a | Agent | p-Value a |
|---|---|---|---|
| Propafenone | 3.59 × 10−6 | Iloprost | 2.08 × 10−4 |
| Clopamide | 6.47 × 10−6 | Alclometasone | 2.56 × 10−4 |
| Fenbufen | 4.89 × 10−5 | Lomustine | 3.40 × 10−4 |
| Etanidazole | 1.00 × 10−4 | Rescinnamine | 3.51 × 10−4 |
| Thiethylperazine | 1.22 × 10−4 | Benzocaine | 4.70 × 10−4 |
| Phenelzine | 1.25 × 10−4 | Proguanil | 5.11 × 10−4 |
| Zalcitabine | 1.32 × 10−4 | Meclocycline | 5.61 × 10−4 |
| Dydrogesterone | 1.62 × 10−4 | Diethylstilbestrol | 7.35 × 10−4 |
| Leflunomide | 1.64 × 10−4 | Midecamycin | 9.59 × 10−4 |
| Albendazole | 1.85 × 10−4 | Pyrvinium | 1.07 × 10−3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, Y.; Zhang, H.-Y.; Xiong, J.-H.; Xu, R.-F.; Gao, M. Heat Diffusion Kernel Algorithm-Based Interpretation of the Disease Intervention Mechanism for DHA. Genes 2020, 11, 754. https://doi.org/10.3390/genes11070754
Quan Y, Zhang H-Y, Xiong J-H, Xu R-F, Gao M. Heat Diffusion Kernel Algorithm-Based Interpretation of the Disease Intervention Mechanism for DHA. Genes. 2020; 11(7):754. https://doi.org/10.3390/genes11070754
Chicago/Turabian StyleQuan, Yuan, Hong-Yu Zhang, Jiang-Hui Xiong, Rui-Feng Xu, and Min Gao. 2020. "Heat Diffusion Kernel Algorithm-Based Interpretation of the Disease Intervention Mechanism for DHA" Genes 11, no. 7: 754. https://doi.org/10.3390/genes11070754
APA StyleQuan, Y., Zhang, H.-Y., Xiong, J.-H., Xu, R.-F., & Gao, M. (2020). Heat Diffusion Kernel Algorithm-Based Interpretation of the Disease Intervention Mechanism for DHA. Genes, 11(7), 754. https://doi.org/10.3390/genes11070754







