Werewolf, There Wolf: Variants in Hairless Associated with Hypotrichia and Roaning in the Lykoi Cat Breed
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Lykoi Samples
2.3. Whole Genome and Variant Calling
2.4. Hairless (HR) Genotyping and Sequencing
3. Results
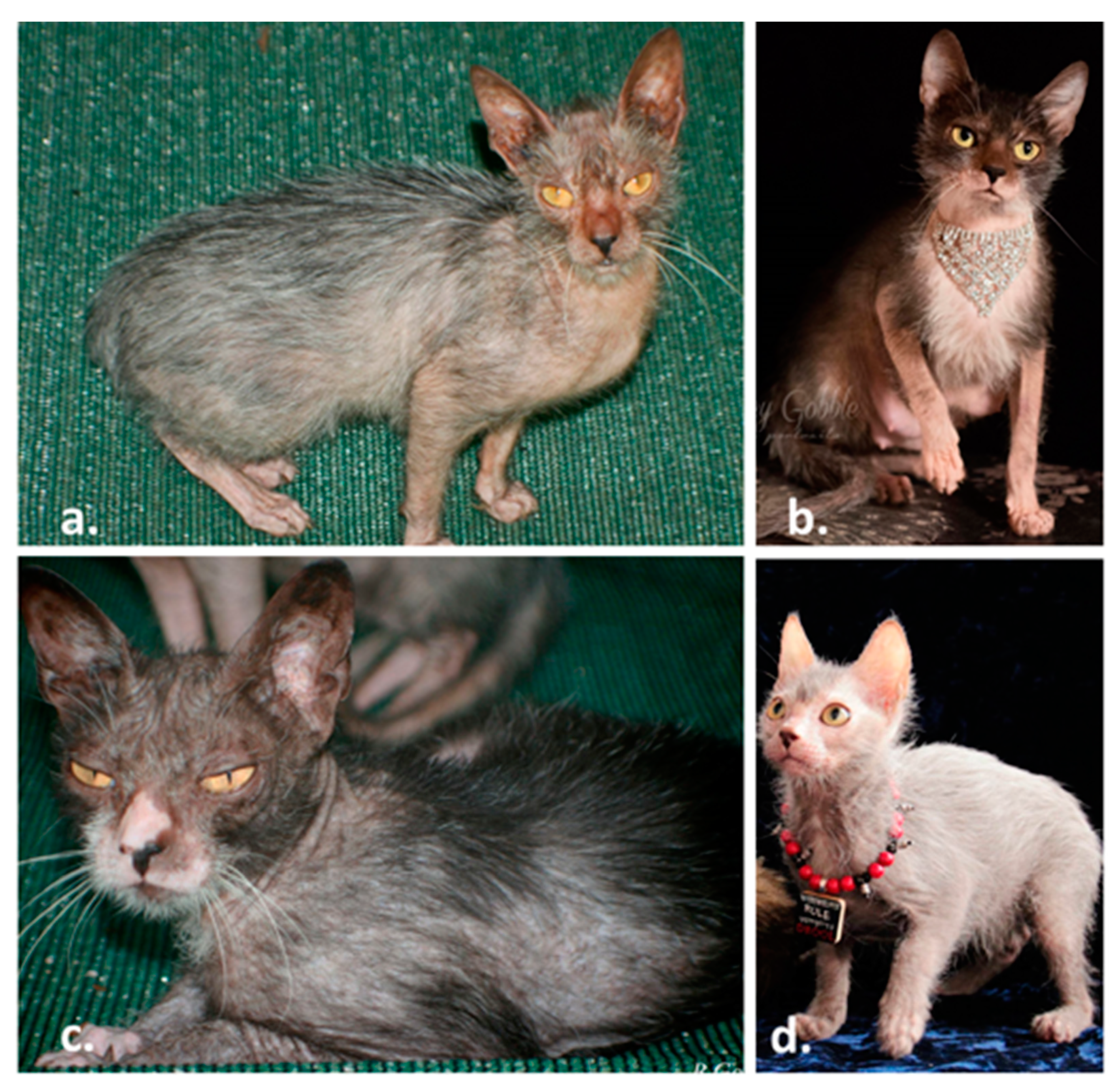
3.1. Lykoi Samples
3.2. Whole Genome Sequencing
3.3. Lykoi Variants in Other Lineages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
- Leslie A. Lyons; Reuben M. Buckley; Department of Veterinary Medicine and Surgery, College of Veterinary Medicine, University of Missouri, Columbia, Missouri, 65211 USA
- Danielle Aberdein; Dorian J.; John S. Munday; Garrick;School of Veterinary Science, Massey University, Palmerston North 4442 New Zealand
- Paulo C. Alves; CIBIO/InBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos/InBIO Associate Lab & Faculdade de Ciências, Universidade do Porto, Campus e Vairão, 4485–661 Vila do Conde, Portugal
- Paulo C. Alves; Wildlife Biology Program, University of Montana, Missoula, Montana, 59812 USA
- Gregory S. Barsh; Christopher B. Kaelin; HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, 35806 USA
- Gregory S. Barsh; Christopher B. Kaelin; Department of Genetics, Stanford University, Stanford, California, 94305 USA
- Rebecca R. Bellone; Veterinary Genetics Laboratory, University of California, Davis, Davis California, 95616 USA
- Tomas F. Bergström; Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, Uppsala, 750 07, Sweden
- Adam R. Boyko; Department of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, New York, 14853 USA
- Jeffrey A. Brockman; Hill’s Pet Nutrition Inc., Topeka, Kansas 66601 USA
- Margret L. Casal; Reproduction, and Pediatrics, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA 19104 USA
- Marta G. Castelhano; Rory J. Todhunter; Elizabeth A. Wilcox; Department of Clinical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, New York, 14853 USA
- sup>∙ Ottmar Distl; Institute for Animal Breeding and Genetics, University of Veterinary Medicine, Hannover, 30559, Hannover, Germany
- Nicholas H. Dodman; Department of Clinical Sciences, Cummings School of Veterinary Medicine, Tufts University, Grafton, MA, 01536 USA
- N. Matthew Ellinwood; Dorian J. Garrick; Max F. Rothschild; Department of Animal Science, College of Agriculture and Life Sciences, Iowa State University, Ames, Iowa, 50011 USA
- Jonathan E. Fogle; College of Veterinary Medicine, North Carolina State University, Raleigh, NC 27607
- Oliver P. Forman; WALTHAM Centre for Pet Nutrition, Freeby Lane, Waltham on the Wolds, Leicestershire, LE14 4RT UK
- Edward I. Ginns; Department of Psychiatry, University of Massachusetts Medical School, Worcester, MA, 01655 USA
- Jens Häggström; Department of Clinical Sciences, Faculty of Veterinary Medicine and Animal Science, Swedish University of Agricultural Sciences, Uppsala, SE-750 07 Sweden
- Robert J. Harvey; School of Health and Sport Sciences, University of the Sunshine Coast, Sippy Downs, QLD, 4558, Australia
- Daisuke Hasegawa; Yoshihiko Yu; Department of Clinical Veterinary Medicine, Nippon Veterinary and Life Science University, Tokyo 180-8602 Japan
- sup>∙ Bianca Haase; Sydney School of Veterinary Science, Faculty of Science, University of Sydney, Sydney, NSW, 2006, Australia
- sup>∙ Christopher R. Helps; Langford Vets, University of Bristol, Langford, Bristol, BS40 5DU UK
- sup>∙ Isabel Hernandez; Pediatrics and Medical Genetics Service, College of Veterinary Medicine, Cornell University, Ithaca, New York, 14853 USA
- Marjo K. Hytönen; Maria Kaukonen; Hannes Lohi; Department of Veterinary Biosciences; Department of Medical Genetics, University of Helsinki and Folkhälsan Research Center, Helsinki 00014 Finland
- Tomoki Kosho; Department of Medical Genetics, Center for Medical Genetics, Shinshu University Hospital, Matsumoto, Nagano 390-8621, Japan
- Emilie Leclerc; SPF - Diana Pet food – Symrise Group – 56250 Elven, France
- Teri L. Lear; Department of Veterinary Science, University of Kentucky - Lexington, Lexington, KY, 40506 USA (In memoriam)
- Tosso Leeb; Vetsuisse Faculty, Institute of Genetics, University of Bern, 3001 Bern, Switzerland
- Ronald H.L. Li; Department of Surgical and Radiological Sciences, School of Veterinary Medicine, University of California Davis, One Shields Ave, Davis, CA, 95616 USA
- Maria Longeri; Dipartimento di Medicina Veterinaria, University of Milan, 20122 Milan, Italy
- Mark A. Magnuson; Departments of Molecular Physiology and Biophysics, Cell and Developmental Biology, and Medicine, Vanderbilt University, School of Medicine, Nashville, Tennessee, 37232 USA
- Richard Malik; Centre for Veterinary Education, University of Sydney, Sydney, NSW, 2006 Australia
- Shrinivasrao P. Mane; Elanco Animal Health, Greenfield, IN 46140 USA
- William J. Murphy; Department of Veterinary Integrative Biosciences, College of Veterinary Medicine, Texas A&M University, College Station, Texas, 77845 USA
- Niels C. Pedersen; Joshua A. Stern; Department of Medicine and Epidemiology, School of Veterinary Medicine, University of California at Davis, Davis, California, 95616 USA
- Simon M. Peterson-Jones; Department of Small Animal Clinical Sciences, Veterinary Medical Center, Michigan State University, East Lansing, Michigan, 48824, USA
- Clare Rusbridge; School of Veterinary Medicine, Faculty of Health & Medical Sciences, Univesity of Surrey, Guildford, Surrey, GU2 7AL, United Kingdom
- Beth Shapiro; Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, Santa Cruz, California 95064 USA
- William F. Swanson; Center for Conservation and Research of Endangered Wildlife (CREW), Cincinnati Zoo & Botanical Garden, Cincinnati, Ohio, 45220 USA
- Karen A. Terio; Zoological Pathology Program, University of Illinois, Brookfield, Illinois 60513 USA
- Wesley C. Warren; Division of Animal Sciences, College of Agriculture, Food and Natural Resources; School of Medicine, University of Missouri, Columbia, Missouri 65211, USA
- Julia H. Wildschutte; Bowling Green State University, Department of Biological Sciences, Bowling Green, Ohio 43403 USA
Conflicts of Interest
Ethical Approval
References
- Crystal Palace—Summer Concert Today Cat Show on July 13. Penny Illustrated Paper and Illustrated Times, Amusement; London, England, 1871; Volume 510, p. 11. Available online: https://www.britishnewspaperarchive.co.uk/titles/penny-illustrated-paper (accessed on 8 June 2020).
- The First Cat Show in America; New York Times: New York, NY, USA, 1881.
- Morris, D. Cat Breeds of the World; Penguin Books: New York, NY, USA, 1999. [Google Scholar]
- Lipinski, M.J.; Froenicke, L.; Baysac, K.C.; Billings, N.C.; Leutenegger, C.M.; Levy, A.M.; Longeri, M.; Niini, T.; Ozpinar, H.; Slater, M.R.; et al. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics 2008, 91, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Kurushima, J.D.; Lipinski, M.J.; Gandolfi, B.; Froenicke, L.; Grahn, J.C.; Grahn, R.A.; Lyons, L.A. Variation of cats under domestication: Genetic assignment of domestic cats to breeds and worldwide random-bred populations. Anim. Genet. 2013, 44, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Animals (OMIA). Available online: https://omia.org/ (accessed on 8 June 2020).
- Lenffer, J.; Nicholas, F.W.; Castle, K.; Rao, A.; Gregory, S.; Poidinger, M.; Mailman, M.D. OMIA (Online Mendelian Inheritance in Animals): An enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 2006, 34, D599–D601. [Google Scholar] [CrossRef]
- Gandolfi, B.; Alhaddad, H.; Affolter, V.K.; Brockman, J.; Haggstrom, J.; Joslin, S.E.K.; Koehne, A.L.; Mullikin, J.C.; Outerbridge, C.A.; Warren, W.C.; et al. To the root of the curl: A signature of a recent selective sweep identifies a mutation that defines the Cornish rex cat breed. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Outerbridge, C.A.; Beresford, L.G.; Myers, J.A.; Pimentel, M.; Alhaddad, H.; Grahn, J.C.; Grahn, R.A.; Lyons, L.A. The naked truth: Sphynx and Devon rex cat breed mutations in KRT71. Mamm. Genome 2010, 21, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Filler, S.; Alhaddad, H.; Gandolfi, B.; Kurushima, J.D.; Cortes, A.; Veit, C.; Lyons, L.A.; Brem, G. Selkirk rex: Morphological and genetic characterization of a new cat breed. J. Hered. 2012, 103, 727–733. [Google Scholar] [CrossRef]
- Gandolfi, B.; Alhaddad, H.; Joslin, S.E.K.; Khan, R.; Filler, S.; Brem, G.; Lyons, L.A. A splice variant in KRT71 is associated with curly coat phenotype of Selkirk rex cats. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Irvine, A.D.; McLean, W.H. Human keratin diseases: The increasing spectrum of disease and subtlety of the phenotype-genotype correlation. Br. J. Dermatol. 1999, 140, 815–828. [Google Scholar] [CrossRef]
- Smith, F.J.D. The molecular genetics of keratin disorders. Am. J. Clin Dermatol. 2003, 4, 347–364. [Google Scholar] [CrossRef]
- Visinoni, A.F.; Lisboa-Costa, T.; Pagnan, N.A.B.; Chautard-Freire-Maia, E.A. Ectodermal dysplasias: Clinical and molecular review. Am. J. Med. Genet. 2009, 149A, 1980–2002. [Google Scholar] [CrossRef]
- Wright, J.T.; Fete, M.; Schenider, H.; Zinser, M.; Koster, M.I.; Clarke, A.J.; Hadj-Rabia, S.; Tadini, G.; Pagnan, N.; Visinoni, A.F.; et al. Ectodermal dysplasias: Classification and organization by phenotype, genotype and molecular pathway. Am. J. Med. Genet. 2019, 179, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Ratterree, M.S.; Panteleyev, A.A.; Aita, V.M.; Sundberg, J.P.; Christiano, A.M. Atrichia with papular lesions resulting from mutations in the rhesus macaque (Macaca mulattta) hairless gene. Lab. Anim. 2002, 36, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Finocchiaro, R.; Portolan, B.; Damiani, G.; Caroli, A.; Budelli, E.; Bolla, P.; Pagnacco, G. The hairless (hr) gene is involved in the congenital hypotrichosis of Valle del Belice sheep. Genet. Sel. Evol. 2003, 35, S147–S156. [Google Scholar] [CrossRef]
- Barlund, C.S.; Clark, E.G.; Leeb, T.; Drogemuller, C.; Palmer, C.W. Congenital hypotrichosis and partial anodontia in a crossbreed beef calf. Can. Vet. J. 2007, 48, 612–614. [Google Scholar] [PubMed]
- Drogemuller, C.; Karlsson, E.K.; Hytonen, M.K.; Perloski, M.; Dolf, G.; Saino, K.; Lohi, H.; Lindblad-Toh, K.; Leeb, T. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science 2008, 321, 1462. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Z.; Xu, S.; Zhou, K.; Yang, G. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol. Biol. 2013, 13, 34. [Google Scholar] [CrossRef]
- Parker, H.G.; Harris, A.; Dreger, D.L.; Davis, B.W.; Ostrander, E.A. The bald and the beautiful: Hairlessness in domestic dog breeds. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Hadji-Rasouliha, S.; Bauer, A.; Dettwiler, M.; Welle, M.M.; Leeb, T. A frameshift variant in the EDA gene in dachshunds with X-linked hypohidrotic ectodermal dysplasia. Anim. Genet. 2018, 49, 651–654. [Google Scholar] [CrossRef]
- Abitbol, M.; Bosse, P.; Thomas, A.; Tiret, L. A deletion in FOXN1 is associated with a syndrome characterized by congenital hypotrichosis and short life expectancy in Birman cats. PLoS ONE 2015, 10, e0120668. [Google Scholar] [CrossRef]
- David, V.A.; Menotti-Raymond, M.; Wallace, A.C.; Roelke, M.; Leighty, R.; Eizirik, E.; Hannah, S.S.; Nelson, G.; Schäffer, A.A.; Connelly, C.J.; et al. Endongenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 Genes Genomes Genet. 2014, 4, 1881–1891. [Google Scholar] [CrossRef]
- Lyons, L.A.; Imes, D.I.; Rah, H.C.; Grahn, R.A. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus). Anim. Genet. 2005, 36, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Imes, D.I.; Geary, L.A.; Grahn, R.A.; Lyons, L.A. Albinism in the domestic cat (Felis catus) is associated with a tyrosinase (TYR) mutation. Anim. Genet. 2006, 37, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Creel, D.; Hendrickson, A.E.; Leventhal, A.G. Retinal projections in tyrosinase-negative albino cats. J. Neurosci. 1982, 2, 907–911. [Google Scholar] [CrossRef]
- LeRoy, M.L.; Senter, D.A.; Kim, D.Y.; Gandolfi, B.; Middleton, J.R.; Bouhan, D.M.; Lyons, L.A. Clinical and histologic description of lykoi cat hair coat and skin. Jpn. J. Vet. Dermatol. 2016, 22, 179–191. [Google Scholar] [CrossRef][Green Version]
- Gobble, J.; Gobble, B. The Lykoi Cat. Available online: http://lykoikitten.com/history/ (accessed on 5 May 2017).
- Lyons, L.A.; Creighton, E.K.; Alhaddad, H.; Beale, H.C.; Grahn, R.A.; Rah, H.; Maggs, D.J.; Helps, C.R.; Gandolfi, B. Whole genome sequencing in cats, identifies new models for blindness in AIPL1 and somite segmentation in HES7. BMC Genom. 2016, 17, 265. [Google Scholar] [CrossRef]
- Mauler, D.A.; Gandolfi, B.; Rineiro, C.R.; O’Brien, D.P.; Spooner, J.L.; Lyons, L.A. Precision medicine in cats: Novel Niemann-Pick type C1 diagnosed by whole-genome sequencing. J. Vet. Intern. Med. 2017, 31, 539–544. [Google Scholar] [CrossRef]
- Oh, A.; Pearce, J.W.; Gandolfi, B.; Creighton, E.K.; Suedmeyer, W.K.; Selig, M.M.; Bosiack, A.P.; Castaner, L.J.; Whiting, R.E.H.; Belknap, E.B.; et al. Early-onset progressive retinal atrophy associated with an IQCB1 variant in African Black-Footed Cats (Felis nigripes). Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Buckley, R.M.; Grahn, R.A.; Gandolfi, B.; Herrick, J.R.; Kittleson, M.D.; Bateman, H.L.; Newsom, J.; Swanson, W.F.; Prieur, D.J.; Lyons, L.A. Assisted reproduction mediated resurrection of a feline model for Chediak-Higashi syndrome caused by a large duplication in LYST. Sci. Rep. 2020, 10, 64. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular Weight DNA from Mammalian Cells Using Proteinase K and Phenol. In Molecular Cloning: A Laboratory Manual, 4th ed.; Green, M.R., Sambrook, J., Eds.; Cold Spring Laboratory Press: Cold Spring Harbor, NY, USA, 2012; pp. 13.47–13.53. [Google Scholar]
- Lipinski, M.J.; Amigues, Y.; Blasi, M.; Broad, T.E.; Cherbonnel, C.; Cho, G.J.; Corley, S.; Daftari, P.; Delattre, D.R.; Dileanis, S.; et al. An international parentage and identification panel for the domestic cat (Felis catus). Anim. Genet. 2007, 38, 371–377. [Google Scholar] [CrossRef]
- Toonen, R.J.; Hughes, S. Increased throughput for fragment analysis on an ABI Prism 377 automated sequencer using a membrane comb and STRand software. Biotechniques 2001, 31, 1320–1324. [Google Scholar]
- Buckley, R.M.; Davis, B.W.; Brashear, W.A.; Farias, F.H.; Kuroki, K.; Graves, T.; Hillier, L.W.; Kremitzki, M.; Li, G.; Middleton, R. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. bioRxiv 2020. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.; Fennell, T.; Carneiro, M.; Van der Auwera, G.; Kling, D.; Gauthier, L.; Levy-Moonshine, A.; Roazen, D. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2017. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Daniel, R.J.; O’Brien, D.P.; Guo, L.T.; Youngs, M.D.; Leach, S.B.; Jones, B.R.; Shelton, G.D.; Lyons, L.A. A novel mutation in CLCN1 associated with feline myotonia congenita. PLoS ONE 2014, 9, e109926. [Google Scholar] [CrossRef] [PubMed]
- Cachon-Gonzalez, M.B.; Fenner, S.; Coffin, J.M.; Moran, C.; Best, S.; Stoye, J.P. Structure and expression of the hairless gene of mice. Proc. Natl. Acad. Sci. USA 1994, 91, 7717–7721. [Google Scholar] [CrossRef]
- Ahmad, W.; ul Haque, M.F.; Brancolini, V.; Tsou, H.C.; ul Haque, S.; Lam, H.; Aita, V.M.; Owen, J.; de Blaquiere, M.; Frank, J.; et al. Alopecia universalis associated with a mutation in the human hairless gene. Science 1998, 279, 720–724. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; David, V.A.; Weir, B.S.; O’Brien, S.J. A population genetic database of cat breeds developed in coordination with a domestic cat STR multiplex. J. Forensic Sci. 2012, 57, 596–601. [Google Scholar] [CrossRef]
- Robinson, R. Devon rex—A third rexoid coat mutation in the cat. Genetica 1969, 40, 597–599. [Google Scholar] [CrossRef]
- Searle, A.G.; Jude, A.C. The ‘rex’ type coat in the domestic cat. J. Genet. 1956, 54, 506–512. [Google Scholar] [CrossRef]
- TICA. The International Cat Association. Available online: https://tica.org/ (accessed on 1 May 2017).
- Bult, C.J.; Eppig, J.T.; Kadin, J.A.; Richardson, J.E.; Blake, J.A.; Group, M.G.D. The mouse genome database (MGD): Mouse biology and model systems. Nucleic Acids Res. 2008, 36, D724–D728. [Google Scholar] [CrossRef] [PubMed]
- Sumner, F.B. Hairless mice. J. Hered. 1924, 15, 475–481. [Google Scholar] [CrossRef]
- Brooke, H.C. Hairless mice. J. Hered. 1926, 17, 173–174. [Google Scholar] [CrossRef]
- Stoye, J.P.; Fenner, S.; Greenoak, G.E.; Moran, C.; Coffin, J.M. Role of endogenous retroviruses as mutagens: The hairless mutation of mice. Cell 1988, 54, 383–391. [Google Scholar] [CrossRef]
- Ahmad, W.; Panteleyev, A.A.; Christiano, A.M. The molecular basis of congential atrichia in humans and mice: Mutations in the hairless gene. J. Investig. Dermatol. Symp. Proc. 1999, 4, 240–243. [Google Scholar] [CrossRef]
- Klein, I.; Bergman, R.; Indelman, M.; Sprecher, E. A novel missense mutation affecting the human hairless thyroid receptor interacting domain 2 causes congenital atrichia. J. Investig. Dermatol. 2002, 119, 920–922. [Google Scholar] [CrossRef][Green Version]
- Moraitis, A.N.; Giguere, V. The co-repressor hairless protects ROR-alpha orphan nuclear receptor from proteasome mediated degradation. J. Biol. Chem. 2003, 278, 52511–52518. [Google Scholar] [CrossRef]
- Hsieh, J.-C.; Sisk, J.M.; Jurutka, P.W.; Haussler, C.A.; Slater, S.A.; Haussler, M.R.; Thompson, C.C. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J. Biol. Chem. 2003, 278, 38665–38674. [Google Scholar] [CrossRef]
- Potter, G.B.; Beaudoin III, G.M.J.; DeRenzo, C.L.; Zarach, J.M.; Chen, S.H.; Thompson, C.C. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001, 15, 2687–2701. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Liu, Y.; Xu, Y.; Zhou, Y.; Hua, R.; Wang, K.; Sun, M.; Li, Y.; Yang, S.; Zhang, X.-J.; et al. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat. Genet. 2009, 41, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1076. [Google Scholar] [CrossRef] [PubMed]
- Aita, V.M.; Ahmad, W.; Panteleyev, A.A.; Kozlowska, U.; Kozlowska, A.; Gilliam, T.C.; Jablonska, S.; Christiano, A.M. A novel missense mutation (C622G) in the zinc-finger domain of the human hairless gene associated with congenital atrichia with papular lesions. Exp. Dermatol. 2001, 9, 157–162. [Google Scholar] [CrossRef]
- Hytonen, M.K.; Lohi, H. A frameshift insertion in SGK3 leads to recessive hairlessness in Scottish Deerhounds: A candidate gene for human alopecia conditions. Hum. Genet. 2019, 138, 535–539. [Google Scholar] [CrossRef]
- Parker, H.G.; Whitaker, D.T.; Harris, A.C.; Ostrander, E.A. Whole genome analysis of a single Scottish deerhound family provides independent corroboration that a SGK3 coding variant leads to hairlessness. G3 Genes Genomes Genet. 2020, 10, 293–297. [Google Scholar] [CrossRef]
- Rishikaysh, P.; Dev, K.; Diaz, D.; Qureshi, W.M.; Filip, S.; Mokry, J. Signaling involved in hair follicle morphogenesis and development. Int. J. Mol. Sci. 2014, 15, 1647–1670. [Google Scholar] [CrossRef]
- Mauro, T.M.; McCormick, J.A.; Wang, J.; Boini, K.M.; Ray, L.; Monks, B.; Birnbaum, M.J.; Lang, F.; Pearce, D. Akt2 and SGK3 are both determinants of postnatal hair follicle development. FASEB J. 2009, 23, 3193–3202. [Google Scholar] [CrossRef]
- Beaudoin, G.M., 3rd; Sisk, J.M.; Coulombe, P.A.; Thompson, C.C. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc. Natl. Acad. Sci. USA 2005, 102, 14653–14658. [Google Scholar] [CrossRef]
- Shirokova, V.; Biggs, L.C.; Jussila, M.; Ohyama, T.; Groves, A.K.; Mikkola, M.L. Foxi3 Deficiency compromises hair follicle stem cell specification and activation. Stem Cells 2016, 34, 1896–1908. [Google Scholar] [CrossRef]
- Drogemuller, C.; Rufenacht, S.; Wichert, B.; Leeb, T. Mutations within the FGF5 gene are associated with hair length in cats. Anim. Genet. 2007, 38, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Kehler, J.S.; David, V.A.; Schaffer, A.A.; Bajema, K.; Eizirik, E.; Ryugo, D.K.; Hannah, S.S.; O’Brien, S.J.; Menotti-Raymond, M. Four independent mutations in the feline Fibroblast Growth Factor 5 gene determine the long-haired phenotype in domestic cats. J. Hered. 2007, 98, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, K.J.; McMillin, M.J.; Brassil, M.M.; Shively, K.M.; Magnaye, K.M.; Cortes, A.; Weinmann, A.S.; Lyons, L.A.; Bamshad, M.J. Multiple mutant T alleles cause haploinsufficiency of brachyury and short tails in Manx cats. Mamm. Genome 2013. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, C.B.; Xu, X.; Hong, L.Z.; David, V.A.; McGowan, K.A.; Schmidt-Kuntzel, A.; Roelke, M.E.; Pino, J.; Pontius, J.; Cooper, G.M.; et al. Specifying and sustaining pigmentation patterns in domestic and wild cats. Science 2012, 337, 1536–1541. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Alamri, S.; Darby, W.G.; Adhikari, B.; Lattimer, J.C.; Malik, R.; Wade, C.M.; Lyons, L.A.; Cheng, J.; Bateman, J.F.; et al. A dominant TRPV4 variant underlies osteochondrodysplasia in Scottish fold cats. Osteoarthr. Cartil. 2016, 24, 1441–1450. [Google Scholar] [CrossRef]
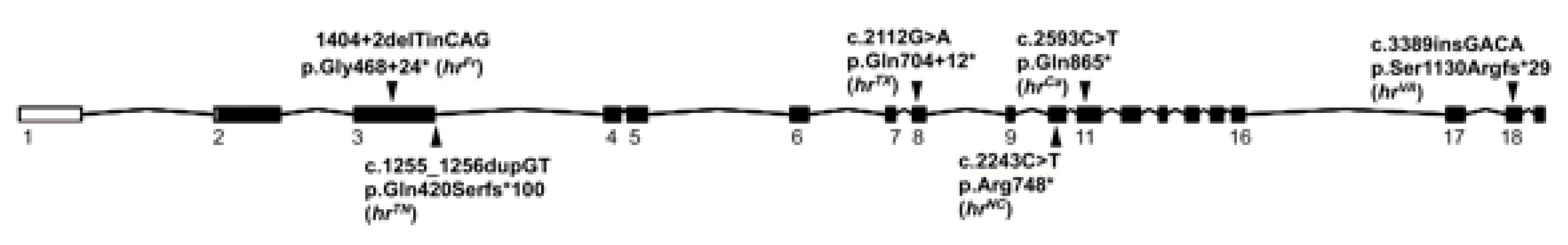
| Founder Lineage | hrTN | hrFr | hrTX | hrNC | hrCa | hrVA | HR Alleles ‡ |
|---|---|---|---|---|---|---|---|
| Exon 3 | Exon 3 | Exon 8 | Exon 10 | Exon 11 | Exon 18 | ||
| c.1255_1256 dupGT | c.1404+2delTinsCAG | c.2112G>A | c.2243 C>T | c.2593 C>T | c.3389ins GACA | ||
| WGS F1 TN/VA * | Wt/dup | G/G | C/C | C/C | Wt/ins | hrTN/hrVA | |
| Tennessee | dup/dup | G/G | C/C | C/C | Wt/Wt | hrTN/hrTN | |
| Virginia | Wt/Wt | G/G | C/C | C/C | ins/ins | hrVA/hrVA | |
| Missouri | dup/dup | G/G | C/C | Wt/Wt | hrTN/hrTN | ||
| France L10, L4 | Wt/Wt | CAG/CAG | G/G | C/C | C/C | Wt/Wt | hrFr/hrFr |
| Texas | Wt/Wt | A/A | C/C | Wt/Wt | hrTX/hrTX | ||
| California 1 | Wt/Wt | A/A | C/C | Wt/Wt | hrTX/hrTX | ||
| California 2 | Wt/Wt | A/A | C/C | C/C | Wt/Wt | hrTX/hrTX | |
| France L12 | Wt/Wt | A/A | C/C | C/C | Wt/Wt | hrTX/hrTX | |
| Georgia | A/A | hrTX/hrTX | |||||
| South Carolina | Wt/Wt | A/A | C/C | C/C | Wt/Wt | hrTX/hrTX | |
| Utah | Wt/Wt | A/A | C/C | Wt/Wt | hrTX/hrTX | ||
| Vermont Gonzalo | Wt/Wt | Wt/Wt | A/A | C/C | C/C | Wt/Wt | hrTX/hrTX |
| North Carolina | Wt/Wt | G/G | T/T | C/C | Wt/Wt | hrNC/hrNC | |
| Canada | Wt/Wt | G/G | C/C | T/T | Wt/Wt | hrCa/hrCa | |
| Florida 1 | Wt/Wt | G/G | C/C | T/T | Wt/Wt | hrCa/hrCa | |
| Florida 2 † | Wt/Wt | G/A | C/C | C/C | Wt/Wt | Wt/hrTX | |
| Protein Change | Q420S | Splice | Splice | R748X | Q865X | S1130R |
| Severity | Effect | No. | Genes | Hairless (HR) |
|---|---|---|---|---|
| High | Frame Shift (LOF) † | 13 | 12 * | 2 |
| Splice donor - acceptor | 0 | 0 | 0 | |
| Stop gained (LOF) | 4 | 5 | 0 | |
| Moderate | Missense | 154 | 146 | 62 |
| Low | Splice region | 31 | 30 | 5 |
| Synonymous | 127 | 120 † | 67 | |
| Other | Intronic | 118 | 103 | 9 |
| Intergenic & UTR | 17 | -- | 7 | |
| Non-coding exon | 94 | 68 | 0 | |
| Total variants | 558 | ~426 ** | 152 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buckley, R.M.; Gandolfi, B.; Creighton, E.K.; Pyne, C.A.; Bouhan, D.M.; LeRoy, M.L.; Senter, D.A.; Gobble, J.R.; Abitbol, M.; Lyons, L.A.; et al. Werewolf, There Wolf: Variants in Hairless Associated with Hypotrichia and Roaning in the Lykoi Cat Breed. Genes 2020, 11, 682. https://doi.org/10.3390/genes11060682
Buckley RM, Gandolfi B, Creighton EK, Pyne CA, Bouhan DM, LeRoy ML, Senter DA, Gobble JR, Abitbol M, Lyons LA, et al. Werewolf, There Wolf: Variants in Hairless Associated with Hypotrichia and Roaning in the Lykoi Cat Breed. Genes. 2020; 11(6):682. https://doi.org/10.3390/genes11060682
Chicago/Turabian StyleBuckley, Reuben M., Barbara Gandolfi, Erica K. Creighton, Connor A. Pyne, Delia M. Bouhan, Michelle L. LeRoy, David A. Senter, Johnny R. Gobble, Marie Abitbol, Leslie A. Lyons, and et al. 2020. "Werewolf, There Wolf: Variants in Hairless Associated with Hypotrichia and Roaning in the Lykoi Cat Breed" Genes 11, no. 6: 682. https://doi.org/10.3390/genes11060682
APA StyleBuckley, R. M., Gandolfi, B., Creighton, E. K., Pyne, C. A., Bouhan, D. M., LeRoy, M. L., Senter, D. A., Gobble, J. R., Abitbol, M., Lyons, L. A., & 99 Lives Consortium. (2020). Werewolf, There Wolf: Variants in Hairless Associated with Hypotrichia and Roaning in the Lykoi Cat Breed. Genes, 11(6), 682. https://doi.org/10.3390/genes11060682





