Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle
Abstract
1. Introduction
2. Material and Methods
2.1. Genotype and Phenotype Data
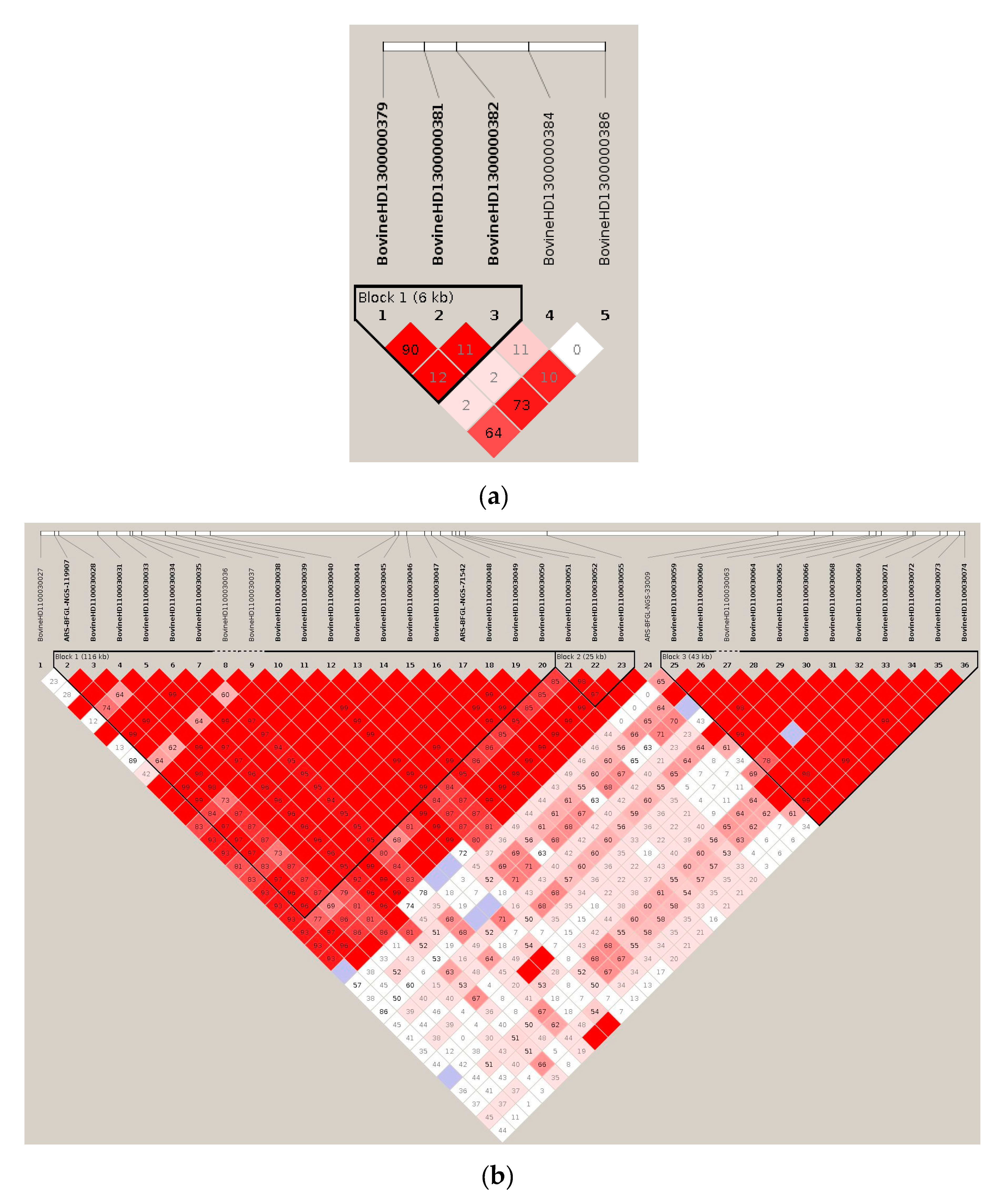
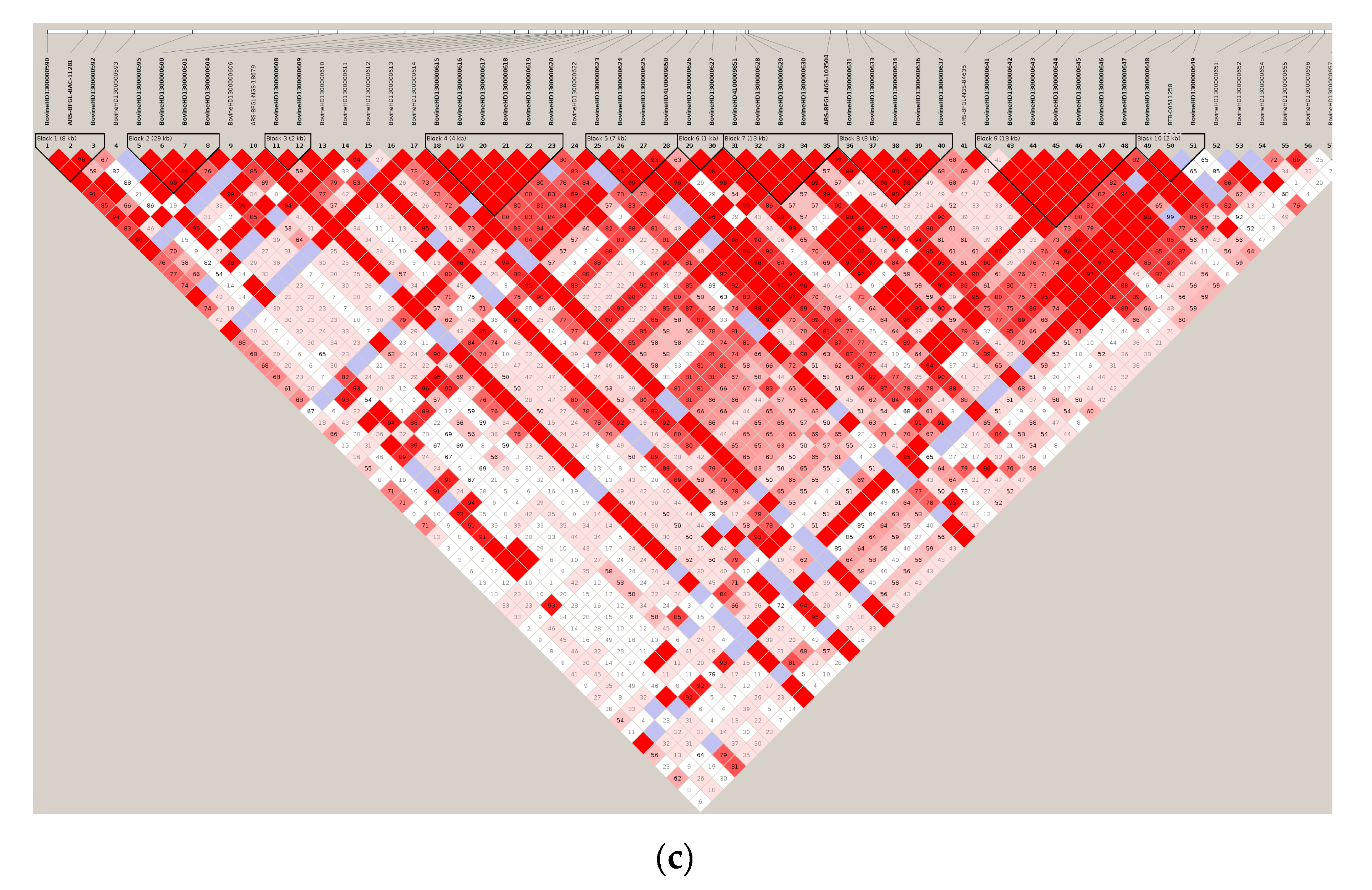
2.2. Construction of Haplotype Blocks
2.2.1. Phasing of Chromosomes
2.2.2. Haplotype Blocks (Haploblocks) Construction
2.3. Genome-Wide Association Study (GWAS)
2.4. Validation
2.5. Gene Identification in Significant Haplotype Blocks
2.6. Gene Functional Enrichment
3. Results
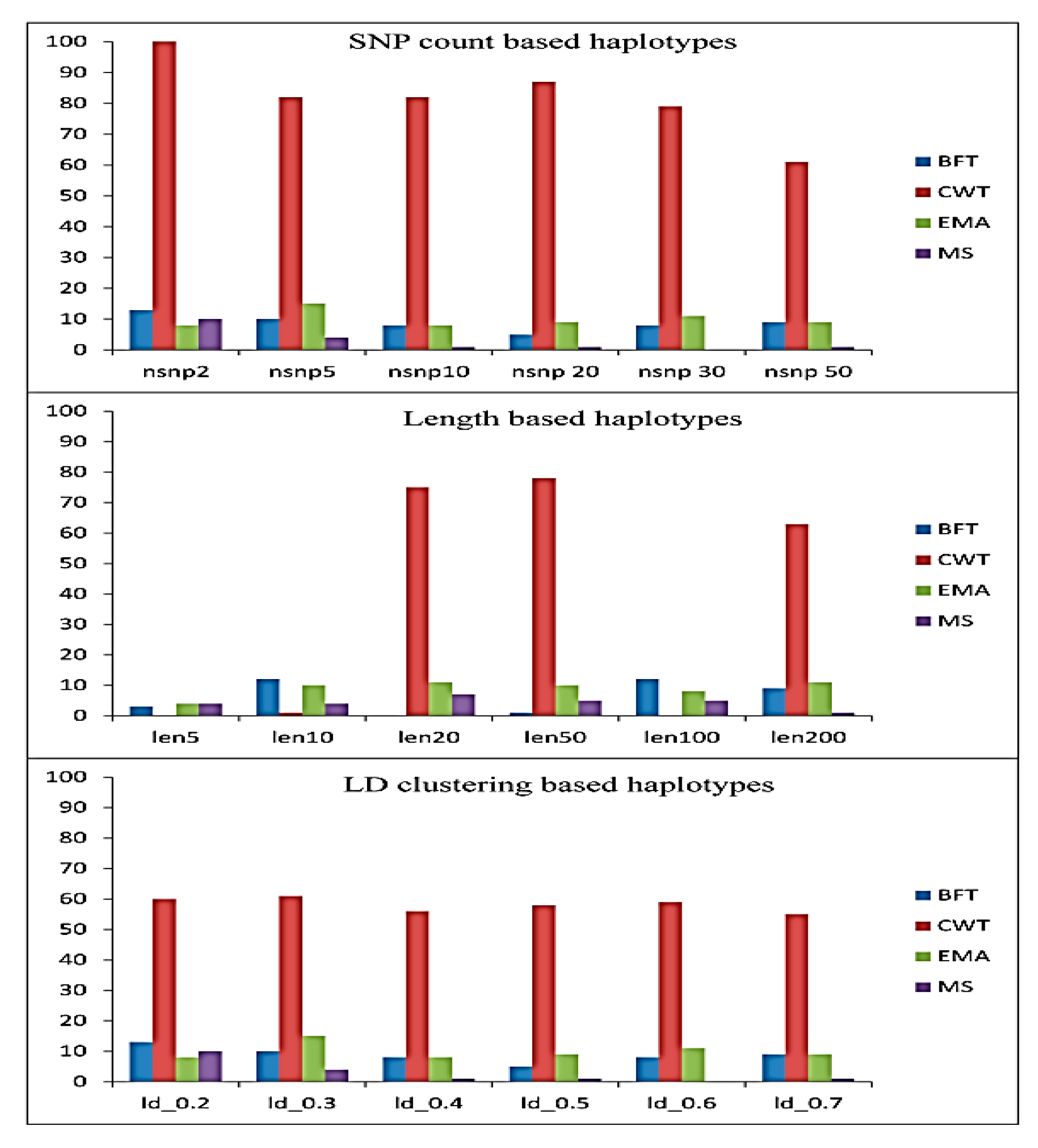
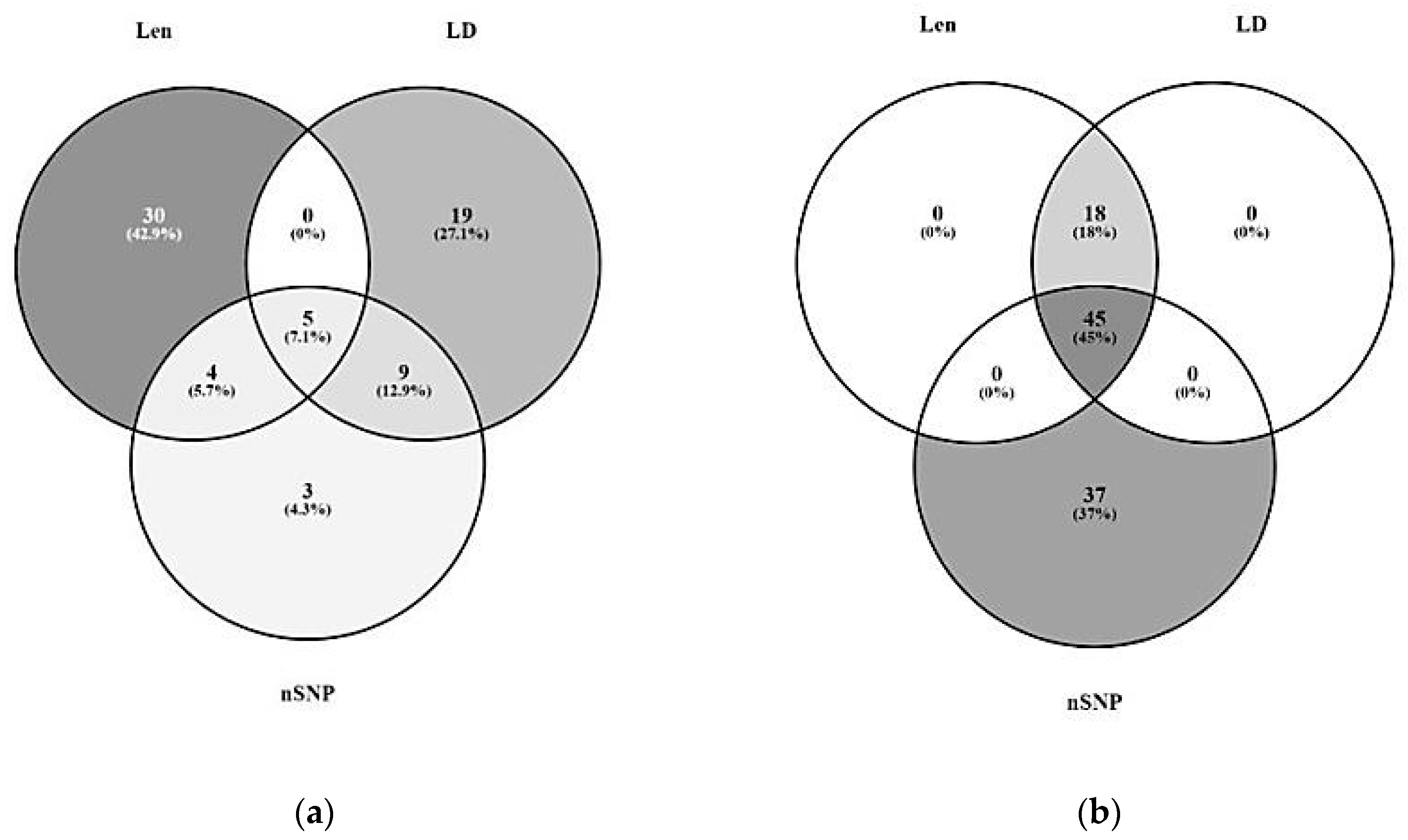
3.1. Significant Haplotype Blocks
3.2. Candidate Gene Identification
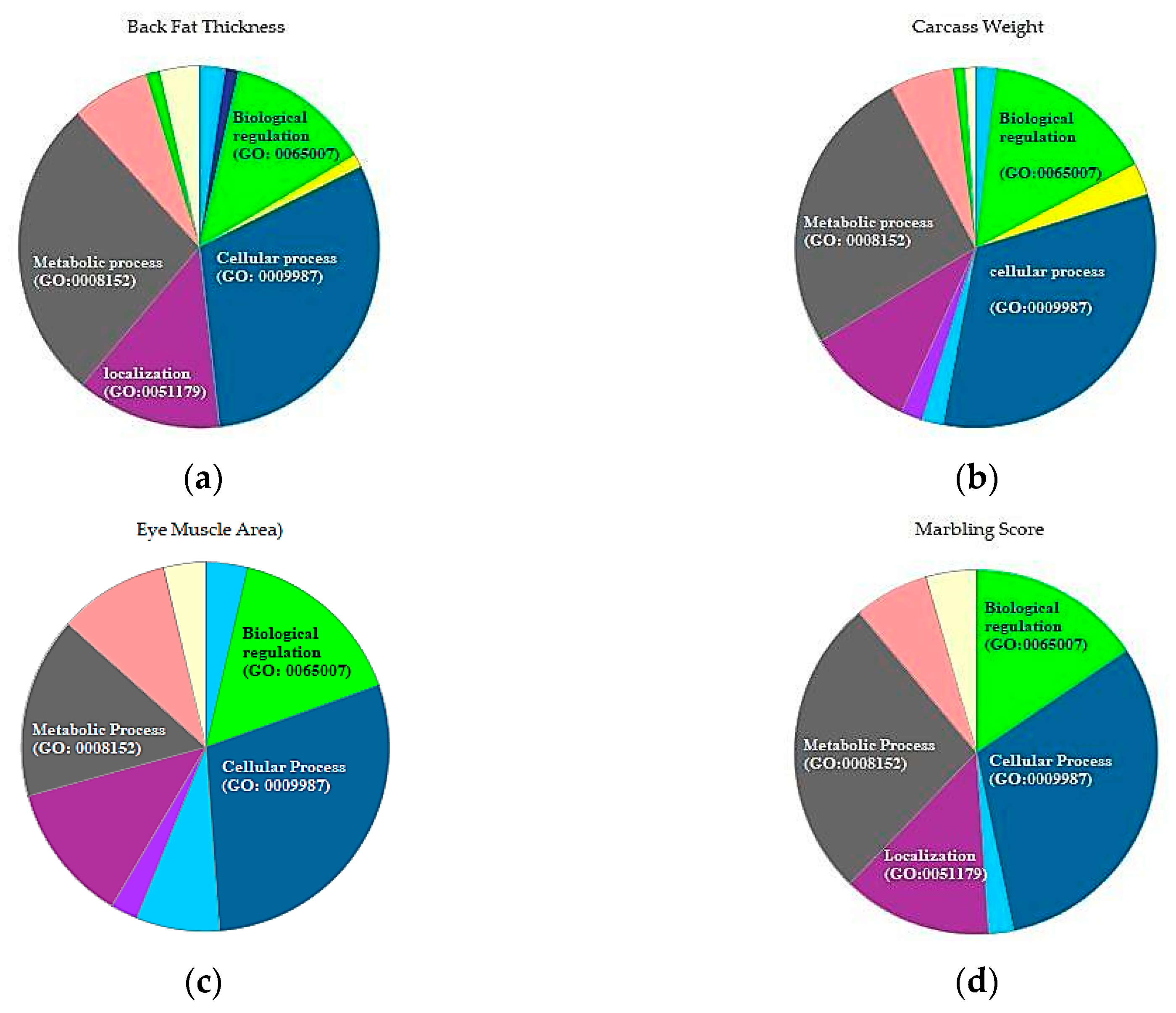
3.3. Gene Function Annotation
4. Discussion
Phospholipase
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Assessing Beef Demand Determinants. Available online: https://www.beefboard.org/wp-content/uploads/2018/01/Assessing-Beef-Demand-Determinants.pdf (accessed on 10 May 2019).
- Koohmaraie, M.K.; Kent, M.P.; Shackelford, S.D.; Veiseth, E.; Wheeler, T.L. Meat tenderness and muscle growth: Is there any relationship? Meat Sci. 2002, 62, 345–352. [Google Scholar] [CrossRef]
- Bhuiyan, M.; Kim, H.; Lee, D.; Lee, S.; Cho, S.; Yang, B.; Kim, S.; Lee, S. Genetic parameters of carcass and meat quality traits in different muscles (longissimus dorsi and semimembranosus) of Hanwoo (Korean cattle). J. Anim. Sci. 2017, 95, 3359–3369. [Google Scholar] [PubMed]
- Lee, S.H.; Van Der Werf, J.; Lee, S.H.; Park, E.W.; Gondro, C.; Yoon, D.; Oh, S.J.; Kim, O.H.; Gibson, J.; Thompson, J. Genome wide QTL mapping to identify candidate genes for carcass traits in Hanwoo (Korean Cattle). Genes Genom. 2012, 34, 43–49. [Google Scholar] [CrossRef]
- Lee, S.H.; Choi, B.H.; Lim, D.; Gondro, C.; Cho, Y.M.; Dang, C.G.; Sharma, A.; Jang, G.W.; Lee, K.T.; Yoon, D. Genome-wide association study identifies major loci for carcass weight on BTA14 in Hanwoo (Korean cattle). PLoS ONE 2013, 8, e74677. [Google Scholar] [CrossRef] [PubMed]
- Hyeong, K.-E.; Lee, Y.-M.; Kim, Y.-S.; Nam, K.; Jo, C.; Lee, K.-H.; Lee, J.-E.; Kim, J.-J. A whole genome association study on meat palatability in hanwoo. Asian-Australas. J. Anim. Sci. 2014, 27, 1219. [Google Scholar] [CrossRef]
- Dang, C.; Cho, S.; Sharma, A.; Kim, H.; Jeon, G.; Yeon, S.; Hong, S.; Park, B.; Kang, H.; Lee, S. Genome-wide association study for Warner-Bratzler shear force and sensory traits in Hanwoo (Korean cattle). Asian-Australas. J. Anim. Sci. 2014, 27, 1328. [Google Scholar] [CrossRef]
- Lee, S.; Choi, B.; Cho, S.; Lim, D.; Choi, T.; Park, B.; Lee, J.; Gondro, C.; Sharma, A.; Dang, C. Genome-wide association study identifies three loci for intramuscular fat in Hanwoo (Korean cattle). Livest. Sci. 2014, 165, 27–32. [Google Scholar] [CrossRef]
- Edea, Z.; Jeoung, Y.H.; Shin, S.-S.; Ku, J.; Seo, S.; Kim, I.-H.; Kim, S.-W.; Kim, K.-S. Genome–wide association study of carcass weight in commercial Hanwoo cattle. Asian-Australas. J. Anim. Sci. 2018, 31, 327. [Google Scholar] [CrossRef]
- Bedhane, M.; van der Werf, J.; Gondro, C.; Duijvesteijn, N.; Lim, D.; Park, B.; Park, M.N.; Hee, R.S.; Clark, S. Genome-Wide Association Study of Meat Quality Traits in Hanwoo Beef Cattle Using Imputed Whole-Genome Sequence Data. Front. Genet. 2019, 10, 1235. [Google Scholar] [CrossRef]
- Srikanth, K.; Lee, S.-H.; Chung, K.-Y.; Park, J.-E.; Jang, G.-W.; Park, M.-R.; Kim, N.Y.; Kim, T.-H.; Chai, H.-H.; Park, W.C. A Gene-Set Enrichment and Protein–Protein Interaction Network-Based GWAS with Regulatory SNPs Identifies Candidate Genes and Pathways Associated with Carcass Traits in Hanwoo Cattle. Genes 2020, 11, 316. [Google Scholar] [CrossRef]
- Khatkar, M.S.; Zenger, K.R.; Hobbs, M.; Hawken, R.J.; Cavanagh, J.A.; Barris, W.; McClintock, A.E.; McClintock, S.; Thomson, P.C.; Tier, B.; et al. A primary assembly of a bovine haplotype block map based on a 15,036-single-nucleotide polymorphism panel genotyped in Holstein–Friesian cattle. Genetics 2007, 176, 763–772. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altshuler, D.; Donnelly, P.; The International HapMap, C. A haplotype map of the human genome. Nature 2005, 437, 1299–1320. [Google Scholar] [CrossRef]
- Wu, Y.; Fan, H.; Wang, Y.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Ren, H.; Gao, H. Genome-wide association studies using haplotypes and individual SNPs in Simmental cattle. PLoS ONE 2014, 9, e109330. [Google Scholar] [CrossRef]
- Chen, Z.; Yao, Y.; Ma, P.; Wang, Q.; Pan, Y. Haplotype-based genome-wide association study identifies loci and candidate genes for milk yield in Holsteins. PLoS ONE 2018, 13, e0192695. [Google Scholar] [CrossRef]
- Barendse, W. Haplotype analysis improved evidence for candidate genes for intramuscular fat percentage from a genome wide association study of cattle. PLoS ONE 2011, 6, e29601. [Google Scholar] [CrossRef]
- Aylor, D.L.; Valdar, W.; Foulds-Mathes, W.; Buus, R.J.; Verdugo, R.A.; Baric, R.S.; Ferris, M.T.; Frelinger, J.A.; Heise, M.; Frieman, M.B. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011, 21, 1213–1222. [Google Scholar] [CrossRef]
- Barsh, G.S.; Copenhaver, G.P.; Gibson, G.; Williams, S.M. Guidelines for genome-wide association studies. PLoS Genet 2012, 8, e1002812. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Wang, S.; Li, H. Progress of genome wide association study in domestic animals. J. Anim. Sci. Biotechnol. 2012, 3, 26. [Google Scholar] [CrossRef]
- Grapes, L.; Dekkers, J.; Rothschild, M.; Fernando, R. Comparing linkage disequilibrium-based methods for fine mapping quantitative trait loci. Genetics 2004, 166, 1561–1570. [Google Scholar] [CrossRef]
- Calus, M.; De Roos, A.; Veerkamp, R. Accuracy of genomic selection using different methods to define haplotypes. Genetics 2008, 178, 553–561. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.C.; Verbyla, K.; Goddard, M.E. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 2009, 41, 51. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Orr, A.; Guernsey, D.L.; Robitaille, J.; Asselin, G.; Samuels, M.E.; Dubé, M.P. Application of homozygosity haplotype analysis to genetic mapping with high-density SNP genotype data. PLoS ONE 2009, 4, e5280. [Google Scholar] [CrossRef]
- Villa-Angulo, R.; Matukumalli, L.K.; Gill, C.A.; Choi, J.; Van Tassell, C.P.; Grefenstette, J.J. High-resolution haplotype block structure in the cattle genome. BMC Genet. 2009, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Mckay, S.D.; Schnabel, R.D.; Murdoch, B.M.; Matukumalli, L.K.; Aerts, J.; Coppieters, W.; Crews, D.; Neto, E.D.; Gill, C.A.; Gao, C. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 2007, 8, 74. [Google Scholar] [CrossRef]
- Hayes, B.; Chamberlain, A.; Mcpartlan, H.; Macleod, I.; Sethuraman, L.; Goddard, M. Accuracy of marker-assisted selection with single markers and marker haplotypes in cattle. Genet. Res. 2007, 89, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Calus, M.P.; Meuwissen, T.H.; Windig, J.J.; Knol, E.F.; Schrooten, C.; Vereijken, A.L.; Veerkamp, R.F. Effects of the number of markers per haplotype and clustering of haplotypes on the accuracy of QTL mapping and prediction of genomic breeding values. Genet. Sel. Evol. 2009, 41, 11. [Google Scholar] [CrossRef] [PubMed]
- Huson, H.J.; Kim, E.-S.; Godfrey, R.W.; Olson, T.A.; McClure, M.C.; Chase, C.C.; Rizzi, R.; O’Brien, A.M.; Van Tassell, C.P.; Garcia, J.F. Genome-wide association study and ancestral origins of the slick-hair coat in tropically adapted cattle. Front. Genet. 2014, 5, 101. [Google Scholar] [CrossRef]
- Do Nascimento, A.V.; da Silva Romero, A.R.; Utsunomiya, Y.T.; Utsunomiya, A.T.H.; Cardoso, D.F.; Neves, H.H.R.; Carvalheiro, R.; Garcia, J.F.; Grisolia, A.B. Genome-wide association study using haplotype alleles for the evaluation of reproductive traits in Nelore cattle. PLoS ONE 2018, 13, e0201876. [Google Scholar] [CrossRef]
- Mateescu, R.G.; Garrick, D.J.; Reecy, J.M. Network analysis reveals putative genes affecting meat quality in Angus cattle. Front. Genet. 2017, 8, 171. [Google Scholar] [CrossRef]
- Mathias, R.A.; Gao, P.; Goldstein, J.L.; Wilson, A.F.; Pugh, E.W.; Furbert-Harris, P.; Dunston, G.M.; Malveaux, F.J.; Togias, A.; Barnes, K.C. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006, 7, 38. [Google Scholar] [CrossRef]
- Contreras-Soto, R.I.; Mora, F.; de Oliveira, M.A.R.; Higashi, W.; Scapim, C.A.; Schuster, I. A Genome-Wide Association Study for Agronomic Traits in Soybean Using SNP Markers and SNP-Based Haplotype Analysis. PLoS ONE 2017, 12, e0171105. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Uemoto, Y.; Kikuchi, T.; Egawa, S.; Kohira, K.; Saito, T.; Sakuma, H.; Miyashita, S.; Arata, S.; Kojima, T.; et al. SNP- and haplotype-based genome-wide association studies for growth, carcass, and meat quality traits in a Duroc multigenerational population. BMC Genet. 2016, 17, 60. [Google Scholar] [CrossRef] [PubMed]
- Cuyabano, B.C.; Su, G.; Lund, M.S. Genomic prediction of genetic merit using LD based haplotypes in the Nordic Holstein population. BMC Genet. 2014, 15, 1171. [Google Scholar] [CrossRef]
- Ayalew, W.; Aliy, M.; Negussie, E. Estimation of genetic parameters of the productive and reproductive traits in Ethiopian Holstein using multi-trait models. Asian-Australas. J. Anim. Sci. 2017, 30, 1550. [Google Scholar] [CrossRef]
- Espigolan, R.; Baldi, F.; Boligon, A.A.; Souza, F.R.; Gordo, D.G.; Tonussi, R.L.; Cardoso, D.F.; Oliveira, H.N.; Tonhati, H.; Sargolzaei, M. Study of whole genome linkage disequilibrium in Nellore cattle. BMC Genom. 2013, 14, 305. [Google Scholar] [CrossRef]
- Gurgul, A.; Semik, E.; Pawlina, K.; Szmatoła, T.; Jasielczuk, I.; Bugno-Poniewierska, M. The application of genome-wide SNP genotyping methods in studies on livestock genomes. J. Appl. Genet. 2014, 55, 197–208. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Alam, M.; Park, M. Breeding initiatives for Hanwoo cattle to thrive as a beef industry–A review study. J. Anim. Breed Genet. 2017, 1, 102–124. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Delaneau, O.; Zagury, J.-F.; Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 2013, 10, 5. [Google Scholar] [CrossRef]
- Chen, Y.; Cunningham, F.; Rios, D.; McLaren, W.M.; Smith, J.; Pritchard, B.; Spudich, G.M.; Brent, S.; Kulesha, E.; Marin-Garcia, P. Ensembl variation resources. BMC Genom. 2010, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An interactive tool for comparing lists with Venn’s diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 22 November 2019).
- Dadousis, C.; Pegolo, S.; Rosa, G.; Gianola, D.; Bittante, G.; Cecchinato, A. Pathway-based genome-wide association analysis of milk coagulation properties, curd firmness, cheese yield, and curd nutrient recovery in dairy cattle. J. Dairy Sci. 2017, 100, 1223–1231. [Google Scholar] [CrossRef]
- Joaquim, L.B.; Chud, T.C.S.; Marchesi, J.A.P.; Savegnago, R.P.; Buzanskas, M.E.; Zanella, R.; Cantão, M.E.; Peixoto, J.O.; Ledur, M.C.; Irgang, R. Genomic structure of a crossbred Landrace pig population. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Hunt, S.E.; McLaren, W.; Gil, L.; Thormann, A.; Schuilenburg, H.; Sheppard, D.; Parton, A.; Armean, I.M.; Trevanion, S.J.; Flicek, P.; et al. Ensembl variation resources. Database 2018, 2018. [Google Scholar] [CrossRef]
- Kolar, M.J.; Kamat, S.S.; Parsons, W.H.; Homan, E.A.; Maher, T.; Peroni, O.D.; Syed, I.; Fjeld, K.; Molven, A.; Kahn, B.B.; et al. Branched Fatty Acid Esters of Hydroxy Fatty Acids Are Preferred Substrates of the MODY8 Protein Carboxyl Ester Lipase. Biochemistry 2016, 55, 4636–4641. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Kuehn, L.A.; Smith, T.P.L.; Ferrell, C.L.; Jenkins, T.G.; Freetly, H.C.; Snelling, W.M. A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle1. Anim. Genet. 2012, 43, 216–219. [Google Scholar] [CrossRef]
- Suzuki, J.; Imanishi, E.; Nagata, S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 2014, 289, 30257–30267. [Google Scholar] [CrossRef] [PubMed]
- Vlahcevic, Z.R.; Pandak, W.M.; Stravitz, R.T. REGULATION OF BILE ACID BIOSYNTHESIS. Gastroenterol. Clin. North Am. 1999, 28, 1–25. [Google Scholar] [CrossRef]
- Pausch, H.; Flisikowski, K.; Jung, S.; Emmerling, R.; Edel, C.; Götz, K.-U.; Fries, R. Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle. Genetics 2011, 187, 289–297. [Google Scholar] [CrossRef]
- Pace, J.M.; Corrado, M.; Missero, C.; Byers, P.H. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol. 2003, 22, 3–14. [Google Scholar] [CrossRef]
- Quick, C.B.; Fisher, R.A.; Harris, H. A kinetic study of the isozymes determined by the three human phosphoglucomutase loci PGM1, PGM2 and PGM3. European J. Biochem. 1974, 42, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Dunham, A.; Matthews, L.; Burton, J.; Ashurst, J.; Howe, K.; Ashcroft, K.; Beare, D.; Burford, D.; Hunt, S.; Griffiths-Jones, S. The DNA sequence and analysis of human chromosome 13. Nature 2004, 428, 522. [Google Scholar] [CrossRef]
- Cho, H.-S.; Hayami, S.; Toyokawa, G.; Maejima, K.; Yamane, Y.; Suzuki, T.; Dohmae, N.; Kogure, M.; Kang, D.; Neal, D.E. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia (New York, NY) 2012, 14, 476. [Google Scholar] [CrossRef]
- Zhao, Y.M.; Basu, U.; Dodson, M.V.; Basarb, J.A. Proteome differences associated with fat accumulation in bovine subcutaneous adipose tissues. Proteome Sci. 2010, 8, 14. [Google Scholar] [CrossRef]
- Li, C.-F.; Liu, T.-T.; Chuang, I.-C.; Chen, Y.-Y.; Fang, F.-M.; Chan, T.-C.; Li, W.-S.; Huang, H.-Y. PLCB4 copy gain and PLCß4 overexpression in primary gastrointestinal stromal tumors: Integrative characterization of a lipid-catabolizing enzyme associated with worse disease-free survival. Oncotarget 2017, 8, 19997. [Google Scholar] [CrossRef]
- Pethick, D.W.; D’souza, D.N.; Dunshea, F.R.; Harper, G.S. Fat metabolism and regional distribution in ruminants and pigs-influences of genetics and nutrition. In Proceedings of the Recent Advances in Animal Nutrition in Australia, University of New England, Armidale, NSW, Australia, 10–13 July 2005; p. 39. [Google Scholar]
- Baker, R.C.; Nikitina, Y.; Subauste, A.R. Chapter Six—Analysis of Adipose Tissue Lipid Using Mass Spectrometry. In Methods in Enzymology; MacDougald, O.A., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 538, pp. 89–105. [Google Scholar]
- Dennis, E.A.; Rhee, S.G.; Billah, M.M.; Hannun, Y.A. Role of phospholipase in generating lipid second messengers in signal transduction. FASEB J. 1991, 5, 2068–2077. [Google Scholar] [CrossRef]
- Lands, W.E. Phospholipases. In Biochemistry of Arachidonic Acid Metabolism. Prostaglandins, Leukotrienes, and Cancer; Springer: Boston, MA, USA, 1985; Volume 1. [Google Scholar]
- Fridlyand, L.E.; Philipson, L.H. Pancreatic Beta Cell G-Protein Coupled Receptors and Second Messenger Interactions: A Systems Biology Computational Analysis. PLoS ONE 2016, 11, e0152869. [Google Scholar] [CrossRef] [PubMed]
- Bergen, W.G.; Mersmann, H.J. Comparative Aspects of Lipid Metabolism: Impact on Contemporary Research and Use of Animal Models. J. Nutr. 2005, 135, 2499–2502. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.W.; Ballard, F.J. The relative significance of acetate and glucose as precursors for lipid synthesis in liver and adipose tissue from ruminants. Biochem. J. 1967, 105, 529–536. [Google Scholar] [CrossRef]
- Smith, S.B.; Crouse, J.D. Relative Contributions of Acetate, Lactate and Glucose to Lipogenesis in Bovine Intramuscular and Subcutaneous Adipose Tissue. J. Nutr. 1984, 114, 792–800. [Google Scholar] [CrossRef] [PubMed]


| Trait | Mean | SD | Min | Max |
|---|---|---|---|---|
| BFT (mm) | 8.92 | 3.65 | 2 | 25 |
| CWT (kg) | 369.16 | 41.36 | 158 | 518 |
| EMA (cm2) | 82.75 | 9.25 | 22 | 130 |
| MS (1–9) | 3.86 | 1.65 | 1 | 9 |
| Trait | BTA | Genes | Detected by Method | Function |
|---|---|---|---|---|
| BFT | 13 | PLCB1 | LD, Len, nsnp | Lipid metabolism [48] |
| 13 | TMX4 | LD, Len, nsnp | Thioredoxin-related trans membrane protein 4, responsible for negative regulation of hormone secretion [49] | |
| 13 | PLCB4 | LD, Len | Phospholipase C Beta 4 [50] involved in lipid metabolism [48] | |
| 11 | CEL | LD | Carboxyl ester lipase (CEL) is a bile-salt-activated lipase. It is also responsible for lipid catabolic process [51] | |
| CWT | 14 | LYPLA1 | LD, Len, nsnp | Hydrolase and lipase activity [52] |
| 14 | TMEM68 | LD, Len, nsnp | Expressed in bovine rumen, abomasum, intestine, and adipose tissue in cattle, and likely affects lipid biosynthetic processes [52] | |
| 14 | XKR9 | LD, Len, nsnp | Belongs to Xkr family possessing caspases recognition site, responsible for apoptosis [52,53] | |
| 14 | PREK2 | LD, Len, nsnp | GTPase activator, an important paralog of PREX1 responsible for increasing carcass weight in Hanwoo cattle. It also aids in meat development with richness in texture and flavor [9] | |
| 14 | KLHL38 | LD, Len, nsnp | Helps in maintaining the balance between intermediate filament stability, a process in maintaining skin integrity. It is an important component of ubiquitin–protein–ligase complex also formed by KLHl2 [30] | |
| 14 | TOX | LD, Len, nsnp | Responsible for the molecular regulation of puberty in Brahman cattle [3,5] | |
| 14 | CYP7B1 | LD, Len, nsnp | Oxysterol 7-hydroxylase, involved in bile acid synthesis [54] | |
| 14 | MYC | nsnp | Myelocytomatosis oncogene, responsible for ribosomal and mitochondrial biogenesis, glucose and glutamine metabolism, lipid synthesis, and cell cycle progression [2,3]. | |
| 13 | FAM110B | nsnp | A paralog of FAM110A gene, which is responsible for cell cycle progression through G1 phase and hence might play a role in increasing carcass weight in Hanwoo cattle by increasing cell number and cell size [5] | |
| 14 | SOX17 | LD, Len, nsnp | It [55] is a positional and functional gene involved in transcriptional regulation via inhibiting Wnt signaling (GO: 0060070). These are also responsible for tyrosine protein kinase and postnatal angiogenesis and hence affect growth-related traits in cattle | |
| 14 | PRKDC | LD, Len, nsnp | Plays an important role in cell cycle progression, apoptosis, and cellular transformation(bta04110) [3] | |
| EMA | 10 | SLC8A3 | LD, Len, nsnp | Promotes easy uptake of protein for muscular building and fibrillar collagen building [56]. |
| 1 | COL18A1 | LD | ||
| 10 | RORA | nsnp | RAR-related orphan receptor, responsible for cholesterol homeostasis (GO: 0042632) and negative regulation of fat cell differentiation (GO: 0045599) | |
| MS | 9 | FYN | LD, Len, nsnp | It belongs to member of Src family of non-receptor tyrosine kinases; This gene plays an important role in increasing adipogenesis [4]. |
| 1 | PARP14 | Len | It belongs to poly (ADP-ribose) polymerase family and, it play key role in regulating adipogenesis [9]. | |
| 6 | PGM2 | LD, nsnp | It is involved in galactose metabolism, which can help in adipogenesis in an organism [57] | |
| 12 | RB1 | LD | It plays an important role in posttranslational modifications and hence regulates cell cycle (GO: 2000134) [58,59] |
| Term | Pathway | Count | % | p-Value | Genes |
|---|---|---|---|---|---|
| Back Fat Thickness | |||||
| GO: 0016042 | Lipid catabolic process | 3 | 4.76 | 0.02 | CEL, PLCB4, PLCB1 |
| bta04911 | Insulin secretion | 3 | 4.76 | 0.03 | PLCB4, PLCB1, SNAP25 |
| bta03015 | mRNA surveillance pathway | 3 | 4.76 | 0.035 | UPF2, MAGOH, MSI1 |
| bta04922 | Glucagon signaling pathway | 3 | 4.76 | 0.039 | LDHA, PLCB4, PLCB1 |
| GO: 0005975 | Carbohydrate metabolic process | 3 | 4.76 | 0.039 | LDHA, GBGT1, GLT6D1 |
| bta04972 | Pancreatic secretion | 3 | 4.76 | 0.04 | CEL, PLCB4, PLCB1 |
| bta01100 | Metabolic pathways | 9 | 14.29 | 0.04 | CEL, LDHA, DHFR, PLCB4, GBGT1, SPTLC3, AOX4, PLCB1, NANP |
| bta04724 | Glutamatergic synapse | 3 | 4.76 | 0.053 | PLCB4, SLC1A7, PLCB1 |
| bta04071 | Sphingolipid signaling pathway | 3 | 4.76 | 0.059 | PLCB4, SPTLC3, PLCB1 |
| Carcass Weight | |||||
| GO: 0031648 | Protein destabilization | 4 | 3.92 | 0.001 | DERL1, RNF139, PRKDC, SOX17 |
| bta04110 | Cell cycle | 4 | 3.92 | 0.021 | PRKDC, CDK6, MCM4, MYC |
| GO: 0071805 | Potassium ion trans membrane transport | 3 | 2.94 | 0.034 | KCNQ3, KCNB2, KCNIP4 |
| GO: 0071498 | Cellular response to fluid shear stress | 2 | 1.96 | 0.051 | MTSS1, HAS2 |
| GO: 0060070 | Canonical Wnt signaling pathway | 3 | 2.94 | 0.059 | SFRP5, SOX17, MYC |
| GO: 0034765 | Regulation of ion trans membrane transport | 2 | 1.96 | 0.089 | KCNQ3, KCNB2 |
| GO: 0090090 | Negative regulation of canonical Wnt signaling pathway | 3 | 2.94 | 0.09 | SFRP5, RGS20, SOX17 |
| Eye Muscle Area | |||||
| GO: 0002092 | Positive regulation of receptor internalization | 3 | 6.25 | 0.001 | SCYL2, WNT3A, SYNJ2BP |
| GO: 0021766 | Hippocampus development | 3 | 6.25 | 0.003 | WNT3A, PAFAH1B1, YWHAE |
| GO: 0010988 | Regulation of low-density Lipoprotein particle clearance | 2 | 4.17 | 0.008 | CNPY2, NR1H4 |
| GO: 2000188 | Regulation of cholesterol Homeostasis (replaced by GO: 0042632) | 2 | 4.17 | 0.018 | RORA, NR1H4 |
| GO: 0070507 | Regulation of microtubule Cytoskeleton organization | 2 | 4.17 | 0.037 | WNT3A, PAFAH1B1 |
| GO: 0006605 | Protein targeting | 2 | 4.17 | 0.04 | SYNJ2BP, YWHAE |
| GO: 0043124 | Negative regulation of I-kappaB Kinase/NF-kappaB signaling | 2 | 4.17 | 0.078 | RORA, NR1H4 |
| GO: 0043087 | Regulation of GTPase activity | 2 | 4.17 | 0.083 | PAFAH1B1, CRK |
| GO: 0045599 | Negative regulation of fat cell differentiation | 2 | 4.17 | 0.083 | WNT3A, RORA |
| Marbling Score | |||||
| bta04110 | Cell cycle | 3 | 8.57 | 0.022 | RB1, CHEK2, CUL1 |
| GO: 2000134 | Negative regulation of G1/S Transition of mitotic cell cycle | 2 | 5.71 | 0.024 | EZH2, RB1 |
| GO: 0006915 | Apoptotic process | 3 | 8.57 | 0.031 | RRAGA, RB1, CUL1 |
| GO: 0042752 | Regulation of circadian rhythm | 2 | 5.71 | 0.043 | EZH2, PPARG |
| Trait | Chromosome | Gene | Gene Position | Method | Haplotype Block ID | Haplotype Block Start | Haplotype Block End | p-Value | Validation Result |
|---|---|---|---|---|---|---|---|---|---|
| BFT | 13 | PLCB1 | NC_037340.1 (933195–1796412) | LD0.2 | 20,062 | 1,440,165 | 1,484,853 | 7.39 × 10−6 | 3.39 × 10−2 |
| LD0.3 | 25,710 | 1,488,714 | 1,520,870 | 7.86 × 10−6 | 8.61 × 10−4 | ||||
| LD0.4 | 30,888 | 1,582,812 | 1,622,566 | 7.29 × 10−6 | 1.16 × 10−3 | ||||
| Len200 | 7071 | 1,400,001 | 1,600,001 | 5.56 × 10−6 | 4.16 × 10−3 | ||||
| Len100 | 13,990 | 1,400,001 | 1,500,001 | 6.01 × 10−6 | 1.77 × 10−2 | ||||
| 13,991 | 1,500,001 | 1,600,001 | 5.79 × 10−6 | 2.87 × 10−3 | |||||
| nsnp10 | 34,050 | 1,555,543 | 1,582,812 | 5.43 × 10−6 | 2.10 × 10−2 | ||||
| 34,051 | 1,588,829 | 1,627,590 | 7.32 × 10−6 | 1.02 × 10−2 | |||||
| nsnp20 | 17,020 | 1,444,917 | 1,504,649 | 5.96 × 10−6 | 3.16 × 10−3 | ||||
| nsnp30 | 11,346 | 1,444,917 | 1,529,072 | 7.2 × 10−6 | 5.62 × 10−3 | ||||
| nsnp5 | 68,104 | 1,564,591 | 1,582,812 | 5.27 × 10−6 | 1.14 × 10−2 | ||||
| nsnp50 | 6806 | 1,299,046 | 1,480,919 | 7.89 × 10−6 | 1.03 × 10−2 | ||||
| 13 | PLCB4 | NC_037340.1 (2021085-2503509) | Len200 | 7021 | 2,200,001 | 2,400,001 | 3.02 × 10−6 | 9.59 × 10−3 | |
| nsnp50 | 6811 | 2,172,478 | 2,350,887 | 3.86 × 10−6 | 9.46 × 10−3 | ||||
| Len100 | 13,990 | 1,400,001 | 1,500,001 | 6.01 × 10−6 | 1.77 × 10−2 | ||||
| 11 | CEL | NC_037338.1 (103058913–103067774) | LD0.2 | 18,600 | 103,051,196 | 103,319,019 | 5.16 × 10−6 | 1.40 × 10−1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, S.; Srikanth, K.; Won, S.; Son, J.-H.; Park, J.-E.; Park, W.; Chai, H.-H.; Lim, D. Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle. Genes 2020, 11, 551. https://doi.org/10.3390/genes11050551
Srivastava S, Srikanth K, Won S, Son J-H, Park J-E, Park W, Chai H-H, Lim D. Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle. Genes. 2020; 11(5):551. https://doi.org/10.3390/genes11050551
Chicago/Turabian StyleSrivastava, Swati, Krishnamoorthy Srikanth, Sohyoung Won, Ju-Hwan Son, Jong-Eun Park, Woncheoul Park, Han-Ha Chai, and Dajeong Lim. 2020. "Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle" Genes 11, no. 5: 551. https://doi.org/10.3390/genes11050551
APA StyleSrivastava, S., Srikanth, K., Won, S., Son, J.-H., Park, J.-E., Park, W., Chai, H.-H., & Lim, D. (2020). Haplotype-Based Genome-Wide Association Study and Identification of Candidate Genes Associated with Carcass Traits in Hanwoo Cattle. Genes, 11(5), 551. https://doi.org/10.3390/genes11050551






