Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GRAS Members in Barley
2.2. Phylogenetic Analysis of GRAS Members
2.3. Chromosomal Localization and Evolution Pressure of HvGRAS
2.4. Analysis of Conserved Motif and Gene Structure
2.5. Expression Analysis of HvGRAS Members
2.6. Plant Materials and Growth
2.7. RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
2.8. HvGRAS Proteins Interaction Network
3. Results
3.1. Identification and Characterization of GRAS Proteins in Barley
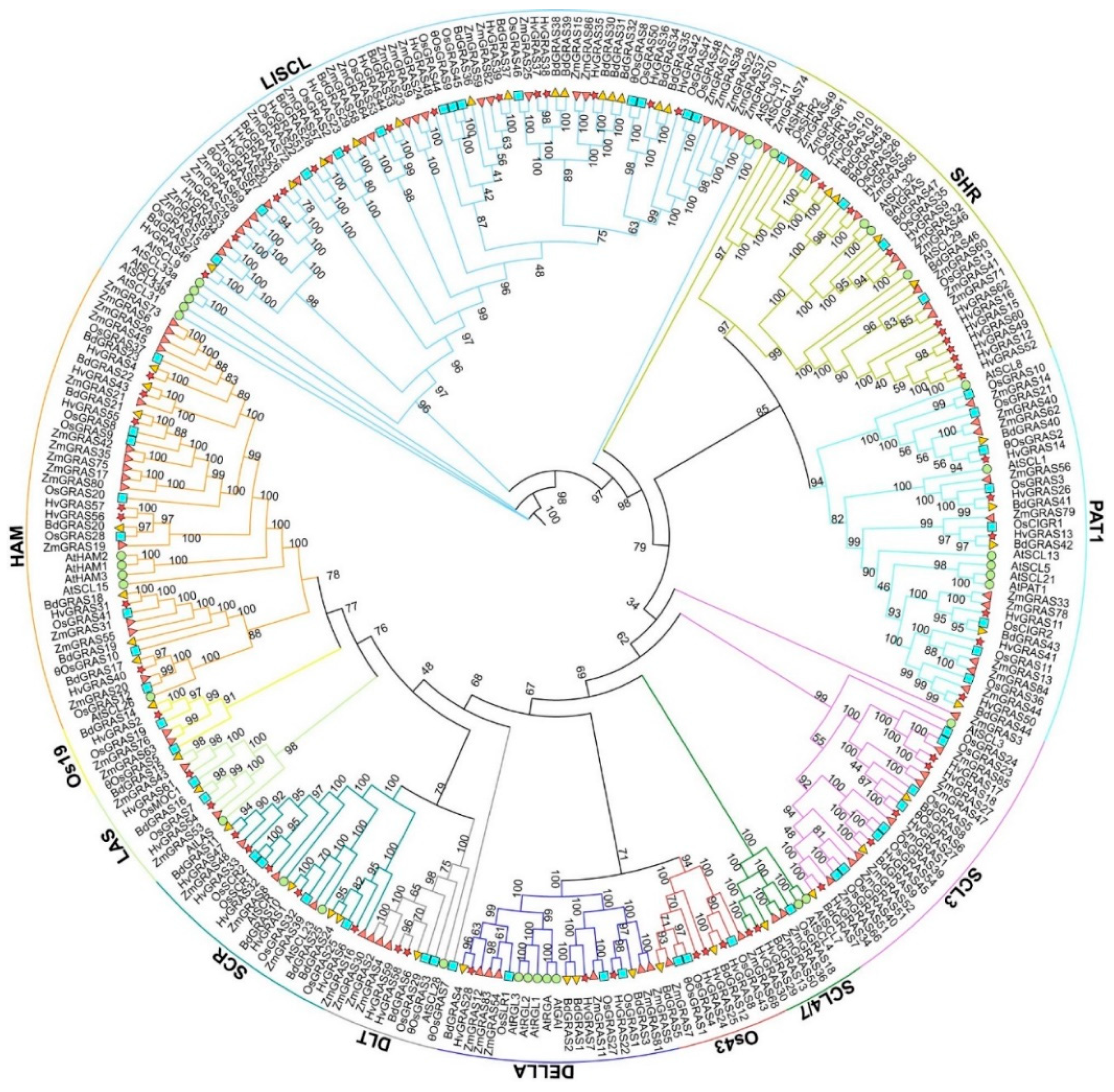
3.2. Phylogenetic Analysis of GRAS Proteins in Barley, Arabidopsis, Rice, Maize, Wheat and Brachypodium Distachyon
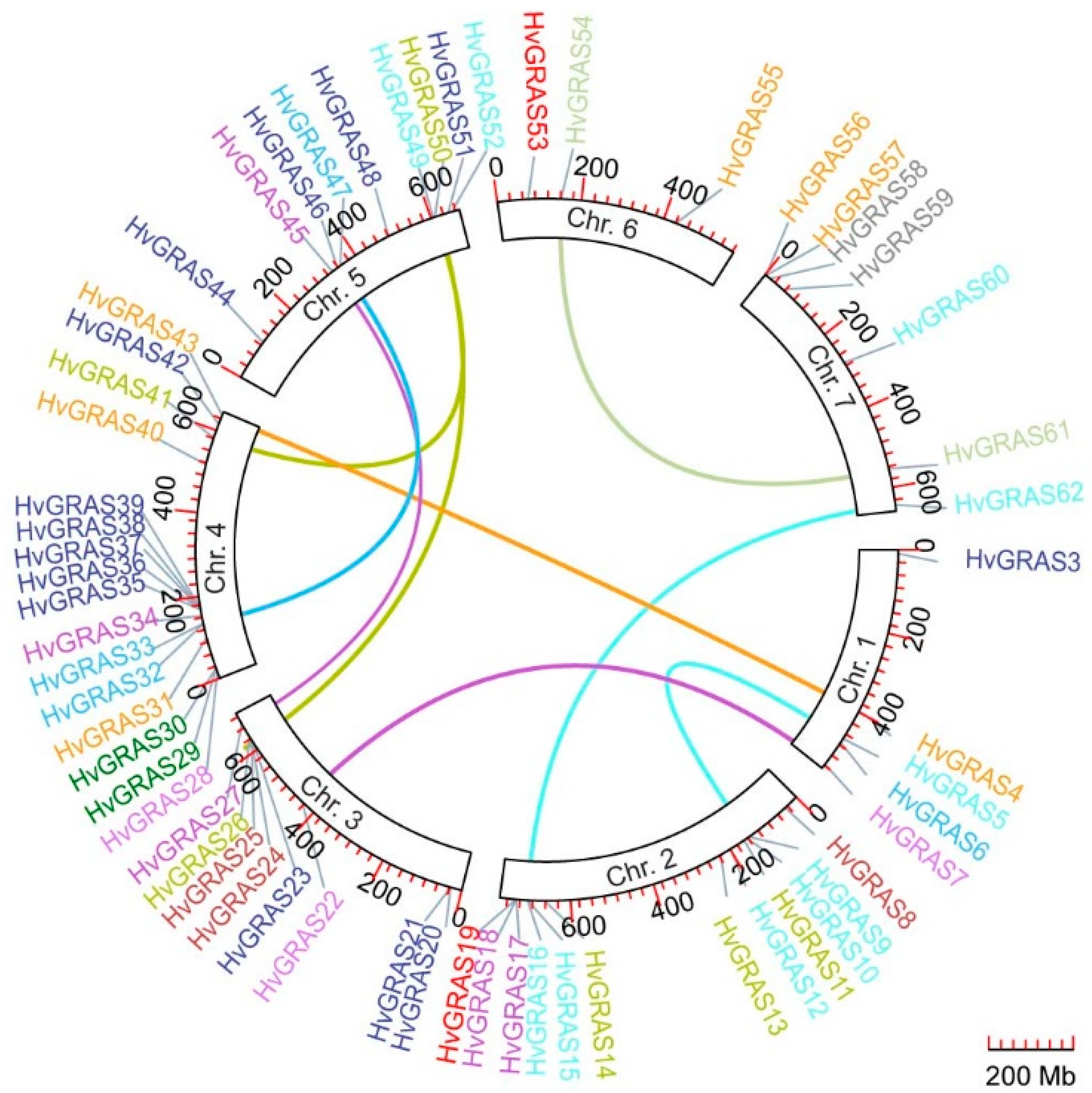
3.3. Chromosomal Location and Evolutionary Analysis of HvGRAS Genes
3.4. Protein Motif and Gene Structure Analysis of HvGRAS Family
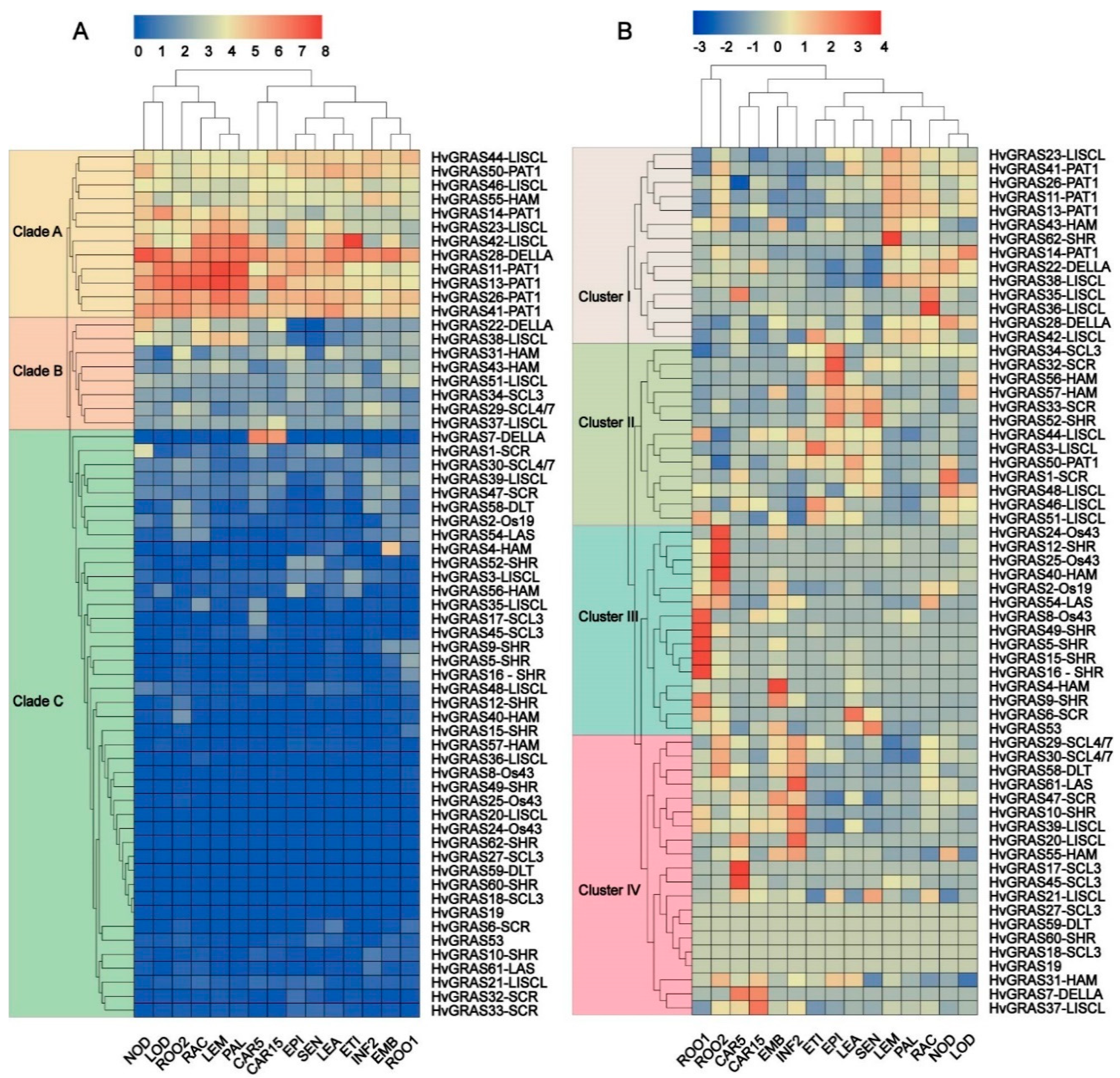
3.5. Expression Profile of HvGRAS Genes in Different Barley Tissue
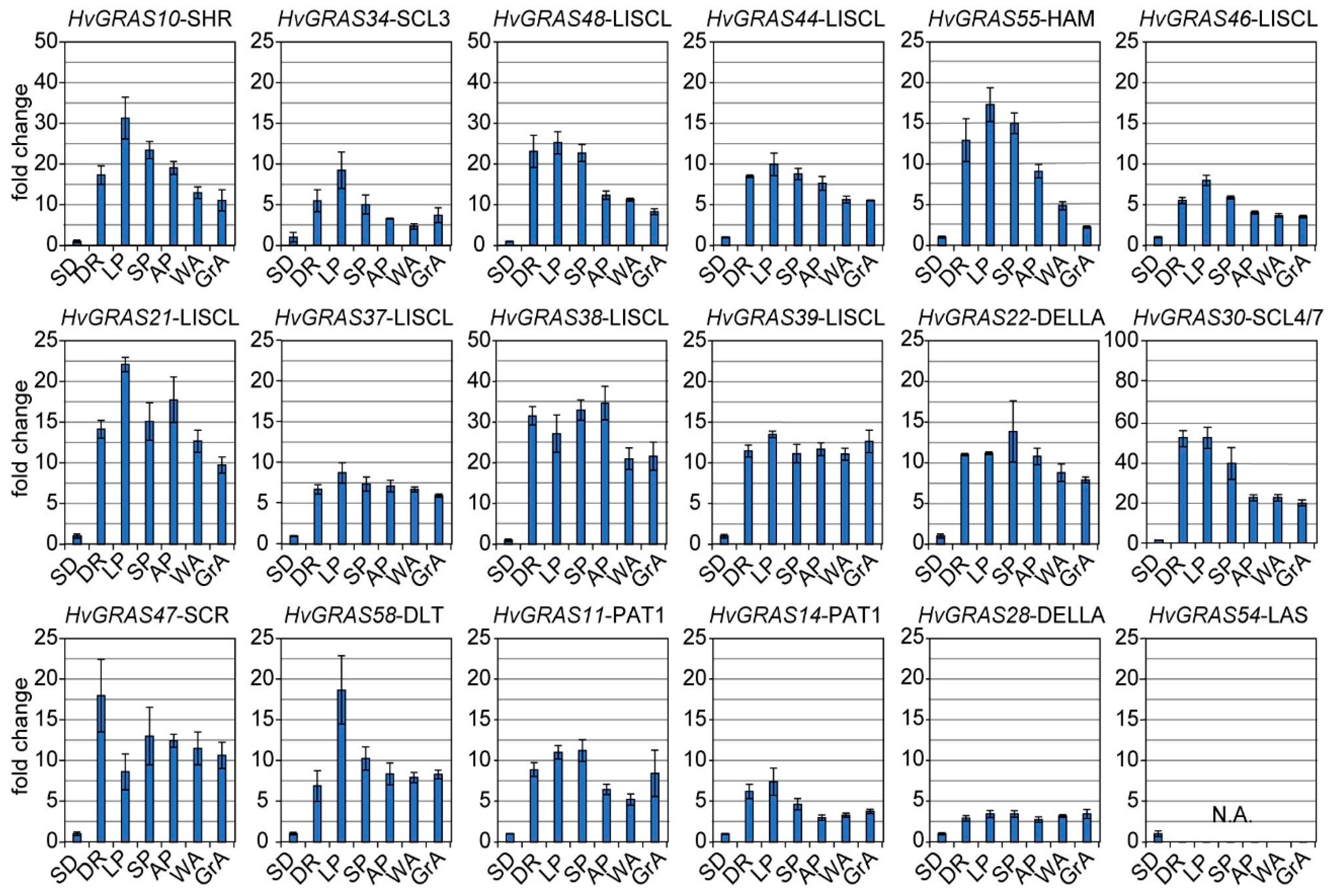
3.6. Expression of 18 HvGRAS Genes during Barley Spike Development by RT-PCR
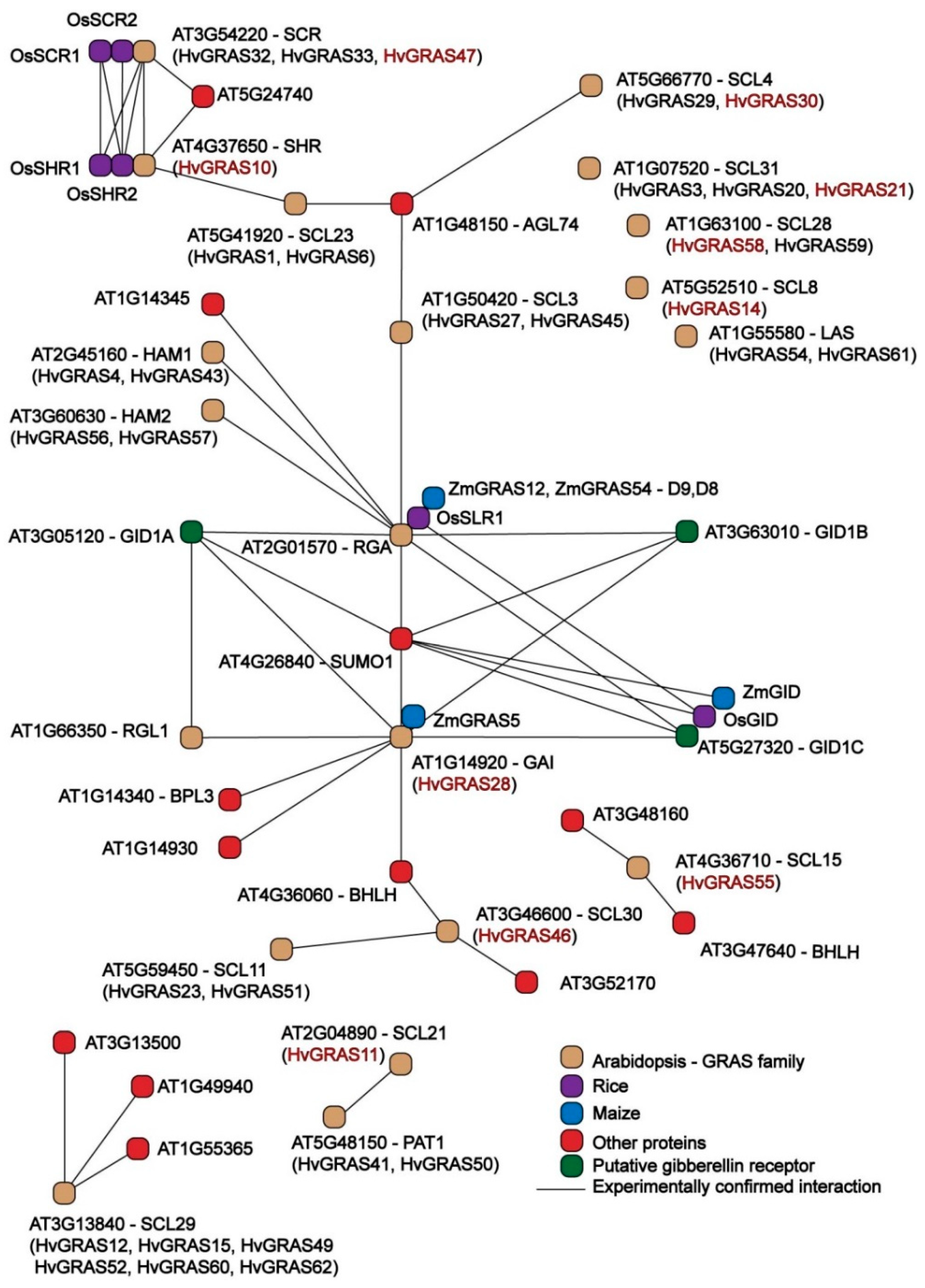
3.7. Interaction Network of HvGRAS Spike Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis sequence characterization and basic expression analysis of the SCARECROW-LIKE. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef]
- Hirano, K.; Asano, K.; Tsuji, H.; Kawamura, M.; Mori, H.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 2010, 22, 2680–2696. [Google Scholar] [CrossRef]
- Zhang, D.; Iyer, L.M.; Aravind, L. Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 2012, 28, 2407–2411. [Google Scholar] [CrossRef]
- Tian, C.G.; Wan, P.; Sun, S.H.; Li, J.Y.; Chen, M.S. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice. Plant Mol. Biol. Rep. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Guo, Y.; We, H.; Li, X.; Li, Q.; Zhao, X.; Duan, X.; An, Y.; Lv, W.; An, H. Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-H.; Kim, B.; Song, S.-K.; Heo, J.-O.; Yu, N.-I.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, E.M. Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal. Behav. 2011, 6, 850–854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.-X.; Liu, Z.-W.; Wu, Z.-J.; Li, H.; Wang, W.-L.; Cui, X.; Zhuang, J. Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, Y.; Shang, C.; Li, J.; Wang, J.; Wu, Z.; Ma, L.; Qi, T.; Fu, C.; Bai, Z.; et al. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS ONE 2017, 12, e0185439. [Google Scholar] [CrossRef] [PubMed]
- Claeys, H.; De Bodt, S.; Inzé, D. Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Itoh, H.; Gomi, K.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Jeong, D.H.; An, G.; Kitano, H.; Ashikari, M.; et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 2003, 299, 1896–1898. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Hirano, Y.; Sun, T.-P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P.; Belfield, E.; Yasumura, Y. The angiosperm Gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 2009, 21, 1328–1339. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef]
- Heo, J.-O.; Chang, K.S.; Kim, I.A.; Lee, M.-H.; Lee, S.A.; Song, S.-K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.-O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.-P. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.-T.; Benfey, P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef]
- Cui, H.; Levesque, M.P.; Vernoux, T.; Jung, J.W.; Paquette, A.J.; Gallagher, K.L.; Wang, J.Y.; Blilou, I.; Scheres, B.; Benfey, P.N. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 2007, 316, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Kalo, P.; Gleason, C.; Edwards, A.; Marsh, J.; Mitra, R.M.; Hirsch, S.; Jakab, J.; Sims, S.; Long, S.R.; Rogers, J.; et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 2005, 308, 1786–1789. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.; Raedts, J.; Portyanko, V.; Debelle, F.; Gough, C.; Bisseling, T.; Geurts, R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 2005, 308, 1789–1791. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef]
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide Research Trends on Wheat and Barley: A Bibliometric Comparative Analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef]
- Malcomber, S.T.; Preston, J.C.; Reinheimer, R.; Kossuth, J.; Kellogg, E.A. Developmental gene evolution and the origin of grass inflorescence diversity. Adv. Bot. Res. 2006, 44, 425–481. [Google Scholar] [CrossRef]
- Niu, X.; Chen, S.; Li, J.; Liu, Y.; Ji, W.; Li, H. Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genom. 2019, 20, 635. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J.; et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 2012, 491, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Himmelbach, A.; Colmsee, C.; Zhang, X.Q.; Barrero, R.A.; Zhang, Q.; Li, L.; Bayer, M.; Bolser, D.; Taudien, S.; et al. Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci. Data 2017, 4, 170044. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. Expasy: Sib bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. Dnasp 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. Tbtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Curaba, J.; Talbot, M.; Li, Z.; Helliwell, C. Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol. 2013, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The barley draft genome explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef]
- Liu, H.; Li, G.; Yang, X.; Kuijer, H.N.J.; Liang, W.; Zhang, D. Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. Crop J. 2019, 8, 71–86. [Google Scholar] [CrossRef]
- Waddington, S.R.; Cartwright, P.M.; Wall, P.C. A Quantitative Scale of Spike Initial and Pistil Development in Barley and Wheat. Ann. Bot. 1983, 51, 119–130. [Google Scholar] [CrossRef]
- Bi, X.; van Esse, W.; Mulki, M.A.; Kirschner, G.; Zhong, J.; Simon, R.; von Korff, M. Centroradialis interacts with flowering locus t-like genes to control floret development and grain number. Plant Physiol. 2019, 180, 1013–1030. [Google Scholar] [CrossRef]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, H.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 2007, 104, 1424–1429. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2018, 47, D529–D541. [Google Scholar] [CrossRef]
- McWhite, C.D.; Papoulas, O.; Drew, K.; Cox, R.M.; June, V.; Dong, O.X.; Kwon, T.; Wan, C.; Salmi, M.L.; Roux, S.J.; et al. A Pan-plant Protein Complex Map Reveals Deep Conservation and Novel Assemblies. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Bari, A.; Orazova, S.; Ivashchenko, A. miR156- and miR171-binding sites in the protein-coding sequences of several plant genes. Biomed. Res. Int. 2013, 2013, 307145. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Schnurbusch, T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 2012, 17, 91–101. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Ait-Ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef]
- Tyler, L.; Thomas, S.G.; Hu, J.H.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Locascio, A.; Blázquez, M.A.; Alabadí, D. Genomic Analysis of DELLA Protein Activity. Plant Cell Physiol. 2013, 54, 1229–1237. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Engstrom, E.M.; Andersen, C.M.; Gumulak-Smith, J.; Hu, J.; Orlova, E.; Sozzani, R.; Bowman, J.L. Arabidopsis homologs of the Petunia HAIRY MERISTEM gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011, 155, 735–750. [Google Scholar] [CrossRef]
- Fan, T.; Li, X.; Yang, W.; Xia, K.; Ouyang, J.; Zhang, M. Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS ONE 2015, 10, e0125833. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, X.; Xu, W.; Chang, J.; Li, A.; Mao, X.; Zhang, X.; Jing, R. Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci. Rep. 2015, 5, 12211. [Google Scholar] [CrossRef] [PubMed]
- Boden, S.A.; Weiss, D.; Ross, J.J.; Davies, N.W.; Trevaskis, B.; Chandler, P.M.; Swain, S.M. Early flowering3 regulates flowering in spring barley by mediating gibberellin production and flowering locus t expression. Plant Cell 2014, 26, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, Y.; Zhao, Z.; Wu, X.; Sun, L.; Liu, Q.; Wu, Y. Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 2016, 28, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Kim, J.; Munoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef]
- Xu, W.; Chen, Z.; Ahmed, N.; Han, B.; Cui, Q.; Liu, A. Genome-wide identification, evolutionary analysis, and stress responses of the GRAS gene family in Castor Beans. Int. J. Mol. Sci. 2016, 17, 1004. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Martis, M.; Hedley, P.E.; Šimková, H.; Liu, H.; Morris, J.A.; Steuernagel, B.; Taudien, S.; Roessner, S.; Gundlach, H.; et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 2011, 23, 1249–1263. [Google Scholar] [CrossRef]
- Song, X.-M.; Liu, T.-K.; Duan, W.-K.; Ma, Q.-H.; Ren, J.; Wang, Z.; Li, Y.; Hou, X.-L. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 2014, 103, 135–146. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2008, 8, 69–78. [Google Scholar] [CrossRef]
- Zou, M.; Guo, B.; He, S. The roles and evolutionary patterns of intronless genes in deuterostomes. Comp. Funct. Genom. 2011, 2011, 8. [Google Scholar] [CrossRef]
- William Roy, S.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Chandler, P.M.; Marion-Poll, A.; Ellis, M.; Gubler, F. Mutants at the Slender1 locus of barley cv himalaya. Molecular and physiological characterization. Plant Physiol. 2002, 129, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Gubler, F.; Chandler, P.M.; White, R.G.; Llewellyn, D.J.; Jacobsen, J.V. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 2002, 129, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Jung, H.-S.; Sun, T.-P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 2001, 98, 14162. [Google Scholar] [CrossRef]
- Levesque, M.P.; Vernoux, T.; Busch, W.; Cui, H.; Wang, J.Y.; Blilou, I.; Hassan, H.; Nakajima, K.; Matsumoto, N.; Lohmann, J.U.; et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006, 4, e143. [Google Scholar] [CrossRef]
- Kim, G.; Dhar, S.; Lim, J. The SHORT-ROOT regulatory network in the endodermis development of Arabidopsis roots and shoots. J. Plant Biol. 2017, 60, 306–313. [Google Scholar] [CrossRef]
- Yoon, E.K.; Dhar, S.; Lee, M.-H.; Song, J.H.; Lee, S.A.; Kim, G.; Jang, S.; Choi, J.W.; Choe, J.-E.; Kim, J.H.; et al. Conservation and diversification of the SHR-SCR-SCL23 regulatory network in the development of the functional endodermis in Arabidopsis shoots. Mol. Plant 2016, 9, 1197–1209. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, G.; Cui, X.; Yan, M.; Xu, T.; Qian, Q.; Xue, Y.; Li, J.; Wang, Y. OSGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol. Plant 2013, 6, 988–991. [Google Scholar] [CrossRef]
- Lin, Z.; Yan, J.; Su, J.; Liu, H.; Hu, C.; Li, G.; Wang, F.; Lin, Y. Novel OsGRAS19 mutant, D26, positively regulates grain shape in rice (Oryza sativa). Funct. Plant Biol. 2019, 46, 857–868. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; Cools, T.; Canher, B.; Shavialenka, S.; Traas, J.; Vercauteren, I.; Van den Daele, H.; Persiau, G.; De Jaeger, G.; Sugimoto, K.; et al. The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants 2016, 2, 16165. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.; Minami, M.; Takase, H.; Hotta, Y.; Hiratsuka, K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 2003, 278, 20865–20873. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.; Vielba, J.M.; Ferro, E.; Covelo, G.; Solé, A.; Abarca, D.; de Mier, B.S.; Díaz-Sala, C. Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 2007, 27, 1459–1470. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef]
- Fiorilli, V.; Volpe, V.; Zanini, S.; Vallino, M.; Abbà, S.; Bonfante, P. A rice GRAS gene has an impact on the success of arbuscular mycorrhizal colonization. Am. J. Plant Sci. 2015, 6, 1905–1915. [Google Scholar] [CrossRef]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OSGRAS23, a rice gras transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 141. [Google Scholar] [CrossRef]
- Zwirek, M.; Waugh, R.; McKim, S.M. Interaction between row-type genes in barley controls meristem determinacy and reveals novel routes to improved grain. New Phytol. 2019, 221, 1950–1965. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
To, V.-T.; Shi, Q.; Zhang, Y.; Shi, J.; Shen, C.; Zhang, D.; Cai, W. Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.). Genes 2020, 11, 553. https://doi.org/10.3390/genes11050553
To V-T, Shi Q, Zhang Y, Shi J, Shen C, Zhang D, Cai W. Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.). Genes. 2020; 11(5):553. https://doi.org/10.3390/genes11050553
Chicago/Turabian StyleTo, Vinh-Trieu, Qi Shi, Yueya Zhang, Jin Shi, Chaoqun Shen, Dabing Zhang, and Wenguo Cai. 2020. "Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.)" Genes 11, no. 5: 553. https://doi.org/10.3390/genes11050553
APA StyleTo, V.-T., Shi, Q., Zhang, Y., Shi, J., Shen, C., Zhang, D., & Cai, W. (2020). Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.). Genes, 11(5), 553. https://doi.org/10.3390/genes11050553




