Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Data Sources
2.2. Identification and Filtration
2.3. Phylogenetic Analysis and Classification
2.4. Gene Structure and Conserved Motif Analysis
2.5. Chromosomal Location, Gene Duplication, and Evolutionary Analysis
2.6. Dimerization Properties of OebZIP Proteins
2.7. Spatiotenporal Expression Analysis of OebZIP Gene Using Transcriptome Data
2.8. Plant Material
2.9. Plant RNA Extraction and qRT-PCR
3. Results
3.1. Identification and Naming of OebZIP
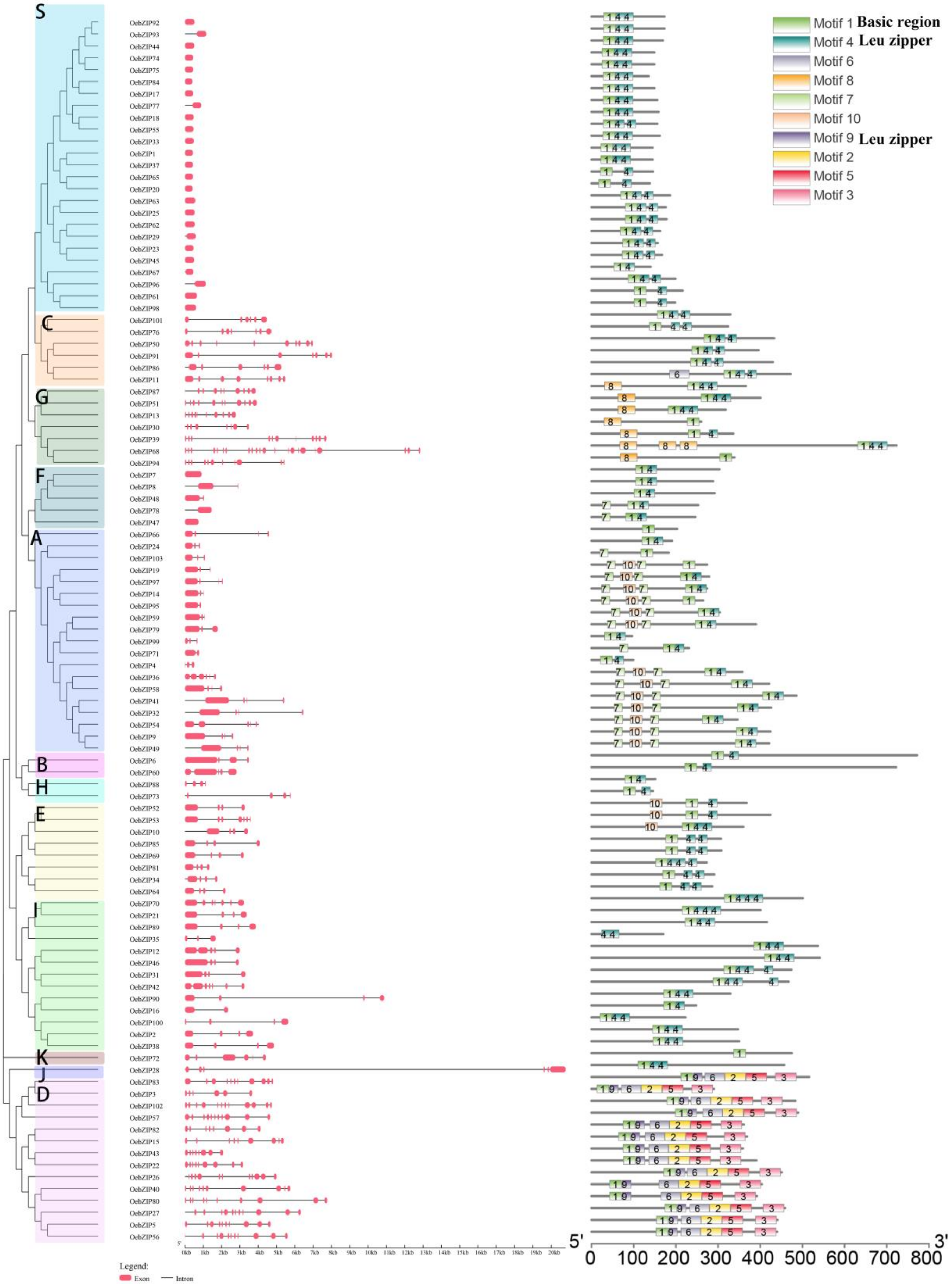
3.2. Phylogenetic Analysis and Classification
3.3. Gene Structure and Motif Composition of Olive bZIP
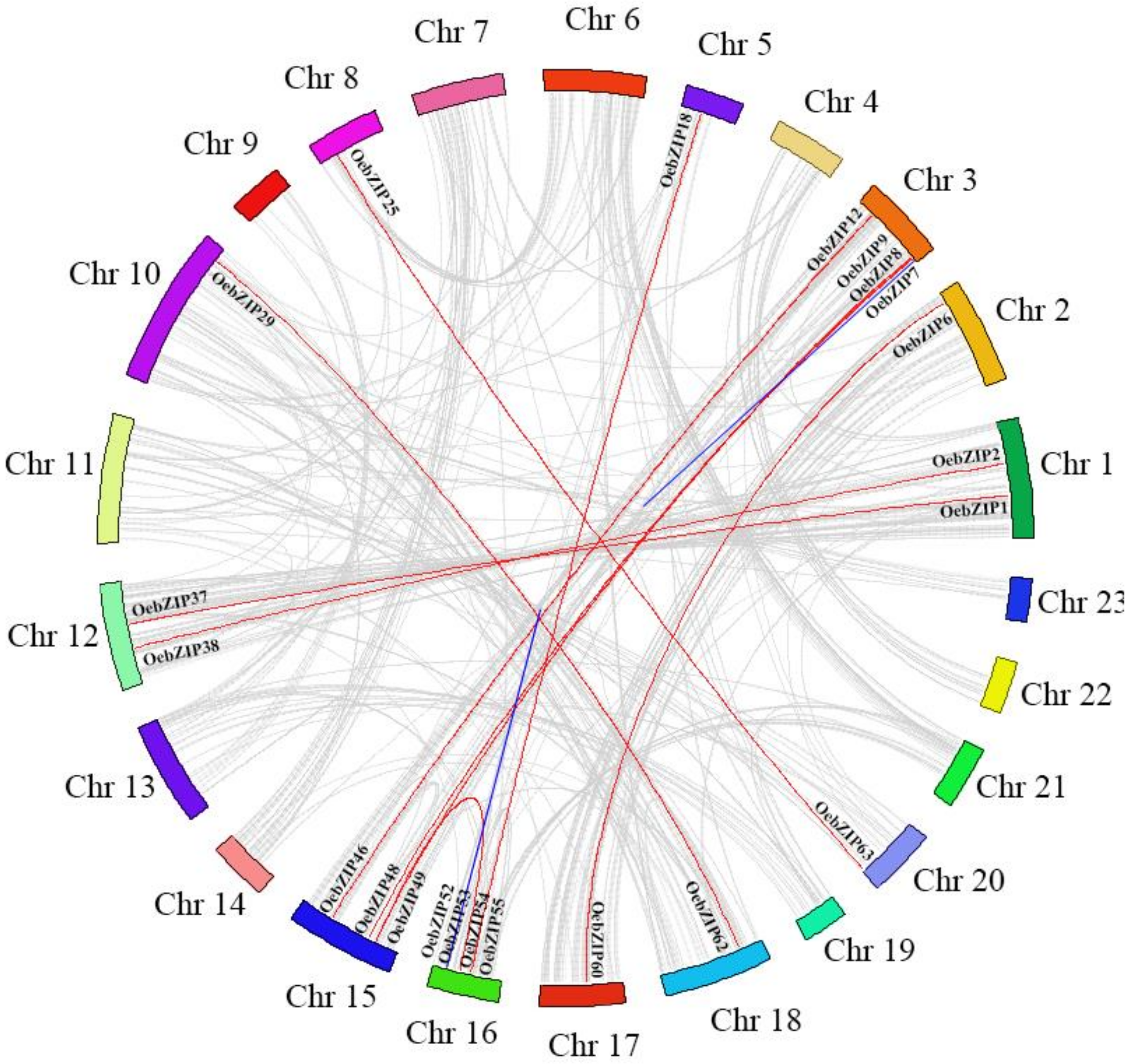
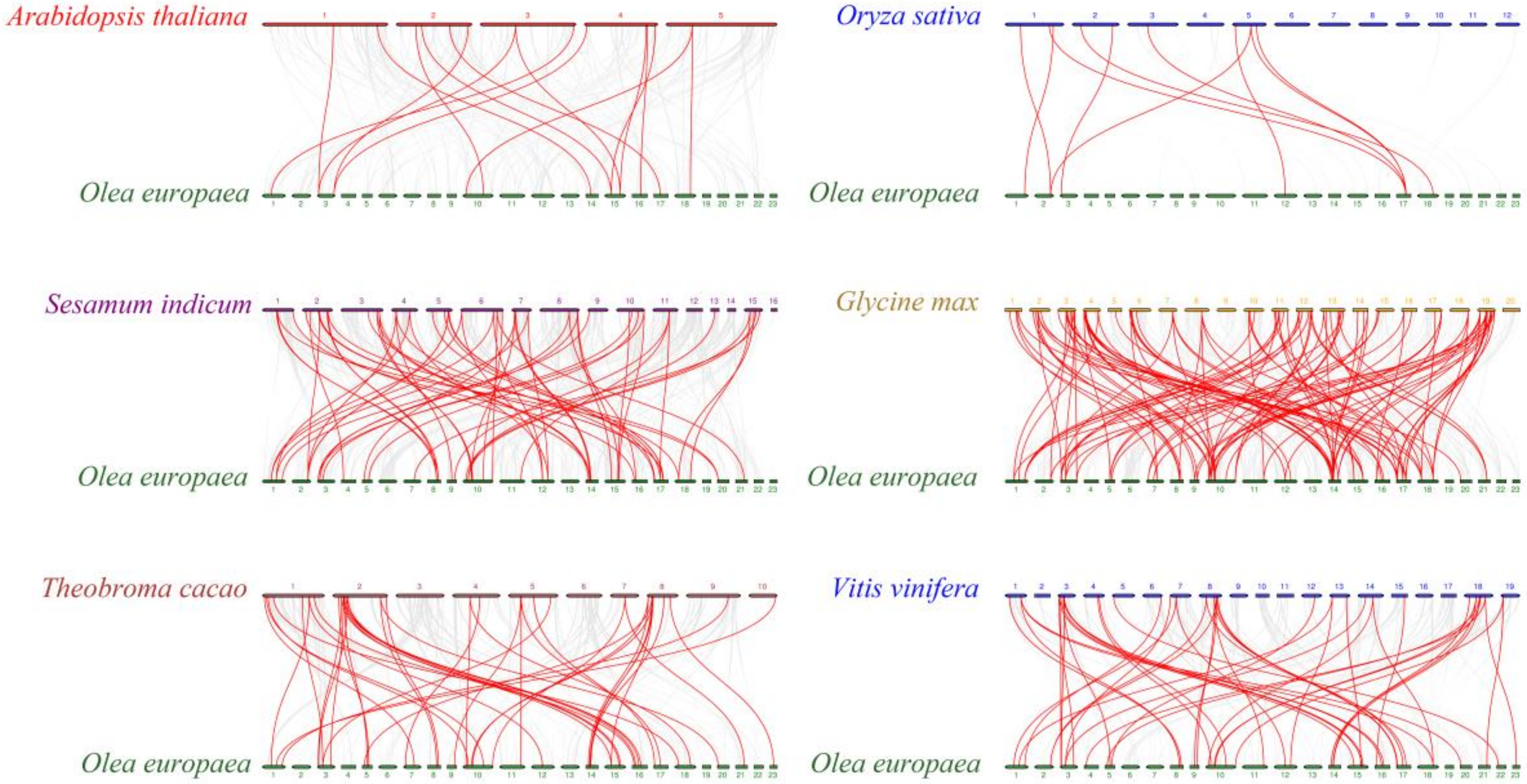
3.4. Chromosomal Locations and Gene Collinearity Analysis of OebZIP
3.5. Prediction of Dimerization Properties
3.6. Gene Expression profiles of OebZIPs
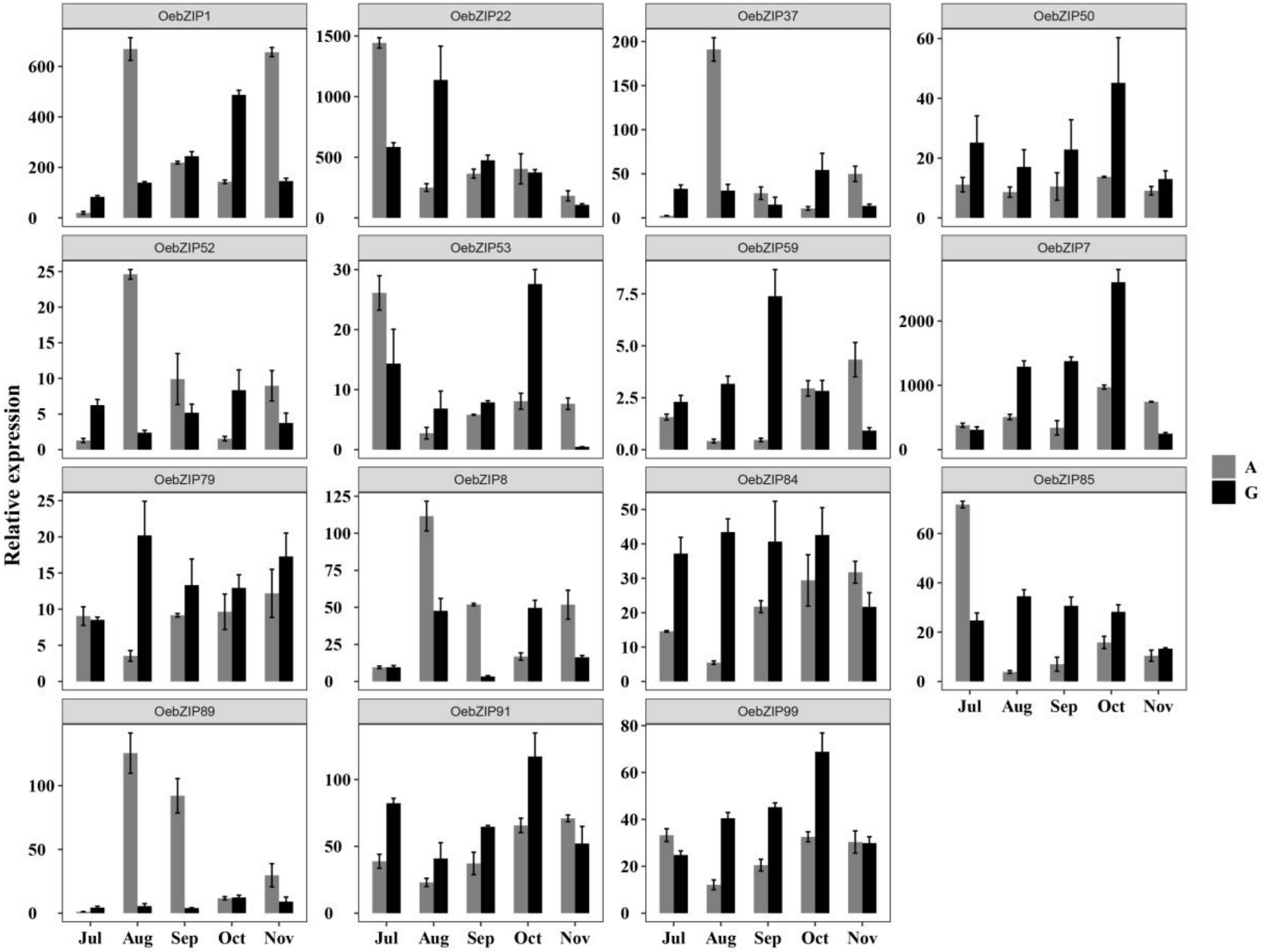
3.6.1. Gene Expression Profiles of OebZIPs on Transcription Data
3.6.2. Expression Patterns of OebZIP Genes in Different Tissues
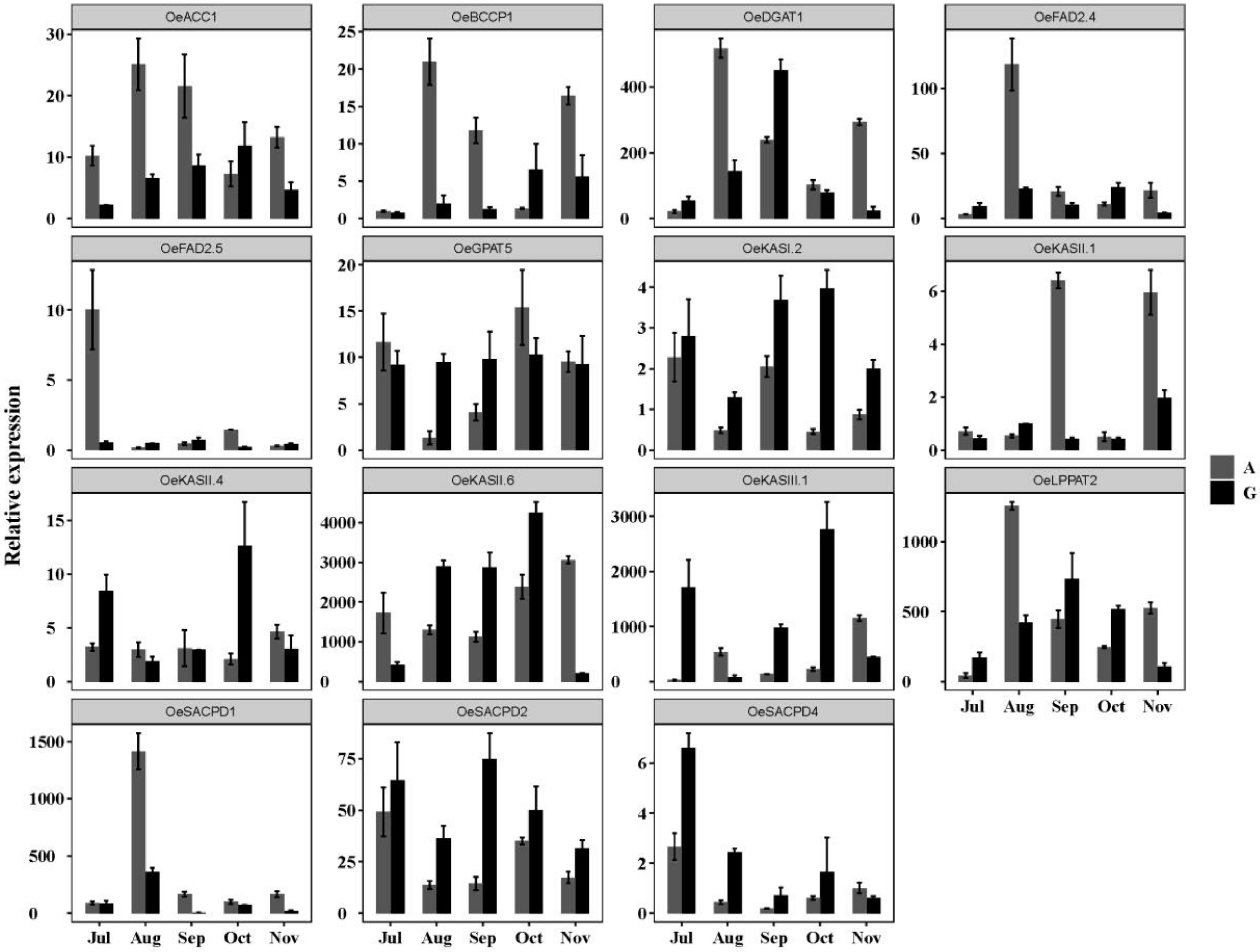
3.6.3. Expression of OebZIP Genes in Different Varieties During Fruit Development
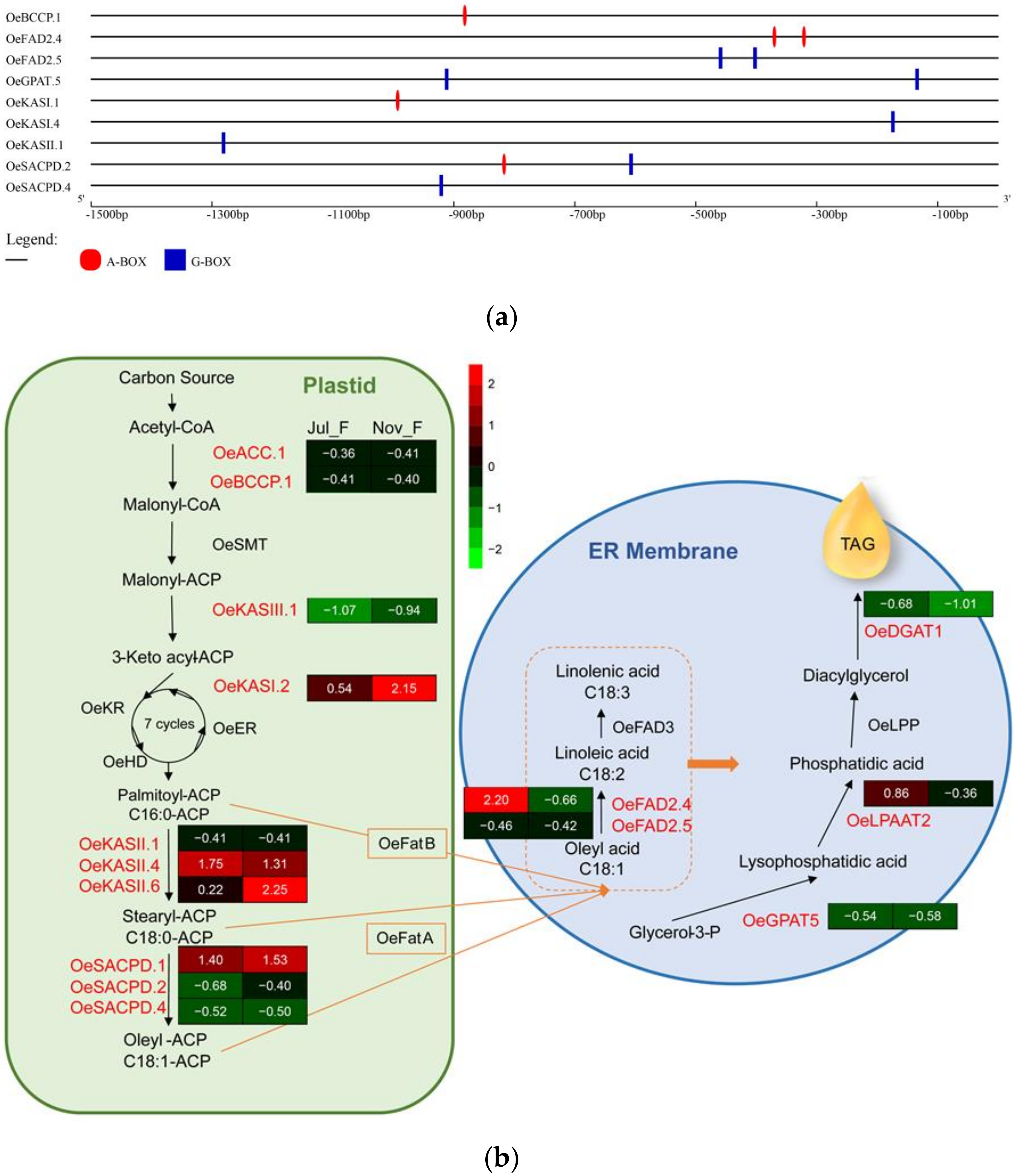
3.7. Predicting Interaction with Lipid Synthesis Genes
3.7.1. Expression Patterns of Lipid Synthesis Genes
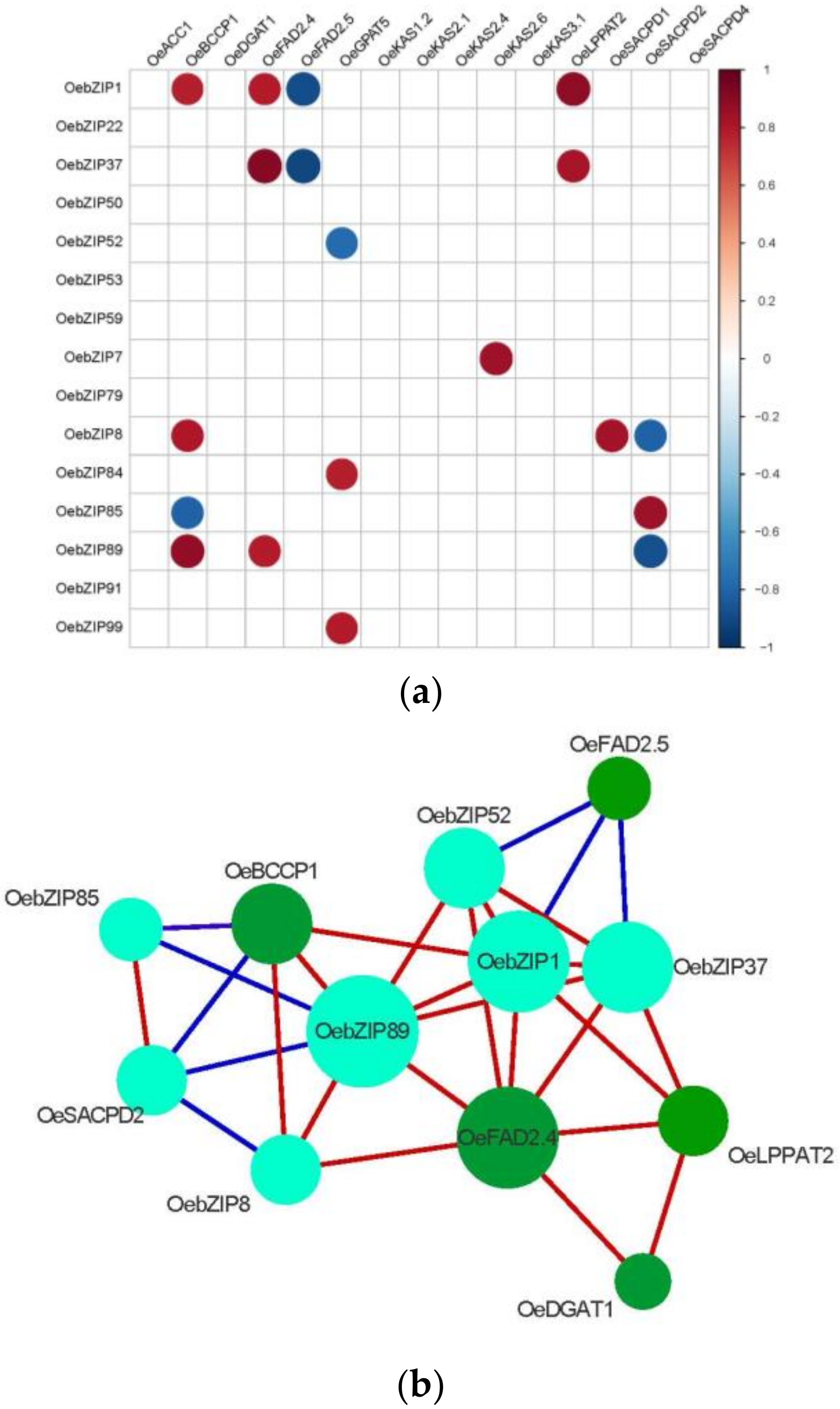
3.7.2. Correlation Analysis of OebZIP with Lipid Synthesis Genes
4. Discussion
4.1. Evolutionary Analysis of the OebZIP Gene Family
4.2. The Expression Pattern of OebZIP was Related to Fruit Development and Lipid Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gouvinhas, I.; Machado, N.; Sobreira, C.; Dominguez-Perles, R.; Gomes, S.; Rosa, E.; Barros, A. Critical Review on the Significance of Olive Phytochemicals in Plant Physiology and Human Health. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, R.; Anwar, F.; Alkharfy, K.M.; Gilani, A.H.; Saari, N. Valuable nutrients and functional bioactives in different parts of olive (Olea europaea L.)—A review. Int. J. Mol. Sci. 2012, 13, 3291–3340. [Google Scholar] [CrossRef] [PubMed]
- El, S.N.; Karakaya, S. Olive tree (Olea europaea) leaves: Potential beneficial effects on human health. Nutr. Rev. 2009, 67, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, S.; Akhlaghi, M.; Sasani, M.R.; Boldaji, R.B. Olive oil improved fatty liver severity independent of cardiometabolic correction in patients with non-alcoholic fatty liver disease, a randomized clinical trial. Nutrition 2019, 57, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.C.; Su, K.P.; Cheng, T.C.; Liu, H.C.; Chang, C.J.; Dewey, M.E.; Stewart, R.; Huang, S.Y. The effects of omega-3 fatty acids monotherapy in Alzheimer’s disease and mild cognitive impairment: A preliminary randomized double-blind placebo-controlled study. Prog. Neuro Psychopharmacol. Biol. Psychiatr. 2008, 32, 1538–1544. [Google Scholar] [CrossRef]
- Manan, S.; Chen, B.; She, G.; Wan, X.; Zhao, J. Transport and transcriptional regulation of oil production in plants. Crit. Rev. Biotechnol. 2017, 37, 641–655. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Breton, G.; Nagel, D.H.; Kang, S.E.; Bonaldi, K.; Doherty, C.J.; Ravelo, S.; Galli, M.; Ecker, J.R.; Kay, S.A. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 2014, 8, 622–632. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Liu, Y.F.; Li, Q.T.; Lu, X.; Song, Q.X.; Lam, S.M.; Zhang, W.K.; Ma, B.; Lin, Q.; Man, W.Q.; Du, W.G.; et al. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014, 14, 23–31. [Google Scholar] [CrossRef]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef]
- Kim, H.U.; Jung, S.J.; Lee, K.R.; Kim, E.H.; Lee, S.M.; Roh, K.H.; Kim, J.B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio. 2013, 4, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sedbrook, J.; Benning, C. Ectopic Expression of WRINKLED1 Affects Fatty Acid Homeostasis in Brachypodium distachyon Vegetative Tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Ibanez-Salazar, A.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Ramirez-Alonso, J.I.; Lara-Hernandez, I.; Hernandez-Torres, A.; Paz-Maldonado, L.M.; Silva-Ramirez, A.S.; Banuelos-Hernandez, B.; Martinez-Salgado, J.L.; et al. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2014, 184, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hao, Q.; Bai, L.; Xu, J.; Yin, W.; Song, L.; Xu, L.; Guo, X.; Fan, C.; Chen, Y.; et al. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 2014, 7, 128–135. [Google Scholar] [CrossRef]
- Song, A.; Gao, T.; Li, P.; Chen, S.; Guan, Z.; Wu, D.; Xin, J.; Fan, Q.; Zhao, K.; Chen, F. Transcriptome-Wide Identification and Expression Profiling of the DOF Transcription Factor Gene Family in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 199. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Toyoshima, R.; Kagaya, M.; Takeda, S.; Hattori, T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. Cell Mol. Biol. 2009, 58, 843–856. [Google Scholar] [CrossRef]
- Gibalova, A.; Renak, D.; Matczuk, K.; Dupl’akova, N.; Chab, D.; Twell, D.; Honys, D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol. Biol. 2009, 70, 581–601. [Google Scholar] [CrossRef]
- Mendes, A.; Kelly, A.A.; van Erp, H.; Shaw, E.; Powers, S.J.; Kurup, S.; Eastmond, P.J. bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 2013, 25, 3104–3116. [Google Scholar] [CrossRef]
- Song, Q.X.; Li, Q.T.; Liu, Y.F.; Zhang, F.X.; Ma, B.; Zhang, W.K.; Man, W.Q.; Du, W.G.; Wang, G.D.; Chen, S.Y.; et al. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 2013, 64, 4329–4341. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Li, S.; Lauri, A.; Ziemann, M.; Busch, A.; Bhave, M.; Zachgo, S. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 2009, 21, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hu, X.; Li, C.; Xu, X.; Su, C.; Li, J.; Song, H.; Zhang, X.; Pan, Y. SlbZIP38, a Tomato bZIP Family Gene Downregulated by Abscisic Acid, Is a Negative Regulator of Drought and Salt Stress Tolerance. Genes 2017, 8. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef]
- Alonso, R.; Oclate-Sclnchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Drcege-Laser, W.J.P.C. A Pivotal Role of the Basic Leucine Zipper Transcription Factor bZIP53 in the Regulation of Arabidopsis Seed Maturation Gene Expression Based on Heterodimerization and Protein Complex Formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Zhou, R.; Dossa, K.; Yu, J.; Li, D.; Liu, A.; Mmadi, M.A.; Zhang, X.; You, J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS ONE 2018, 13, e0200850. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Chen, X.; Yang, T.B.; Cheng, Q.; Cheng, Z.M. Genome-Wide Identification of bZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca). Int. J. Genom. 2017, 2017, 3981031. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Meng, D.; Li, M.; Cheng, L. Genomes. Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica). Tree Genet. 2016, 12, 82. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8. [Google Scholar] [CrossRef]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 9413–9422. [Google Scholar] [CrossRef] [PubMed]
- Droge-Laser, W.; Weiste, C. The C/S1 bZIP Network: A Regulatory Hub Orchestrating Plant Energy Homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Sanchez, J. Lipid photosynthesis in olive fruit. Prog. Lipid Res. 1994, 33, 97–104. [Google Scholar] [CrossRef]
- Connor, D.; Fereres, E. The Physiology of Adaptation and Yield Expression in Olive. Hortic. Rev. 2010, 31, 155–229. [Google Scholar] [CrossRef]
- Olive Genome Olive Genome Consortium. Available online: http://olivegenome.org/ (accessed on 4 May 2020).
- National Institute of Agrobiological Sciences. Available online: https://www.nig.ac.jp/nig/ja/ (accessed on 4 May 2020).
- Arabidopsis Information Resource (TAIR). Available online: https://www.arabidopsis.org/index.jsp (accessed on 4 May 2020).
- Joint Genome Institute. Available online: https://jgi.doe.gov/our-projects/ (accessed on 4 May 2020).
- CIRAD. Available online: https://www.cirad.fr/en/ (accessed on 4 May 2020).
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Wang, L.; Yu, J.; Li, D.; Zhang, X. Sinbase: An integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2015, 56, e2. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Deppmann, C.D.; Acharya, A.; Rishi, V.; Wobbes, B.; Smeekens, S.; Taparowsky, E.J.; Vinson, C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: A comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004, 32, 3435–3445. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef]
- Ünel, N.; Cetin, F.; Karaca, Y.; Celik, A.Y.; Baloglu, M. Comparative identification, characterization, and expression analysis of bZIP gene family members in watermelon and melon genomes. Plant Growth Regul. 2018, 87. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. Int. J. Rapid Publ. Rep. Genes Genom. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Popova, O.V.; Suthoff, U.; Luking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Tsugama, D.; Liu, S.; Takano, T. A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol. 2012, 159, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Gibalova, A.; Steinbachova, L.; Hafidh, S.; Blahova, V.; Gadiou, Z.; Michailidis, C.; Muller, K.; Pleskot, R.; Duplakova, N.; Honys, D. Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod. 2017, 30, 1–17. [Google Scholar] [CrossRef]
- Van Leene, J.; Blomme, J.; Kulkarni, S.R.; Cannoot, B.; De Winne, N.; Eeckhout, D.; Persiau, G.; Van De Slijke, E.; Vercruysse, L.; Vanden Bossche, R.; et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J. Exp. Bot. 2016, 67, 5825–5840. [Google Scholar] [CrossRef]
- Maurya, J.P.; Sethi, V.; Gangappa, S.N.; Gupta, N.; Chattopadhyay, S. Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J. Cell Mol. Biol. 2015, 83, 439–450. [Google Scholar] [CrossRef]
- Chuang, C.F.; Running, M.P.; Williams, R.W.; Meyerowitz, E.M. The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 1999, 13, 334–344. [Google Scholar] [CrossRef]
- Lara, P.; Onate-Sanchez, L.; Abraham, Z.; Ferrandiz, C.; Diaz, I.; Carbonero, P.; Vicente-Carbajosa, J. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 2003, 278, 21003–21011. [Google Scholar] [CrossRef]
- Hernandez, M.L.; Padilla, M.N.; Mancha, M.; Martinez-Rivas, J.M. Expression analysis identifies FAD2-2 as the olive oleate desaturase gene mainly responsible for the linoleic acid content in virgin olive oil. J. Agric. Food Chem. 2009, 57, 6199–6206. [Google Scholar] [CrossRef]
- Hernández, M.L.; Sicardo, M.D.; Martínez-Rivas, J.M. Differential Contribution of Endoplasmic Reticulum and Chloroplast ω-3 Fatty Acid Desaturase Genes to the Linolenic Acid Content of Olive (Olea europaea) Fruit. Plant Cell Physiol. 2015, 57, 138–151. [Google Scholar] [CrossRef]
- Parvini, F.; Sicardo, M.D.; Hosseini-Mazinani, M.; Martinez-Rivas, J.M.; Hernandez, M.L. Transcriptional Analysis of Stearoyl-Acyl Carrier Protein Desaturase Genes from Olive (Olea europaea) in Relation to the Oleic Acid Content of the Virgin Olive Oil. J. Agric. Food Chem. 2016, 64, 7770–7781. [Google Scholar] [CrossRef] [PubMed]
- Banilas, G.; Karampelias, M.; Makariti, I.; Kourti, A.; Hatzopoulos, P. The olive DGAT2 gene is developmentally regulated and shares overlapping but distinct expression patterns with DGAT1. J. Exp. Bot. 2011, 62, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhan, M.; Yang, Z.; Zumstein, K.; Chen, H.; Huang, Q. The Major Qualitative Characteristics of Olive (Olea europaea L.) Cultivated in Southwest China. Front. Plant Sci. 2017, 8, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Niu, E.; Shi, A.; Mou, B. Genetic Diversity Analysis of Olive Germplasm (Olea europaea L.) With Genotyping-by-Sequencing Technology. Front. Genet. 2019, 10, 53–61. [Google Scholar] [CrossRef]
- Reboredo-Rodriguez, P.; Gonzalez-Barreiro, C.; Cancho-Grande, B.; Fregapane, G.; Salvador, M.D.; Simal-Gandara, J. Characterisation of extra virgin olive oils from Galician autochthonous varieties and their co-crushings with Arbequina and Picual cv. Food Chem. 2015, 176, 493–503. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Ennouri, K.; Ercisli, S.; Ben Hlima, H.; Hanana, M.; Smaoui, S.; Rebai, A.; Moreau, F. First study of correlation between oleic acid content and SAD gene polymorphism in olive oil samples through statistical and bayesian modeling analyses. Lipids Health Dis. 2018, 17, 74. [Google Scholar] [CrossRef]
- Lodolini, E.M.; Polverigiani, S.; Ali, S.; Mutawea, M.; Qutub, M.; Arabasi, T.; Pierini, F.; Abed, M.; Neri, D. Oil Characteristics of Four Palestinian Olive Varieties. J. Oleo Sci. 2017, 66, 435–441. [Google Scholar] [CrossRef]
- Bruno, L.; Picardi, E.; Pacenza, M.; Chiappetta, A.; Muto, A.; Gagliardi, O.; Muzzalupo, I.; Pesole, G.; Bitonti, M.B. Changes in gene expression and metabolic profile of drupes of Olea europaea L. cv Carolea in relation to maturation stage and cultivation area. BMC Plant Biol. 2019, 19, 428. [Google Scholar] [CrossRef]
- Hernandez, M.L.; Sicardo, M.D.; Alfonso, M.; Martinez-Rivas, J.M. Transcriptional Regulation of Stearoyl-Acyl Carrier Protein Desaturase Genes in Response to Abiotic Stresses Leads to Changes in the Unsaturated Fatty Acids Composition of Olive Mesocarp. Front. Plant Sci. 2019, 10, 251. [Google Scholar] [CrossRef]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rong, S.; Wu, Z.; Cheng, Z.; Zhang, S.; Liu, H.; Huang, Q. Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.). Genes 2020, 11, 510. https://doi.org/10.3390/genes11050510
Rong S, Wu Z, Cheng Z, Zhang S, Liu H, Huang Q. Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.). Genes. 2020; 11(5):510. https://doi.org/10.3390/genes11050510
Chicago/Turabian StyleRong, Siyu, Zhiyang Wu, Zizhang Cheng, Shan Zhang, Huan Liu, and Qianming Huang. 2020. "Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.)" Genes 11, no. 5: 510. https://doi.org/10.3390/genes11050510
APA StyleRong, S., Wu, Z., Cheng, Z., Zhang, S., Liu, H., & Huang, Q. (2020). Genome-Wide Identification, Evolutionary Patterns, and Expression Analysis of bZIP Gene Family in Olive (Olea europaea L.). Genes, 11(5), 510. https://doi.org/10.3390/genes11050510



