Abstract
Bradyrhizobium elkanii USDA61 possesses a functional type III secretion system (T3SS) that controls host-specific symbioses with legumes. Here, we demonstrated that B. elkanii T3SS is essential for the nodulation of several southern Asiatic Vigna mungo cultivars. Strikingly, inactivation of either Nod factor synthesis or T3SS in B. elkanii abolished nodulation of the V. mungo plants. Among the effectors, NopL was identified as a key determinant for T3SS-dependent symbiosis. Mutations of other effector genes, such as innB, nopP2, and bel2-5, also impacted symbiotic effectiveness, depending on host genotypes. The nopL deletion mutant formed no nodules on V. mungo, but infection thread formation was still maintained, thereby suggesting its pivotal role in nodule organogenesis. Phylogenetic analyses revealed that NopL was exclusively conserved among Bradyrhizobium and Sinorhizobium (Ensifer) species and showed a different phylogenetic lineage from T3SS. These findings suggest that V. mungo evolved a unique symbiotic signaling cascade that requires both NFs and T3Es (NopL).
1. Introduction
Bacteria have continuously evolved a multitude of strategies to promote infections in their hosts, including the direct injection of proteins via protein secretion systems. Numerous Gram-negative pathogenic bacteria employ a type III secretion system (T3SS) to translocate their virulence effector proteins (hereafter T3Es) directly into host cells to promote infection and pathogenesis [1]. Similarly, rhizobia, nitrogen-fixing bacteria inducing nodules on leguminous plants, have adopted such a system for symbiotic purposes [2,3,4,5,6]. Rhizobial T3SS-secreted proteins, called nodulation outer proteins (Nops), are injected into host cells as T3Es depending on an extracellular secretion apparatus [7]. Once translocated, some T3Es modulate host functions toward promoting infection and symbiosis with legumes [6], while several T3Es may be recognized as asymbiotic factors directly or indirectly via host resistance (R) proteins or specific receptors, consequently triggering immune responses that restrict nodulation [7].
Bradyrhizobium elkanii USDA61 (hereafter USDA61), first isolated from the soybean [8], possesses a functional T3SS controlling host-specific symbiosis with legumes [9]. Among the T3SS-secreted proteins, at least eight, including NopA, NopB, NopC, NopX, NopL, NopP, Bel2-5, and InnB, have been confirmed by extracellular protein and immunodetection analyses [9,10,11]. Three of them (NopA, B, and X) are required for T3SS components, protein secretions, and translocations [12]. In contrast, NopC, NopL, NopP, InnB, and Bel2-5 are characterized among rhizobial strains as functioning T3Es in plant cells [9,10,13]. Our previous studies demonstrate that InnB is a novel rhizobium-specific effector that functions negatively or positively in symbioses with Vigna radiata and Vigna mungo, respectively. However, the mutation of innB resulted in symbiotic phenotypes significantly different to those of the T3SS-deficient mutant [9]. The innB mutant induced fewer nodules and less plant biomass on V. mungo cv. PI173934 than USDA61 did, whereas the inactivation of T3SS more fully abolished nodulation [9]. However, symbiotic implications of B. elkanii T3SS among V. mungo cultivars and other Vigna species remain unclear. In this study, we further characterized the roles of bradyrhizobial T3SS as a determinant for symbiosis with V. mungo cultivars. Our results revealed that B. elkanii T3SS is essential for nodulating at least two South Asiatic V. mungo cultivars. Furthermore, T3SS-triggered symbiosis relied on a T3E cocktail, in which NopL served as the key player required for establishing early infections and nodule organogenesis.
2. Materials and Methods
2.1. Microbiological and Molecular Techniques
The bacterial strains used in this study are shown in Table 1 and Table S1. Rhizobia and Escherichia coli were grown as described previously [9]. Briefly, rhizobial and Escherichia coli strains were cultured using arabinose–gluconate (AG) [14] and Luria–Bertani (LB) media, respectively. Antibiotics were supplemented to the media or agar plates with appropriate concentrations when required for the culture and selection of mutants. For rhizobial strains, polymyxin (Pol) at 50 µg mL−1, kanamycin (Km) and streptomycin (Sm) at 200 µg mL−1, and spectinomycin (Sp) at 100 µg mL−1. For E. coli strains, Km, Sm, and Sp at 50 µg mL−1.

Table 1.
Bradyrhiozbium elkanii strains used for inoculation tests.
The primer sets used for rhizobial mutant constructions are detailed in Table S2. Single-crossover homologous recombination was conducted as described previously [15]. For double-crossover homologous recombination [16], the single deletion mutants of the effector genes were constructed by overlap extension PCR using two primer pairs amplifying the upstream and downstream fragments flanking the genes of interest, respectively. The PCR fragments were then cloned into the plasmid pK18mobsac derived from pK18mob [17] and linearized by EcoRI and HindIII restriction enzymes. The plasmid derivatives were transformed into rhizobia via triparental conjugations [18]. Similarly, double gene deletion mutants were constructed using the single deletion mutants as the backgrounds. The mutants were finally verified by antibiotic resistance, PCR, and inoculation assays.
2.2. Plant Assays
The seeds of Vigna species used in this study are shown in Table S3. Seeds of Vigna species were surface sterilized and germinated as described previously [15]. To optimize seed germination, Vigna seeds were sanded prior to surface sterilization. One day after transplantation, seedlings were inoculated with B. elkanii strains (1 mL of 107 cells mL−1 per seedling). The plants were grown in a plant growth cabinet (LPH-410SP; NK Systems, Co. Ltd., Osaka, Japan) at 25 °C and 70% humidity under a day/night regimen of 16/8 h. The plants used for microscopic analysis were grown using seed pouches (Daiki Rika Co. Ltd., Saitama, Japan) supplemented with nitrogen-free Broughton and Dilworth (B&D) solution [20], with two plants per pouch [21]. The symbiotic phenotypes, including nodule number, fresh nodule weight, and whole fresh plant weight, were examined 35 days post-inoculation (dpi).
2.3. GUS Assay and Microscopic Analysis
The GUS-tagged B. elkanii strains were constructed by chromosomal integration of the plasmids pCAM120 [22] via conjugations. Single colonies of the constructed mutants were verified by antibiotic resistance, PCR, and GUS assays using X-Glc (Wako Pure Chemical Industries, Osaka, Japan) in dimethylformamide as a substrate. To avoid unexpected effects possibly caused by transposon random insertions, at least two different GUS-tagged mutants were used for inoculation assays. GUS staining and microscopic analysis were performed as described previously [23]. The basal regions of lateral roots were sampled (~1 cm) to observe infection threads (ITs) and nodule organogenesis events [9,21]. Microscopic observations were conducted under a stereoscopic microscope (BX43; Olympus Corporation, Tokyo, Japan).
2.4. Bioinformatics and Statistical Analyses
The amino acid sequences were aligned using MultAlin [24] or Clustal Omega algorithms. Nuclear localization signals (NLSs) of rhizobial NopLs were predicted using cNLS Mapper software [25]. Phosphorylation sites were computationally identified using the NETPHOS 2 program (3.1 Server) [26]. Gene annotation, genome analyses, and tts box searches were conducted as previously described by Nguyen et al. [15]. For phylogenetic analyses, NopL, RhcJ, and TtsI homologs were BLASTX searched against rhizobial and pathogenic bacterial genomic databases. For RhcJ and TtsI, at least five to ten homologs from each species, and five to ten different bacterial species from each genus were selected to construct phylogenetic trees. The amino acid sequences were aligned using the MUSCLE algorithm, and phylogenetic trees were constructed using the neighbor-joining method based on the Poisson model in MEGA version 7.0 with 1000 or 10,000 bootstrap replications. For statistical analyses, one-way analysis of variance (ANOVA) followed by post hoc tests (Fisher’s or Tukey’s tests at p ≤ 0.05) were conducted using Minitab statistical software version 16.0 for multiple test sample comparisons. A two-tailed Student’s t-test was also performed by Microsoft Excel for pairwise comparisons. The p-values < 0.05 was considered statistically significant. The sample sizes and replications are detailed in figure and table legends.
3. Results
3.1. T3SS of B. elkanii USDA61 Played Pivotal Roles in Host-Specific Symbioses with Different Vigna Species and V. mungo Cultivars
Vigna species, such as V. mungo, V. radiata, and V. unguiculata, were efficiently nodulated by Bradyrhizobium spp., including B. elkanii [9], B. japonicum [27], and B. yuanmingense [28]. We previously reported that T3SS of B. elkanii USDA61 abolished V. radiata nodulation but highly promoted symbiosis with V. mungo, suggesting its important roles in controlling host-species-specific symbiotic interactions [9]. To further understand the diverse and distinct symbiotic functions of T3SS in symbiosis among other Vigna species, we inoculated several selected Vigna species with the wild-type USDA61 and T3SS-deficient mutant BErhcJ. Interestingly, USDA61 T3SS promoted symbioses with V. unguiculata and V. angularis, but impaired V. aconitifolia and V. trinervia (Table 1). However, the T3SS-induced symbiotic effects on these Vigna species were not as strong as previously observed in V. radiata and V. mungo [9], since plant weights were not significantly different between USDA61 and BErhcJ strains (Figure S1).
To further explore the T3SS-induced cultivar-specific nodulation, we conducted inoculation assays using a collection of V. mungo cultivars from different countries (Table S3). The results revealed that USDA61 T3SS exhibited compatibility and incompatibility depending on the V. mungo cultivars. Indeed, T3SS was responsible for negative effects on the nodulation of V. mungo cultivars VM3003, U-THONG2, and CQ5785, but promoted symbiosis with PI173934, MASH, IBPGR2775-3, MAFF2002M3, and CQ5785 (Table 2, Figure S2). Strikingly, among these V. mungo plants, inactivation of the T3SS abolished nodulation on IBPGR2775-3 and MASH occurred (Figure 1 and Figure 2). Furthermore, mutations on T3SS or nodC exhibited similar nodulation phenotypes, comparable to those of the ttsI/nodC double mutant BEttsInod, suggesting that T3SS and NFs were both key determinants in host-specific symbiosis with V. mungo.

Table 2.
Symbiotic properties of V. unguiculata, V. trinervia, V. angularis, V. aconitifolia, and V. mungo varieties inoculated with B. elkanii strains a.
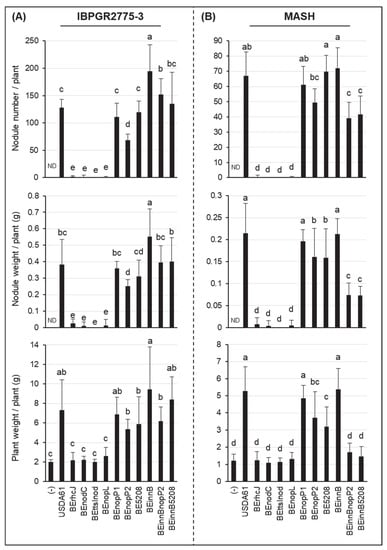
Figure 1.
Symbiotic properties of Vigna mungo cv. IBPGR2775-3 (A) and MASH (B) inoculated with B. elkanii strains. The data shown are the means of 20 to 30 from five or six independent inoculation assays at 35 dpi. The error bars indicate standard deviations. Means followed by different letters are significantly different at the 5% level (p ≤ 0.05 by Tukey’s tests). ND, not detected.
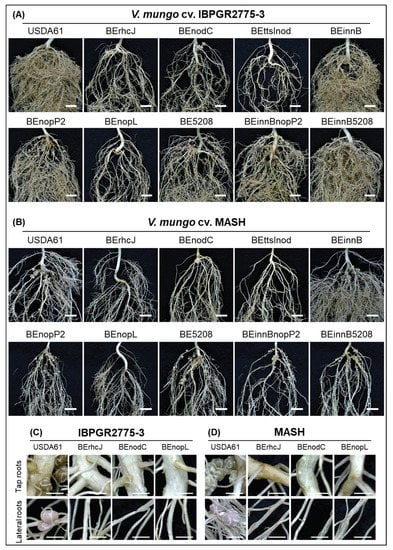
Figure 2.
Roots of V. mungo cv. IBPGR2775-3 (A) and MASH (B) inoculated with the B. elkanii strains at 35 dpi. Nodulation properties IBPGR2775-3 (C) and MASH (D). Scale bars: 1 cm for (A,B) and 0.25 cm for (C,D).
3.2. Identification of B. elkanii USDA61 T3Es Involved in Determining Symbiotic Efficiency on V. mungo
To further identify T3Es determining V. mungo symbiosis, symbiotic phenotypes of the T3E mutants were investigated using inoculation assays. B. elkanii possessed at least two nopP gene copies, designated nopP1 (locus tag BE61_77110) and nopP2 (locus tag BE61_80730) (Data S1). NopP1 likely did not function in symbiosis, whereas NopP2 promoted nodulation on IBPGR2775-3 and MASH, as the deletion of nopP2 significantly reduced nodulation and plant biomass (Figure 1 and Figure 2). Moreover, we identified Bel2-5 as another T3E functioning positively in symbiosis with V. mungo. However, its positive effects were likely weak for PI173934 (Figure S3), compared to IBPGR2775-3 (Figure 1A). Noticeably, double deletions of innB/nopP2 or innB/bel2-5 enhanced nodulation of IBPGR2775-3 and significantly reduced MASH nodulation, as compared to the single mutants of nopP2 and bel2-5. Although nopP2, bel2-5, and innB are beneficial for nodulation on PI173934, their double deletion mutants could not restore the T3SS mutant-induced phenotypes (Figure S3).
Although B. elkanii USDA61 could efficiently nodulate V. mungo, a mixture of pink and white nodules was typically observed on the roots of IBPGR2775-3 at 35 dpi—the tap (white) and lateral (pink) root nodules (Figure 2C). In MASH, nodules were primarily formed around the tap roots, but most exhibited a white color observed at 35 dpi, indicating that early nodulation and nitrogen fixation or weak nitrogenase activities probably occurred (Figure 2D). Such a phenomenon was also observed in the PI173934 roots inoculated with B. elkanii strains (Figure S3). Exceptionally, the formation of the lateral root nodules was highly reduced in IBPGR2775-3 plants inoculated with the nopP2 or bel2-5 mutant backgrounds (Figure 2A).
3.3. NopL of B. elkanii USDA61 is a Key Determinant for T3SS-Dependent Nodulation in South Asiatic V. mungo Cultivars
Among the T3Es of B. elkanii USDA61, NopL served as a key determinant for the T3SS-triggered nodulation of V. mungo plants. Impressively, the deletion of nopL mimics the T3SS mutant in MASH and IBPGR2775-3 cultivars (Figure 1 and Figure 2). Likewise, the nopL and T3SS mutants exhibited comparable symbiotic phenotypes on PI173934 (Figure S3). Noticeably, a few sporadic nodules were formed occasionally on the MASH and IBPGR2775-3 roots inoculated with T3SS, nopL, and nodC mutants. However, these induced nodules were white or black, indicating inefficient infection. The formation of bump-like structures or immature nodules was also not observed on the roots inoculated with T3SS or nopL mutants at 30 dpi (Figure 2).
3.4. B. elkanii USDA61 NopL is Required for Early Nodule Development
To further explore the NopL functions in the nodulation process, we followed the early infection of V. mungo cv. MASH inoculated with GUS-tagged strains of USDA61 (USDA61G), nodC mutant (BEnodCG), and nopL mutant (BEnopLG) (Figure 3). A number of infected young nodules, infection threads (ITs), and nodule primordia were observed in the roots inoculated with USDA61G. BEnodCG and BEnopLG induced numerous rhizobial colonization sites; however, both failed to induce infected nodule primordia and young nodules as USDA61G. The ITs were not observed in the roots inoculated with BEnodCG; however, BEnopLG could induce IT formation at similar levels as USDA61G. Impressively, several nodule bump-like structures were formed on the roots inoculated with the BEnodCG mutant (Figure 3A), consequently developing young nodules occasionally observed at 14 dpi (Figure 3B). However, such organogenesis events were almost absent in the roots inoculated with the BEnopLG mutant.
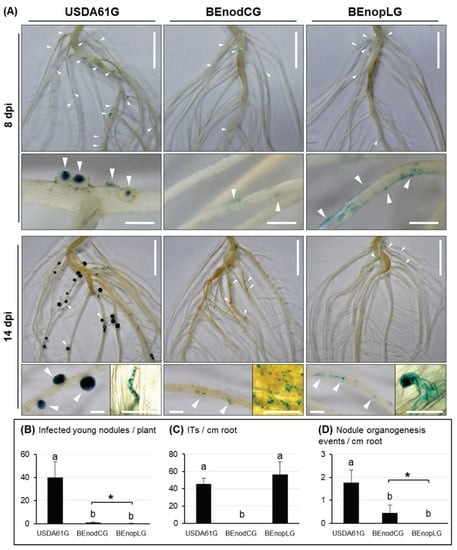
Figure 3.
Infection and nodulation properties of V. mungo cv. MASH inoculated with the GUS-tagged B. elkanii strains. (A) Photos of the entire roots, infection threads (ITs), infected nodule primordia, and young nodules at 8 and 14 dpi, respectively. The rhizobial colonization and nodule organogenesis events are marked with white triangles. Scale bars: 1 cm, entire roots; 1 mm, nodule primordia and young nodules; and 50 µm, ITs and rhizobial colonization. Numbers of infected young nodules per plant (B), ITs (C), and nodule organogenesis events (D) induced per cm of the basal regions in lateral roots at 14 dpi. The data shown are means of six or seven plants (three root samples per plant) and the error bars indicate standard deviations. Means followed by different letters are significantly different at the 5% level (p ≤ 0.05 by Tukey’s test). “*”, p < 0.05 by Student’s t-test.
3.5. Structural Features of B. elkanii USDA61 NopL
We conducted in silico analyses using several NopL homologs among rhizobia as references. Like other sinorhizobial NopLs, a number of putative phosphorylation sites were also predicted in USDA61 NopL, including seven serine–proline (SP) motifs typical for phosphorylation sites in MAPK (mitogen-activated protein kinase) substrates (Figure S6A) [7]. These phosphorylation sites were primarily distributed in the N-terminal and internal portions, in which the serine (S) residues are dominant (Figure S6A and Data S4). Intriguingly, USDA61 NopL also contained two SP-containing repeats (SPQPDS and SPQPGS) specifically conserved in NopLs of Sinorhizobium but not B. japonicum and B. diazoefficiens (Figure S6A and Data S4). We also identified at least three tandem repeat motifs (3 × SQAGP) in the internal region of USDA61 NopL (Figure S6A and Data S4). Interestingly, alignment analysis verified that these tandem repeat motifs exhibited high similarities to the two catalytic motifs previously identified in S. fredii NGR234 NopL [29] (Figure S6B). In addition, the USDA61 NopL, like NGR234 NopL, possessed a nuclear localization signal (NLS) motif situated at the N-terminal Serine-rich region, whereas NopLs of B. diazoefficiens USDA110, B. japonicum USDA6, and S. fredii HH103 did not (Data S3). Notably, NopLs of USDA110 and USDA6 were likely truncated versions lacking the C-terminal portion and functional catalytic domains evolved in the NopLs of USDA61 and NGR234 (Data S4).
3.6. Phylogenetic Analyses of Rhizobial NopLs
To further understand the evolutionary history of NopL, phylogenetic analysis using NopL homologs identified among bacterial groups was performed. Of interest, NopL were exclusively conserved rhizobia, dominantly bradyrhizobia, whereas several were identified in Sinorhizobium, including S. fredii, Ensifer, and Microvirga (Figure 4). Noticeably, these rhizobia could nodulate efficiently and/or be the main symbionts of Vigna plants. No NopL homolog has been identified among Mesorhizobium spp., Rhizobium spp., or B. elkanii strains USDA76 and USDA94, the two closest relatives of USDA61. Surprisingly, the USDA61 NopL group was phylogenetically related to the NopLs of S. fredii, Ensifer, and Microvirga. In contrast, NopL homologs of the group comprised of B. diazoefficiens, B. japonicum, and B. liaoningense were separated from another B. elkanii NopL group excluding the USDA61 NopL. Among the homologs, USDA61 NopL was relatively close to NopLs of B. elkanii NBRC14791 (a soybean symbiont) and B. vorense sp. nov. CI-1B (a symbiont of Cajanus cajan, pigeon pea).
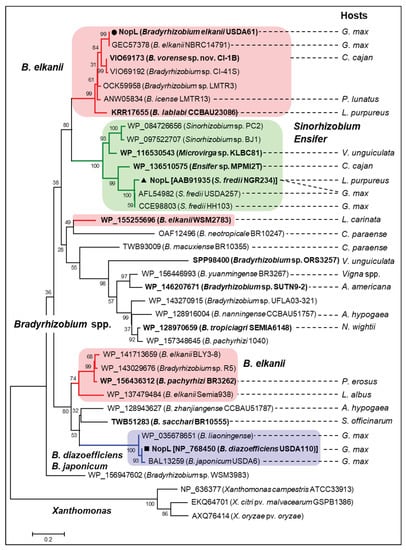
Figure 4.
Phylogenetic analysis of B. elkanii USDA61 NopL and its homologs among rhizobia. Bootstrap values are expressed as percentages of 10,000 replications. The groups of B. elkanii (red), B. japonicum/B. diazoefficiens (blue), and Sinorhizobium/Ensifer (green) NopLs are highlighted. The Xanthomonas DNA polymerase III subunit γ/τ is used as an outgroup. The main hosts or hosts where rhizobial strains were first isolated are shown: A. americana, Aeschynomene americana; A. hypogaea, Arachis hypogaea (peanut); C. cajan, Cajanus cajan (pigeon pea); C. paraense, Centrolobium paraense; G. max, Glycine max (soybean); L. albus, Lupinus albus (lupin); L. carinata, Leobordea carinata (leptis); L. purpureus, Lablab purpureus; N. wightii, Neonotonia wightii (perennial soybean), P. erosus, Pachyrhizus erosus (jicama/Mexican yam bean); P. lunatus, Phaseolus lunatus (lima bean); S. officinarum, Saccharum officinarum (sugarcane); and V. unguiculata, Vigna unguiculata (cowpea).
4. Discussion
In this study, we characterized symbiotic interactions between B. elkanii and V. mungo where both T3SS and NFs play essential roles, and the absence of NFs or T3SS abolished nodulation. Of particular interest, NopL of USDA61 triggered early infection and nodule organogenesis on V. mungo in an NF-dependent manner (Table 2). The NFs induced IT formation, whereas NopL ensured efficient early infection and nodule formation in an NF-dependent manner (Figure 3). Notably, nodule primordia-like structures occasionally formed on V. mungo roots inoculated with the nodC mutant, but not the nopL mutant (Figure 3A), suggesting that NopL might also be capable of triggering nodulation in the absence of NFs, although with very low efficiency.
Previous studies have reported that NopL of S. fredii NGR234 phosphorylated by host MAPK in vitro [30] interfered with MAPK signaling in planta and suppressed premature nodule senescence [29]. In addition, rhizobial NopLs have also exhibited weak similarities to DNA polymerase III subunits γ and τ domain (PRK07764) [7]. However, how NopLs are involved in DNA interactions remain unknown. Our in silico analyses revealed high similarities between NopLs of USDA61 and NGR234, especially in phosphorylation sites/motifs and catalytic domains (Figure S6, Data S4A,B), suggesting their evolutionarily conserved functions. Notably, inactivation of nopL excluded nodulation on IBPGR2775-3 and MASH (Figure 1 and Figure 2) but largely reduced nodule numbers of PI173934 (Figure S3). These results suggest that USDA61 NopL, possibly together with other T3Es, suppresses MAPK-triggered responses and/or modulates NF-dependent symbiosis signaling toward determining the nodulation of V. mungo, depending on host cultivars (Figure S7). Therefore, NopL might act toward other T3Es or be involved in key steps of the symbiotic process. B. diazoefficiens USDA110 and B. japonicum USDA6 also recruit at least one NopL copy. However, these NopLs are remarkably different to NopLs of USDA61 and NGR234 in terms of the phylogenetic relationship, length, and evolved functional motifs (Figure 4 and Data S4C), partly reflecting their differentiated functions.
Inactivation of other T3Es, such as NopP2, Bel2-5, and InnB, also impacts symbiotic effectiveness depending on host cultivar (Figure 1, Figure 2 and Figure S3). Previous studies have revealed phosphorylation activities of the sinorhizobial NopP in vitro [31]. Moreover, at least three NopP2 spots with different pIs were previously detected in the extracellular proteins of USDA61 (Data S2) [11], indicating that NopP2 was possibly phosphorylated in rhizobial cultures post secretion. Hypothetically, NopP2 may be phosphorylated by an as-yet-unknown host kinase to suppress host defense responses and/or promote later nodulation of V. mungo (Figure S7). Unlike NopP2, Bel2-5 contains a C-terminal ubiquitin-like protease (Ulp) domain exhibiting similarity to Xanthomonas XopDs [10], suggesting that protease activities might be involved in Bel2-5 symbiotic functions. The ability to promote nodulation on V. mungo cultivars indicated that Bel2-5 may modulate host symbiosis signaling and/or suppress host defense responses at different levels, depending on host genotypes (Figure S7). Differing from NopP2 and Bel2-5, InnB was a specific “double-edged sword” in symbiosis with Vigna species and V. mungo cultivars. Certain dual effects of InnB were also highly dependent on host genotypes and correlated with the presence/absence of NopP2 and Bel2-5. In S. fredii HH103, NopL and NopP were detrimental to symbiosis with soybean. Conversely, NopL exhibited a negative effect on nodulation on cowpeas, whereas NopP promoted nodulation [32]. However, double deletion of HH103 nopL and nopP largely caused a decline in nodulation on both soybean and cowpea [32]. Similarly, our results represent the distinct and diverse functions of a T3E cocktail in plant cells (Figure S7); thus, the absence of a T3E may affect T3E-triggered signaling pathways directly or indirectly, resulting in enhanced or reduced nodulation.
Although exhibiting T3SS-determined nodulation, differences in nodulation properties and morphological characteristics of the seed, seedling, and stipule were observed among the three South Asiatic V. mungo cultivars (Figure S5). PI173934 and IBPGR2775-3 exhibited some similarities in plant and seed features; however, their stipule morphologies were remarkably different. Interestingly, MASH possessed morphological features of seeds and seedlings that differed from those of PI173934 and IBPGR2775-3. However, the stipule features of MASH were morphologically similar to those of IBPGR2775-3 (Figure S5). These morphological variations may partly reflect the common and distinct traits evolved among these V. mungo plants, potentially related to their symbiotic relationships with specific rhizobial strains and T3Es. Our observations of nodulation properties also strengthened the knowledge of the distinct differences among these cultivars (Figure 2 and Figure S3). Genome analyses of these V. mungo cultivars will reveal relationships among domestication, morphological evolution, and symbiotic phenotypes of V. mungo cultivars.
Phylogenetic analyses revealed that the T3SS of USDA61 is relatively close to those of B. elkanii, B. pachyrhizi, and B. jicamae (Figure S4); the latter two species are novel bradyrhizobia isolated from Pachyrhizus erosus (Yam bean/Jicama) [33]. Although co-localized in T3SS, USDA61 NopL was phylogenetically close to its homologs in S. fredii (Ensifer) (Figure 4), representing their shared functional features and/or similar origins. Surprisingly, NopL homologs have not been identified among genomic databases of Mesorhizobium spp., Rhizobium spp., or B. elkanii strains USDA76 and USDA94, suggesting that NopL is likely not required for the symbiosis of these rhizobia. Our results found that the USDA61-type T3SS was probably horizontally transferred among B. elkanii, B. pachyrhizi, and B. jicamae species as the closest relatives [33]. On the other hand, nopL and other T3E genes might be recruited separately during evolution events and symbiosis with different host legumes.
Rhizobial T3SS (T3Es) and NFs were found to be the key determinants for alternative symbiosis with V. mungo plants. For decades, it has been believed that rhizobial NFs play an essential role in activating infection and symbiosis signaling pathways for nodulation on legumes [34]. We previously reported that B. elkanii T3SS activated nodulation signaling in soybean [19]. Interestingly, the NF-deficient B. elkanii mutant induced bump-like structures that were not evident on plants inoculated with T3SS mutants [19]. The findings in this study advance our understanding that both T3SSs and NFs serve as key determinants for efficient symbiosis with V. mungo plants (Figure 2 and Figure 4). Such T3SS-triggered symbiosis has not previously been observed in G. max [19], V. unguiculata [32], V. radiata [11,15], L. japonicus [35], or Aeschynomene spp. [36]. Furthermore, expressions of the T3SS-related, T3E, and nod genes were highly regulated by the common activator NodD and legume flavonoids [18], showing that T3SS (T3Es) and NFs might be simultaneously co-expressed during symbiotic interactions with host legumes. Collectively, these findings suggest that T3SS and NFs might be mutualistically co-evolved in rhizobia, regulated by symbiosis-related common regulators, and act synergistically toward triggering and maintaining efficient nodulation.
5. Conclusions
Here, we demonstrated diverse roles of B. elkanii T3SS in symbiosis with several Vigna species. Of particular interest, we demonstrated that the T3SS of B. elkanii USDA61 is essential for symbiosis with at least two cultivars of V. mungo. Among the T3Es, NopL served as a key player for inducing nodule primordia. Meanwhile, NF synthesis was required for infection thread formation. These findings provide useful insights into the genetic basis of host specificity in the rhizobium-legume symbiosis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/5/474/s1. Table S1: Bacterial strains and plasmids used in this study. Table S2: DNA oligonucleotide primers used in this study. Table S3: Legume seeds used in this study. Figure S1: Symbiotic properties of several Vigna species inoculated with B. elkanii strains. Figure S2: Symbiotic properties of several V. mungo varieties inoculated with the B. elkanii strains. Figure S3: Roots (A) and symbiotic properties (B) of V. mungo cv. PI173934 inoculated with the B. elkanii strains. Figure S4: Phylogenetic analysis of the T3SS structural component protein RhcJ (A) and transcriptional regulator TtsI (B) of B. elkanii USDA61. Figure S5: Morphological characteristics of V. mungo cv. PI173934, IBPGR2775-3, and MASH. Figure S6: Structural features of B. elkanii NopL. Figure S7: Putative model of T3E-triggered symbiotic interactions with V. mungo. Data S1: Nucleotide sequences of nopL, nopPs, rhcJ, and ttsI in the B. elkanii USDA61 genome. Data S2: Alignment of the NopP1, NopP2, and NopP peptide fragments previously detected in the extracellular proteins of B. elkanii USDA61. Data S3: Predictions of nuclear localization signals (NLSs) among rhizobial NopLs. Data S4: Alignment analysis of B. elkanii USDA61 NopL and its homologs among bradyrhizobia and sinorhizobia (A), sinorhizobia (B), B. diazoefficiens/B. japonicum (C), and several selected rhizobia (D).
Author Contributions
H.P.N. and S.O. designed the study and wrote the manuscript; H.P.N. and S.T.N.R. performed research; H.P.N. coordinated experiments and analyzed data; and M.Y., N.T., and S.O. provided analytic tools/materials. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by JSPS Kakenhi (19H02860 and 19K22303).
Acknowledgments
We would like to thank K. Suzuki and Wint Phoo (Tokyo University of Agriculture and Technology (TUAT), Tokyo, Japan) for their technical support; Michael Göttfert (Technische Universität Dresden, Dresden, Germany) for his critical and helpful discussions; and Editage (www.editage.com) for English language editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Deakin, W.J.; Broughton, W.J. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Kimbrel, J.A.; Thomas, W.J.; Jiang, Y.; Creason, A.L.; Thireault, C.A.; Sachs, J.L.; Chang, J.H. Mutualistic co-evolution of type III effector genes in Sinorhizobium fredii and Bradyrhizobium japonicum. PLoS Pathog. 2013, 9, e1003204. [Google Scholar] [CrossRef] [PubMed]
- Angus, A.A.; Agapakis, C.M.; Fong, S.; Yerrapragada, S.; Estrada-de los Santos, P.; Yang, P.; Song, N.; Kano, S.; Caballero-Mellado, J.; de Faria, S.M.; et al. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 2014, 9, e83779. [Google Scholar] [CrossRef] [PubMed]
- Viprey, V.; Del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Tampakaki, A.P. Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front. Plant Sci. 2014, 5, 1–19. [Google Scholar] [CrossRef]
- Nelson, M.S.; Sadowsky, M.J. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef]
- Staehelin, C.; Krishnan, H.B. Nodulation outer proteins: Double-edged swords of symbiotic rhizobia. Biochem. J. 2015, 470, 263–274. [Google Scholar] [CrossRef]
- Keyser, H.H.; van Berkum, P.; Weber, D.F. A comparative study of the physiology of symbioses formed by Rhizobium japonicum with Glycine max, Vigna unguiculata, and Macroptilium atropurpurem. Plant Physiol. 1982, 70, 1626–1630. [Google Scholar] [CrossRef]
- Nguyen, H.P.; Ratu, S.T.N.; Yasuda, M.; Göttfert, M.; Okazaki, S. InnB, a novel Type III effector of Bradyrhizobium elkanii USDA61, controls symbiosis with Vigna species. Front. Microbiol. 2018, 9, 3155. [Google Scholar] [CrossRef]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef]
- Okazaki, S.; Zehner, S.; Hempel, J.; Lang, K.; Göttfert, M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009, 295, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Deakin, W.J.; Marie, C.; Saad, M.M.; Krishnan, H.B.; Broughton, W.J. NopA is associated with cell surface appendages produced by the type III secretion system of Rhizobium sp. strain NGR234. Mol. Plant-Microbe Interact. 2005, 18, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC is a Rhizobium-specific Type 3 secretion system effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Tully, R.E.; Cregan, P.B.; Keyser, H.H. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl. Environ. Microbiol. 1987, 53, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Miwa, H.; Kaneko, T.; Sato, S.; Okazaki, S. Identification of Bradyrhizobium elkanii genes involved in incompatibility with Vigna radiata. Genes (Basel) 2017, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, Y.; Sakanaka, M.; Fukuma, H.; Murayama, H.; Kano, Y.; Fukiya, S.; Yokota, A. Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: Functional analysis of the α-galactosidase gene for raffinose assimilation. Appl. Environ. Microbiol. 2012, 78, 4984–4994. [Google Scholar] [CrossRef] [PubMed]
- Schafer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Puhler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 2002, 15, 1228–1235. [Google Scholar] [CrossRef]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef]
- Broughton, W.J.; Dilworth, M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef]
- Yasuda, M.; Miwa, H.; Masuda, S.; Takebayashi, Y.; Sakakibara, H.; Okazaki, S. Effector-triggered immunity determines host genotype-specific incompatibility in legume-Rhizobium symbiosis. Plant Cell Physiol. 2016, 57, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.J.; Sessitsch, A.; Corbo, J.C.; Giller, K.E.; Akkermans, A.D.L.; Jefferson, R.A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 1995, 141, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Hattori, Y.; Saeki, K. The Mesorhizobium loti purB gene is involved in infection thread formation and nodule development in Lotus japonicus. J. Bacteriol. 2007, 189, 8347–8352. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Wang, E.T.; Tian, C.F.; Wang, F.Q.; Han, L.L.; Chen, W.F.; Chen, W.X. Bradyrhizobium elkanii, Bradyrhizobium yuanmingense and Bradyrhizobium japonicum are the main rhizobia associated with Vigna unguiculata and Vigna radiata in the subtropical region of China. FEMS Microbiol. Lett. 2008, 285, 146–154. [Google Scholar] [CrossRef]
- Appunu, C.; N’Zoue, A.; Moulin, L.; Depret, G.; Laguerre, G. Vigna mungo, V. radiata and V. unguiculata plants sampled in different agronomical–ecological–climatic regions of India are nodulated by Bradyrhizobium yuanmingense. Syst. Appl. Microbiol. 2009, 32, 460–470. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, X.-J.; Lu, H.-B.; Xie, Z.-P.; Staehelin, C. Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: Symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 2011, 286, 32178–32187. [Google Scholar] [CrossRef]
- Ge, Y.-Y.; Xiang, Q.-W.; Wagner, C.; Zhang, D.; Xie, Z.-P.; Staehelin, C. The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 2016, 67, 2483–2494. [Google Scholar] [CrossRef]
- Skorpil, P.; Saad, M.M.; Boukli, N.M.; Kobayashi, H.; Ares-Orpel, F.; Broughton, W.J.; Deakin, W.J. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 2005, 57, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 nodulation outer protein NopI is a determinant for efficient nodulation of soybean and cowpea plants. Appl. Environ. Microbiol. 2017, 83, e02770-16. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Bahena, M.H.; Peix, A.; Rivas, R.; Camacho, M.; Rodríguez-Navarro, D.N.; Mateos, P.F.; Martínez-Molina, E.; Willems, A.; Velázquez, E. Bradyrhizobium pachyrhizi sp. nov. and Bradyrhizobium jicamae sp. nov., isolated from effective nodules of Pachyrhizus erosus. Int. J. Syst. Evolut. Microbiol. 2009, 59, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Okabe, S.; Higashi, M.; Shimoda, Y.; Sato, S.; Tabata, S.; Hashiguchi, M.; Akashi, R.; Göttfert, M.; Saeki, K. Identification and functional analysis of type III effector proteins in Mesorhizobium loti. Mol. Plant-Microbe Interact. 2010, 23, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Moulin, L.; Vallenet, D.; Barbe, V.; Cytryn, E.; Avarre, J.-C.; Jaubert, M.; Simon, D.; Cartieaux, F.; Prin, Y.; et al. Legumes symbioses: Absence of Nod genes in photosynthetic bradyrhizobia. Science 2007, 316, 1307–1312. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).