The Peptidoglycan Biosynthesis Gene murC in Frankia: Actinorhizal vs. Plant Type
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Gene Expression Analysis using Real-Time Quantitative PCR (RT-qPCR)
2.3. Phylogenetic and Synteny Analysis
2.4. Sequence Similarity Analysis
2.5. Tetranucleotide Frequency
2.6. Protein Modelling
3. Results
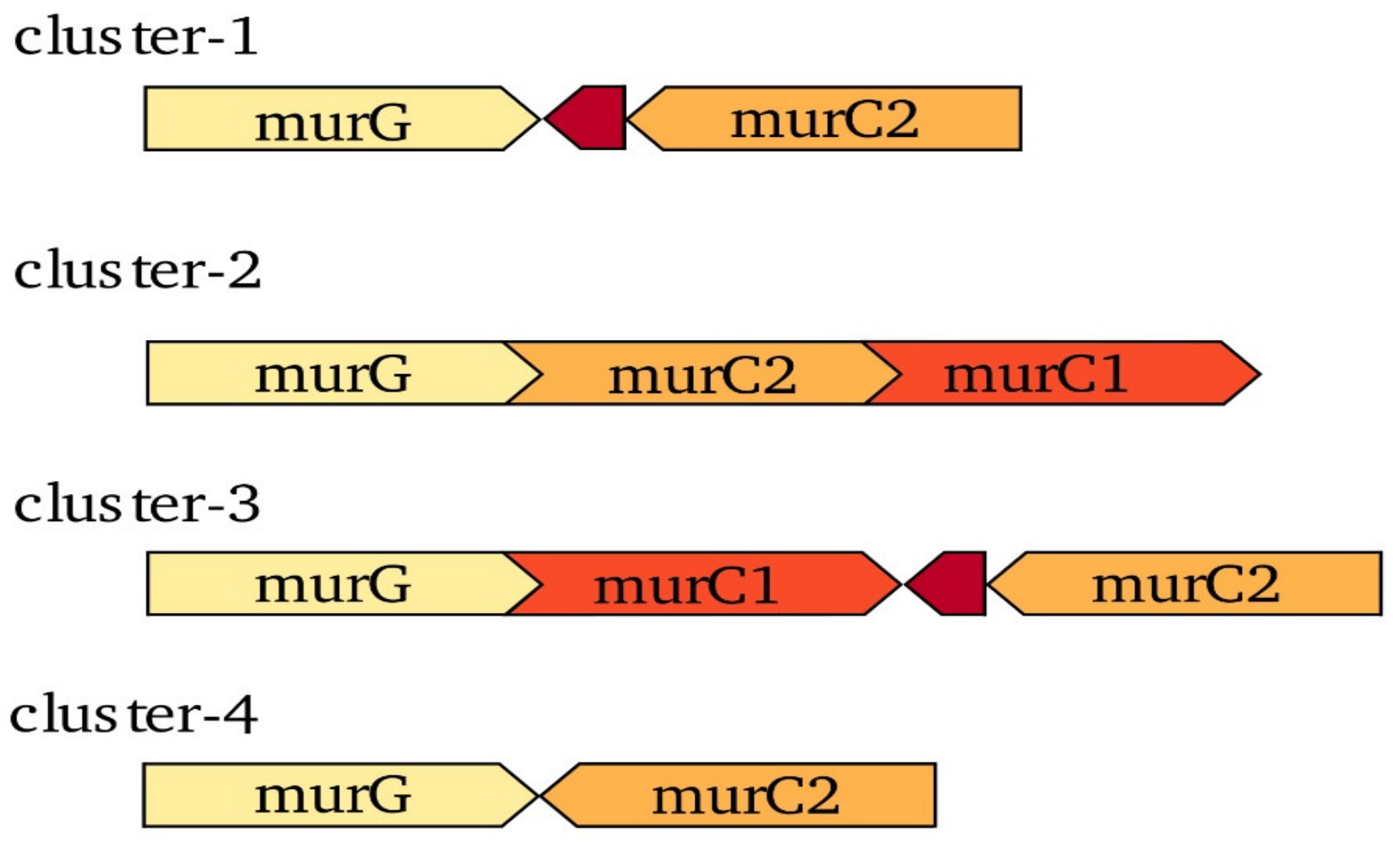
3.1. Synteny Analysis Indicates Differences in Gene Organization of murC in the Four Frankia Clusters
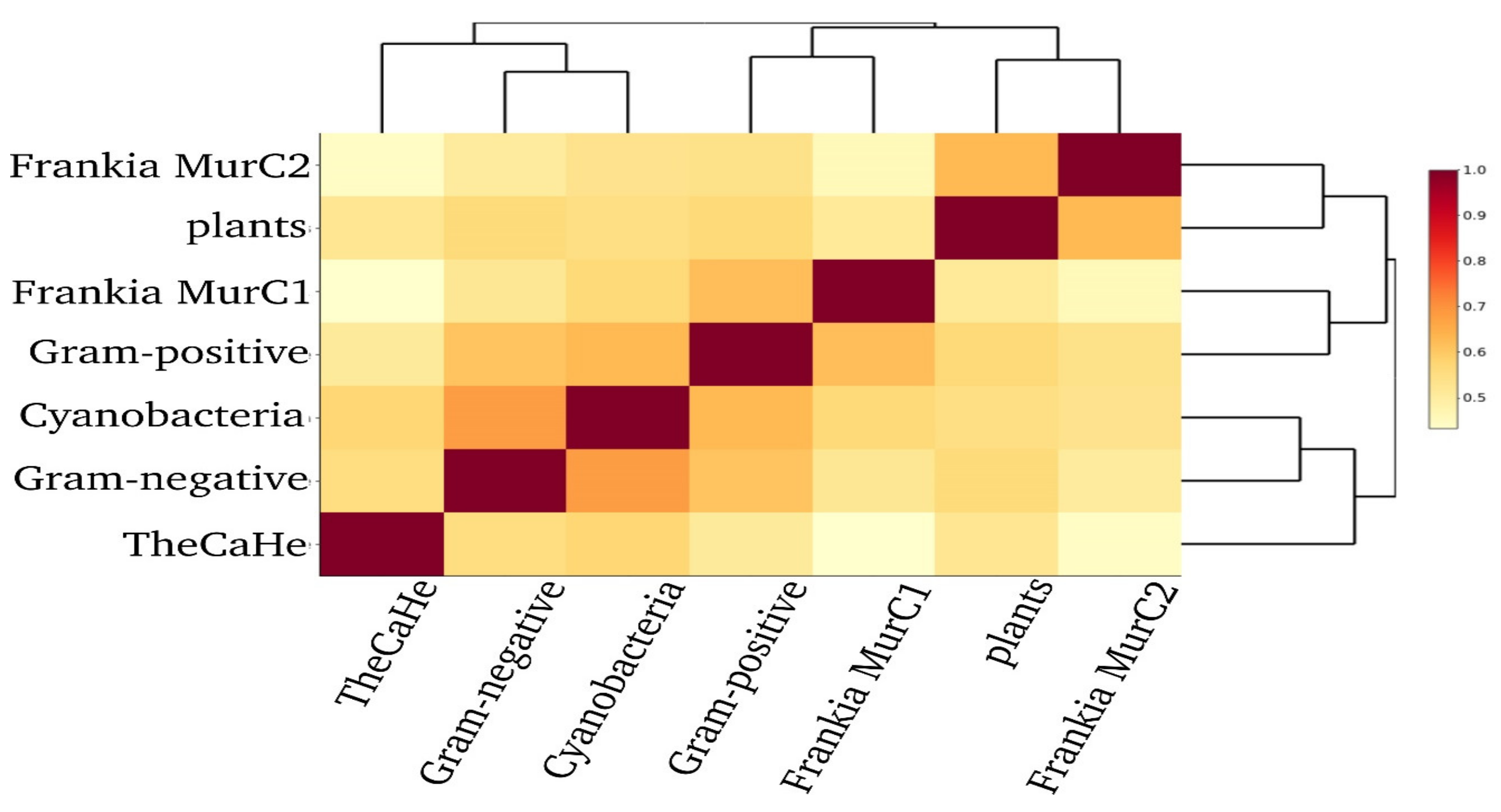
3.2. Consensus Amino Acid Sequences of Frankia MurC2 Shows Higher Similarity to MurC from Plants than from other Actinobacteria
3.3. No Sequence Evidence for Horizontal Gene Transfer (HGT) could be Found
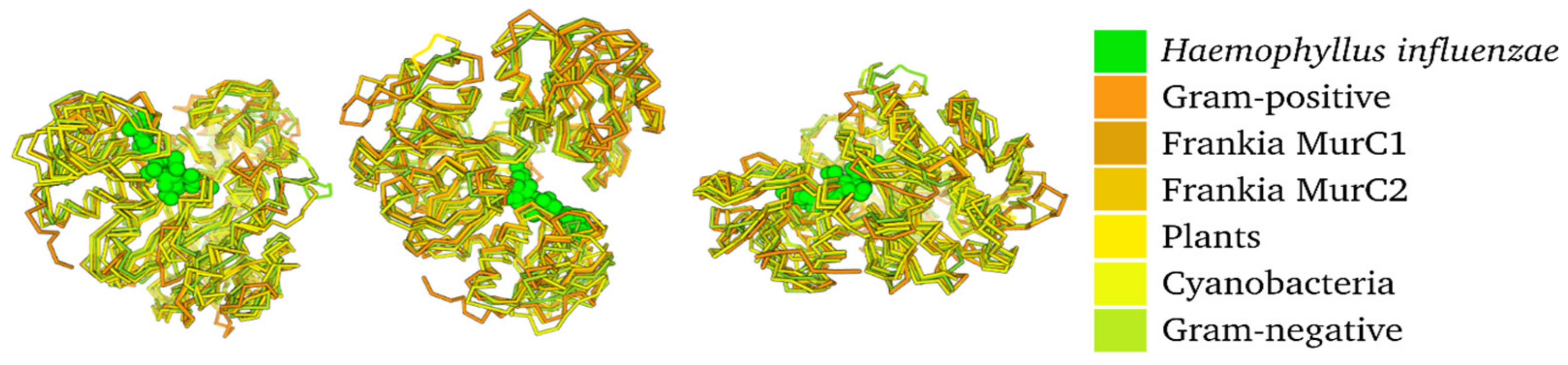
3.4. Protein Modelling Reveals Little Structural Difference among Different Groups of MurC Sequences
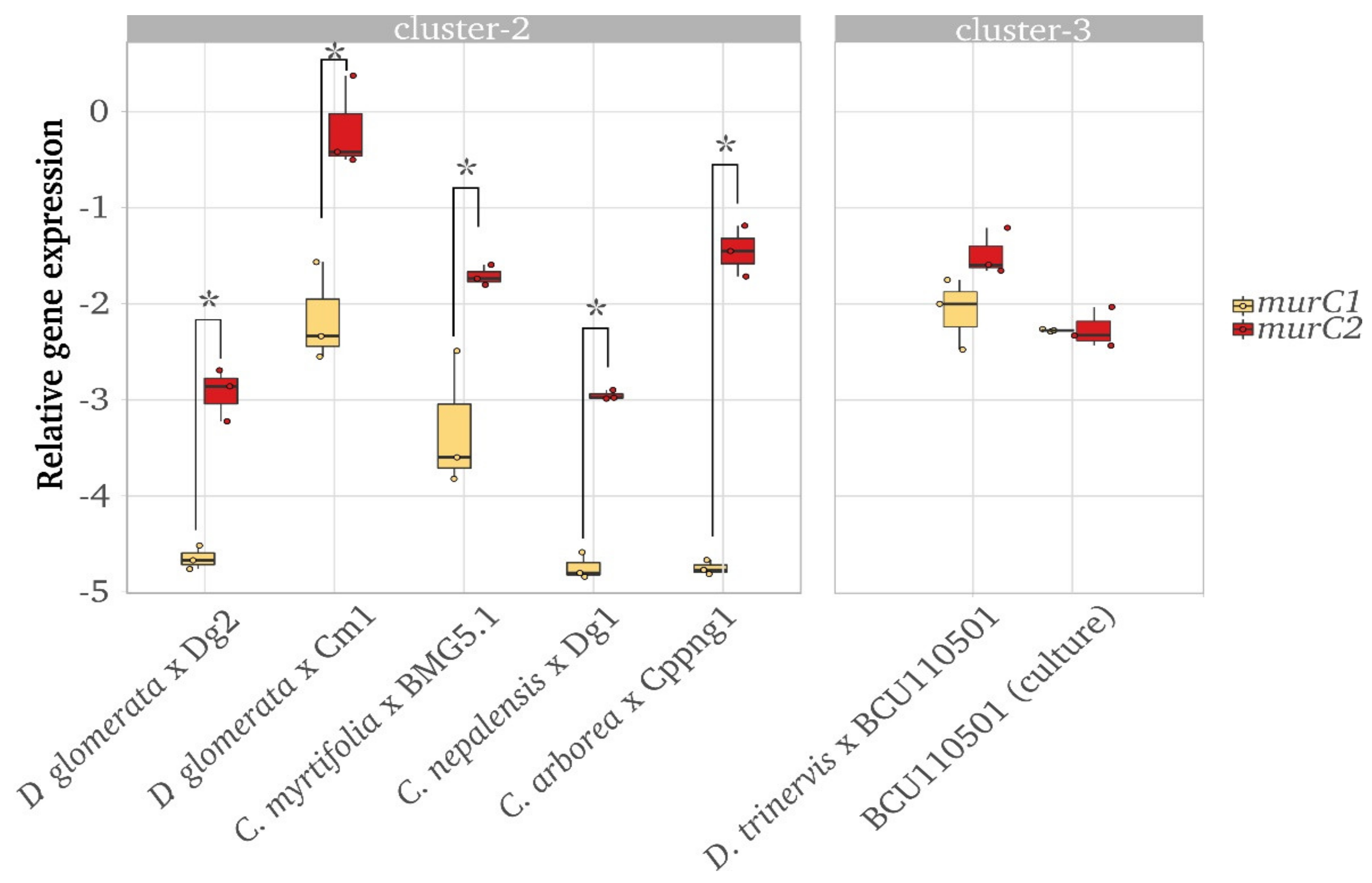
3.5. Gene Expression Data Shows Differential Expression of murC1 and murC2 in Frankia Cluster-2 in Nodules, but not in Frankia Cluster-3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gantt, E. Supramolecular membrane organization. In The Molecular Biology of Cyanobacteria; Bryant, D.A., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994; pp. 119–138. [Google Scholar]
- Takano, H.; Takechi, K. Plastid peptidoglycan. Biochim. Biophys. Acta. 2010, 18, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Seligman, S.J. Architecture of peptidoglycan: More data and more models. Trends Microbiol. 2009, 18, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Toyoshima, M.; Tajima, N.; Takechi, K.; Takano, H. Single-pixel densitometry revealed the presence of peptidoglycan in the intermembrane space of the moss chloroplast envelope in conventional electron micrographs. Plant Cell Physiol. 2017, 58, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef]
- Munshi, T.; Gupta, A.; Evangelopoulos, D.; Guzman, J.D.; Gibbons, S.; Keep, N.H.; Bhakta, S. Characterisation of ATP-dependent mur ligases involved in the biogenesis of cell wall peptidoglycan in Mycobacterium tuberculosis. PLoS ONE 2013, 8, e60143. [Google Scholar] [CrossRef]
- Videau, P.; Rivers, O.S.; Ushijima, B.; Oshiro, R.T.; Kim, M.J.; Philmus, B.; Cozy, L.M. Mutation of the murC and murB genes impairs heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 2016, 198, 1196–1206. [Google Scholar] [CrossRef]
- Garcia, M.; Myouga, F.; Takechi, K.; Sato, H.; Nabeshima, K.; Nagata, N.; Takio, S.; Shinozaki, K.; Takano, H. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Cell. Mol. Biol. 2008, 53, 924–934. [Google Scholar] [CrossRef]
- Pawlowski, K.; Demchenko, K.N. The diversity of actinorhizal symbiosis. Protoplasma 2012, 249, 967–979. [Google Scholar] [CrossRef]
- Normand, P.; Orso, S.; Cournoyer, B.; Jeannin, P.; Chapelon, C.; Dawson, J.; Evtushenko, L.; Misra, A.K. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int. J. Syst. Bacteriol. 1996, 46, 1–9. [Google Scholar] [CrossRef]
- Clawson, M.L.; Bourret, A.; Benson, D.R. Assessing the phylogeny of Frankia-actinorhizal plant nitrogen-fixing root nodule symbiosis with Frankia 16S rRNA and glutamine synthetase gene sequences. Mol. Phylogenetics Evol. 2004, 31, 131–138. [Google Scholar] [CrossRef]
- Maréchal, J.; Clement, B.; Nalin, R.; Gandon, C.; Orso, S.; Cvejic, J.H.; Bruneteau, M.; Berry, A.M.; Normand, P. A recA gene phylogenetic analysis confirms the close proximity of Frankia to Acidothermus. Int. J. Syst. Evol. Microbiol. 2000, 50, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Wibberg, D.; Battenberg, K.; Blom, J.; Vanden Heuvel, B.; Berry, A.M.; Kalinowski, J.; Pawlowski, K. An assemblage of Frankia cluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. BMC Genomics. 2016, 17, 796. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Wibberg, D.; Vigil-Stenman, T.; Berckx, F.; Battenberg, K.; Demchenko, K.; Blom, J.; Fernandez, M.P.; Yamanaka, T.; Berry, A.M.; et al. Frankia-enriched metagenomes from the earliest divergent symbiotic Frankia cluster: They come in teams. Genome Biol. Evol. 2019, 11, 2273–2291. [Google Scholar] [CrossRef] [PubMed]
- Persson, T.; Battenberg, K.; Demina, I.V.; Vigil-Stenman, T.; Vanden Heuvel, B.; Pujic, P.; Facciotti, M.T.; Wilbanks, E.G.; O’Brien, A.; Fournier, P.; et al. Candidatus Frankia Datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS ONE 2015, 10, e0127630. [Google Scholar] [CrossRef]
- Pozzi, A.C.; Bautista-Guerrero, H.H.; Abby, S.S.; Herrera-Belaroussi, A.; Abrouk, D.; Normand, P.; Menu, F.; Fernandez, M.P. Robust Frankia phylogeny, species delineation and intraspecies diversity based on Multi-Locus Sequence Analysis (MLSA) and Single-Locus Strain Typing (SLST) adapted to a large sample size. Sys. App. Microbiol. 2018, 41, 311–323. [Google Scholar] [CrossRef]
- Gtari, M.; Ghodhbane-Gtari, F.; Nouioui, I.; Ktari, A.; Hezbri, K.; Mimouni, W.; Sbissi, I.; Ayari, A.; Yamanaka, T.; Normand, P.; et al. Cultivating the uncultured: Growing the recalcitrant cluster-2 Frankia strains. Sci. Rep. 2015, 5, 13112. [Google Scholar] [CrossRef]
- Gueddou, A.; Swanson, E.; Hezbri, K.; Nouioui, I.; Ktari, A.; Simpson, S.; Morris, K.; Kelley Thomas, W.; Ghodhbane-Gtari, F.; Gtari, M.; et al. Draft genome sequence of the symbiotic Frankia sp. strain BMG5.30 isolated from root nodules of Coriaria myrtifolia in Tunisia. Antonie Van Leeuwenhoek 2019, 112, 67–74. [Google Scholar] [CrossRef]
- Hurles, M. Gene duplication: The genomic trade in spare parts. PLoS Biol. 2004, 2, 900–904. [Google Scholar] [CrossRef]
- Sato, N. Complex origins of chloroplast membranes with photosynthetic machineries: Multiple transfers of genes from divergent organisms at different times or a single endosymbiotic event? J. Plant Res. 2020, 133, 15–33. [Google Scholar] [CrossRef]
- Normand, P.; Nguyen, T.V.; Battenberg, K.; Berry, A.M.; van den Heuvel, B.; Fernandez, M.P.; Pawlowski, K. Proposal of ‘Candidatus Frankia californiensis’, the uncultured symbiont in nitrogen-fixing root nodules of a phylogenetically broad group of hosts endemic to western North America. Int. J. Syst. Evol. Microbiol. 2017, 67, 3706–3715. [Google Scholar] [CrossRef]
- Nouioui, I.; Del Carmen Montero-Calasanz, M.; Ghodhbane-Gtari, F.; Rohde, M.; Tisa, L.S.; Klenk, H.P.; Gtari, M. Frankia discariae sp. nov.: An infective and effective microsymbiont isolated from the root nodule of Discaria trinervis. Arch Microbiol. 2017, 199, 641–647. [Google Scholar] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; University of California, College of Agriculture, Agricultural Experiment Station: Oakland, CA, USA, 1938. [Google Scholar]
- Schwencke, J. Rapid, exponential growth and increased biomass yield of some Frankia strains in buffered and stirred mineral medium (BAP) with phosphatidyl choline. Plant Soil. 1991, 137, 37–41. [Google Scholar] [CrossRef]
- Benchling [Biology Software]. Available online: https://benchling.com (accessed on 21 May 2019).
- Alloisio, N.; Queiroux, C.; Fournier, P.; Pujic, P.; Normand, P.; Vallenet, D.; Médigue, C.; Yamaura, M.; Kakoi, K.; Kucho, K.-I. The Frankia alni symbiotic transcriptome. Mol. Plant Microbe Interact. 2010, 23, 593–607. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. Rstudio: Integrated Development; Rstudio, Inc.: Boston, MA, USA, 2015; Available online: Hppt://www.rstudio.com/ (accessed on 15 Marc 2018).
- Meyer, F.; Goesmann, A.; McHardy, A.C.; Bartels, D.; Bekel, T.; Clausen, J.; Kalinowski, J.; Linke, B.; Rupp, O.; Giegerich, R.; et al. GenDB--an Open Source Genome Annotation System for prokaryote genomes. Nucleic Acids Res. 2003, 31, 2187–2195. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Chen, I.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The Integrated Microbial Genomes Database and Comparative Analysis System. Nucleic Acids Res 2012, 40, D115–D122. [Google Scholar] [CrossRef]
- Wong, G.K.; Soltis, D.E.; Leebens-Mack, J.; Wickett, N.J.; Barker, M.S.; Van de Peer, Y.; Graham, S.W.; Melkonian, M. Sequencing and analyzing the transcriptomes of a thousand species across the tree of life for green plants. Annu. Rev. Plant Biol. 2019. [Google Scholar] [CrossRef]
- Carpenter, E.J.; Matasci, N.; Ayyampalayam, S.; Wu, S.; Sun, J.; Yu, J.; Jimenez Vieira, F.R.; Bowler, C.; Dorrell, R.G.; Gitzendanner, M.A.; et al. Access to RNA-sequencing data from 1,173 plant species: The 1000 Plant transcriptomes initiative (1KP). GigaScience 2019, 8, giz126. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. The UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 45–52. [Google Scholar]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 2019, 35, 5326–5327. [Google Scholar] [CrossRef]
- Mol, D.C.; Brooun, A.; Dougan, D.R.; Hilgers, M.T.; Tari, L.W.; Wijnands, R.A.; Knuth, M.W.; McRee, D.E.; Swanson, R.V. Crystal structures of active fully assembled substrate- and product-bound complexes of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) from Haemophilus influenzae. J. Bacteriol. 2003, 185, 4152–4162. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef]
- Celniker, G.; Nimrod, G.; Ashkenazy, H.; Glaser, F.; Martz, E.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. ISR J Chem. 2013, 53, 199–206. [Google Scholar] [CrossRef]
- Van Velzen, R.; Doyle, J.J.; Geurts, R. A resurrected scenario: Single gain and massive loss of nitrogen-fixing nodulation. Trends Plant Sci. 2019, 24, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Takano, H. Diverse origins of enzymes involved in the biosynthesis of chloroplast peptidoglycan. Plant Res. 2017, 130, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Falk, S.P.; Weisblum, B. Phosphorylation of the Streptococcus pneumoniae cell wall biosynthesis enzyme MurC by a eukaryotic-like Ser/Thr kinase. FEMS Microbiol. Lett. 2013, 340, 19–23. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berckx, F.; Wibberg, D.; Kalinowski, J.; Pawlowski, K. The Peptidoglycan Biosynthesis Gene murC in Frankia: Actinorhizal vs. Plant Type. Genes 2020, 11, 432. https://doi.org/10.3390/genes11040432
Berckx F, Wibberg D, Kalinowski J, Pawlowski K. The Peptidoglycan Biosynthesis Gene murC in Frankia: Actinorhizal vs. Plant Type. Genes. 2020; 11(4):432. https://doi.org/10.3390/genes11040432
Chicago/Turabian StyleBerckx, Fede, Daniel Wibberg, Jörn Kalinowski, and Katharina Pawlowski. 2020. "The Peptidoglycan Biosynthesis Gene murC in Frankia: Actinorhizal vs. Plant Type" Genes 11, no. 4: 432. https://doi.org/10.3390/genes11040432
APA StyleBerckx, F., Wibberg, D., Kalinowski, J., & Pawlowski, K. (2020). The Peptidoglycan Biosynthesis Gene murC in Frankia: Actinorhizal vs. Plant Type. Genes, 11(4), 432. https://doi.org/10.3390/genes11040432





