A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Medium
2.2. Basic Local Alignment Search Tool (BLAST) Search
2.3. Off-Target Efficiency
2.4. Vector Assembly
2.5. Cloning
2.6. Conjugation into C. autoethanogenum
2.7. Colony Polymerase Chain Reaction (PCR)
2.8. Plasmid Loss and Cryopreservation
3. Results
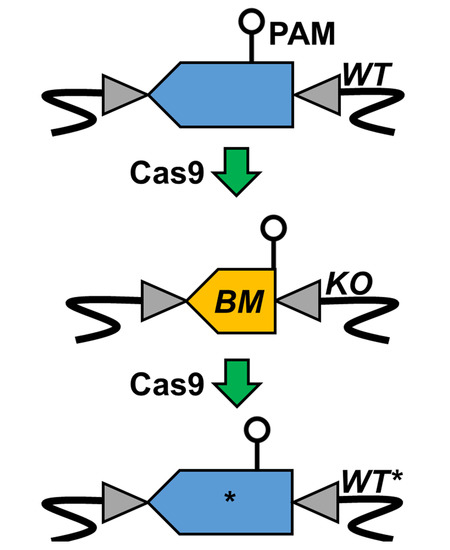
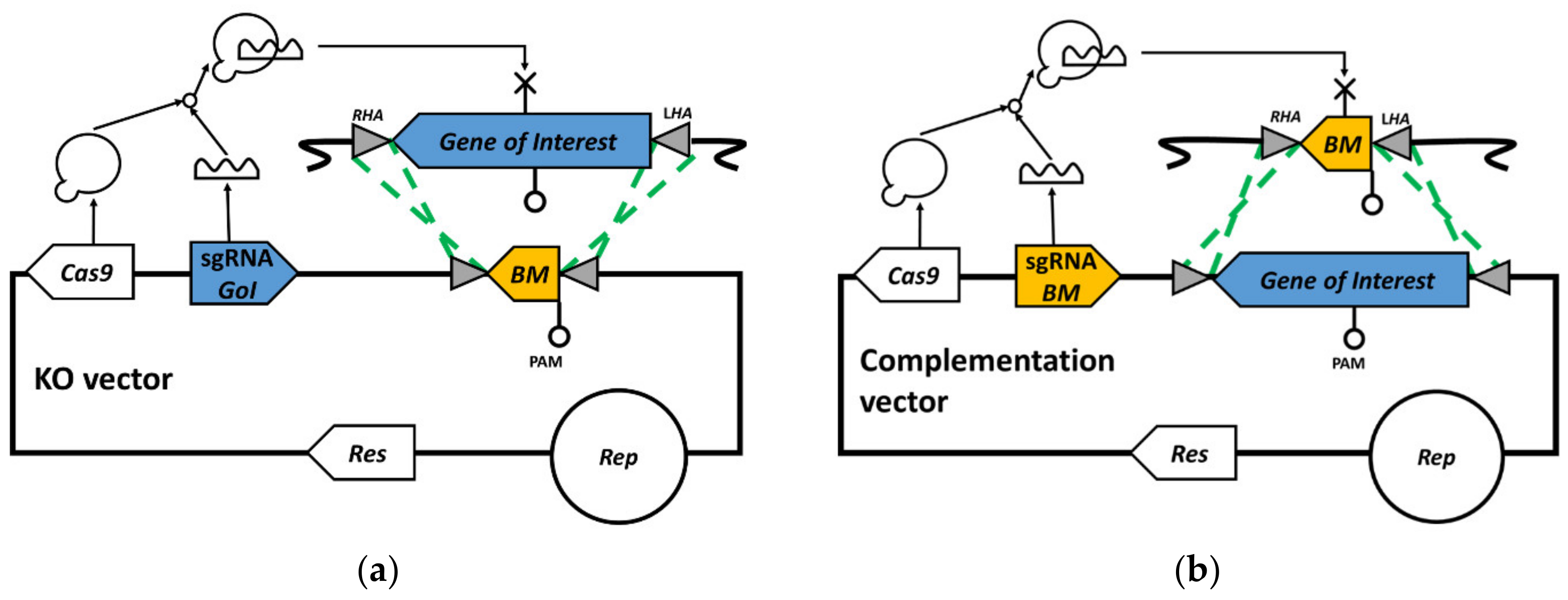
3.1. Bookmark Design
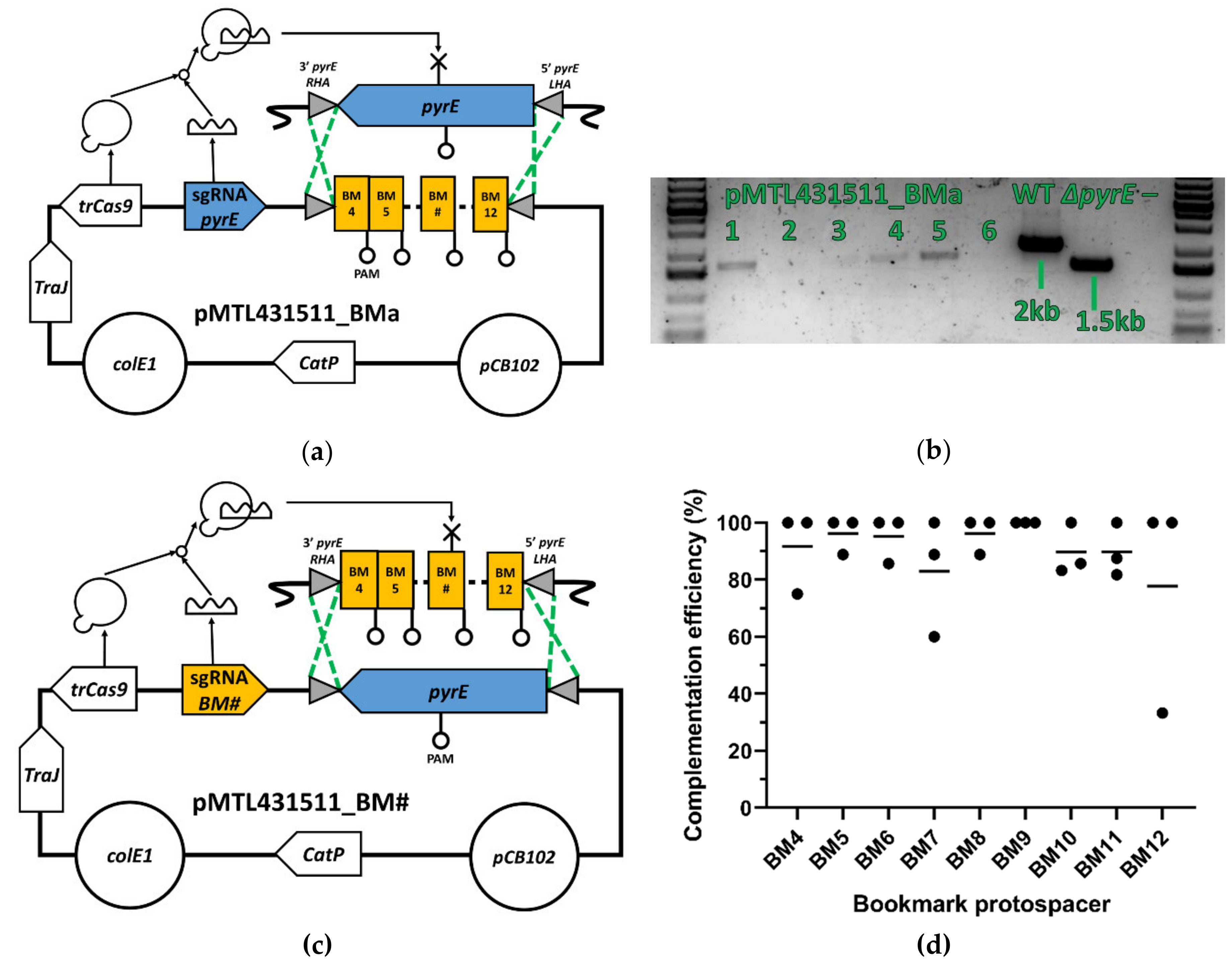
3.2. pyrE Knock-Out and Bookmarks Knock-In
3.3. Bookmark Complementation of pyrE
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Prim. 2016, 2, 16021. [Google Scholar] [CrossRef] [PubMed]
- Peck, M. Bacteria: Clostridium botulinum. In Encyclopedia of Food Safety; Motarjemi, Y., Ed.; Academic Press: Waltham, MA, USA, 2014; Volume 1, pp. 381–394. [Google Scholar] [CrossRef]
- Dürre, P. Fermentative production of butanol—the academic perspective. Curr. Opin. Biotechnol. 2011, 22, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Bengelsdorf, F.R.; Straub, M.; Dürre, P. Bacterial synthesis gas (syngas) fermentation. Environ. Technol. 2013, 34, 1639–1651. [Google Scholar] [CrossRef] [PubMed]
- Cañadas, I.C.; Groothuis, D.; Zygouropoulou, M.; Rodrigues, R.; Minton, N.P. RiboCas: A universal CRISPR-Based editing tool for Clostridium. ACS Synth. Boil. 2019, 8, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chai, C.; Li, N.; Rowe, P.; Minton, N.P.; Yang, S.; Jiang, W.; Gu, Y. CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth. Boil. 2016, 5, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Ingle, P.; Groothuis, D.; Rowe, P.; Huang, H.; Cockayne, A.; Kuehne, S.A.; Jiang, W.; Gu, Y.; Humphreys, C.M.; Minton, N.P. Generation of a fully erythromycin-sensitive strain of Clostridioides difficile using a novel CRISPR-Cas9 genome editing system. Sci. Rep. 2019, 9, 8123. [Google Scholar] [CrossRef] [PubMed]
- Minton, N.P.; Ehsaan, M.; Humphreys, C.M.; Little, G.; Baker, J.; Henstra, A.M.; Liew, F.; Kelly, M.L.; Sheng, L.; Schwarz, K.; et al. A roadmap for gene system development in Clostridium. Anaerobe 2016, 41, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, A.; Jacob, F.; Monod, J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the β-galactosidase structural gene of Escherichia coli. J. Mol. Boil. 1967, 24, 339–343. [Google Scholar] [CrossRef]
- Ng, Y.K.; Ehsaan, M.; Philip, S.; Collery, M.M.; Janoir, C.; Collignon, A.; Cartman, S.T.; Minton, N.P. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile Genome: Allelic exchange using pyrE alleles. PLoS ONE 2013, 8, e56051. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Bosma, E.F.; De Vos, W.M.; Van Kranenburg, R.; Van Der Oost, J. Next generation prokaryotic engineering: The CRISPR-Cas toolkit. Trends Biotechnol. 2016, 34, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.; A Scott, D.; A Weinstein, J.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, C.M.; McLean, S.; Schatschneider, S.; Millat, T.; Henstra, A.M.; Annan, F.J.; Breitkopf, R.; Pander, B.; Piatek, P.; Rowe, P.; et al. Whole genome sequence and manual annotation of Clostridium autoethanogenum, an industrially relevant bacterium. BMC Genom. 2015, 16, 1085. [Google Scholar] [CrossRef] [PubMed]
- Heap, J.T.; Pennington, O.J.; Cartman, S.T.; Minton, N.P. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 2009, 78, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.; Humphreys, C.M.; Rodrigues, R.M.; Ingle, P.; Rowe, P.; Henstra, A.M.; Köpke, M.; Simpson, S.D.; Winzer, K.; Minton, N.P. A novel conjugal donor strain for improved DNA transfer into Clostridium spp. Anaerobe 2019, 59, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Purdy, D.; O’Keeffe, T.A.T.; Elmore, M.; Herbert, M.; McLeod, A.; Bokori-Brown, M.; Ostrowski, A.; Minton, N.P. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 2002, 46, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; A Marraffini, L. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Altenbuchner, J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2016, 82, 5421–5427. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-H.; Van Pijkeren, J.-P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, G.C.; Thorner, J. mCAL: A new approach for versatile multiplex action of Cas9 using one sgRNA and loci flanked by a programmed target sequence. Genes Genomes Genet. 2016, 6, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
| Search Parameter | Value |
|---|---|
| Program | BLASTn |
| Word size | 7 |
| Expect value | 1000 |
| Hitlist size | 100 |
| Match/Mismatch scores | 1, –1 |
| Gapcosts | 5, 2 |
| Filter string | F |
| Genetic Code | 1 |
| Database | |
| Posted date | Mar 24, 2020 9:05 AM |
| Number of letters | 8,771,469 |
| Number of sequences | 18 |
| Entrez query | Includes: Clostridium autoethanogenum DSM 10061 (taxid:1341692) |
| Parameter | Value |
|---|---|
| Reference genome | GCA_000484505.1 (Clostridium autoethanogenum DSM 10061) |
| Design type | Single guide |
| Guide length | 20 bp |
| PAM | NGG (SpCas9, 3’side) |
| Bookmark | Bookmark Sequence (24 nt) | Orientation | Origin | |||
|---|---|---|---|---|---|---|
| Extra nt | Protospacer (20 nt) | PAM | Extra nt | |||
| BM4 | G | AGGGTTGTGGGTTGTACGGA | AGG | / | +/− | S. pneumonia |
| BM5 | / | ATTTCTGATATTACTGTCAC | AGG | A | +/− | S. pneumoniae |
| BM6 | / | ACCGATACCGTTTACGAAAT | AGG | A | +/− | S. pneumoniae |
| BM7 | G | TGAAGATCAGGCTATCACTG | AGG | / | + | B. subtilis |
| BM8 | G | TCCGGAGCTCCGATAAAAAA | TGG | / | +/− | B. subtilis |
| BM9 | G | TATTGATTCTCTTCAAGTAG | AGG | / | − | B. subtilis |
| BM10 | / | CCATTGTACTATCATGCTAG | AGG | A | +/− | L. reuteri |
| BM11 | G | ATGCAGTCGGCTGTAGAAAG | AGG | / | +/− | L. reuteri |
| BM12 | G | CGACTGCATTTTATTATGTA | AGG | / | +/− | L. reuteri |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seys, F.M.; Rowe, P.; Bolt, E.L.; Humphreys, C.M.; Minton, N.P. A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences. Genes 2020, 11, 458. https://doi.org/10.3390/genes11040458
Seys FM, Rowe P, Bolt EL, Humphreys CM, Minton NP. A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences. Genes. 2020; 11(4):458. https://doi.org/10.3390/genes11040458
Chicago/Turabian StyleSeys, François M., Peter Rowe, Edward L. Bolt, Christopher M. Humphreys, and Nigel P. Minton. 2020. "A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences" Genes 11, no. 4: 458. https://doi.org/10.3390/genes11040458
APA StyleSeys, F. M., Rowe, P., Bolt, E. L., Humphreys, C. M., & Minton, N. P. (2020). A Gold Standard, CRISPR/Cas9-Based Complementation Strategy Reliant on 24 Nucleotide Bookmark Sequences. Genes, 11(4), 458. https://doi.org/10.3390/genes11040458