An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes)
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Chromosome Preparations and Ideograms
2.3. Probes for Comparative Genomic Hybridization (CGH)
2.4. Fluorescence in Situ Hybridization (FISH) for CGH
3. Results
3.1. Chromosomal Distribution of the rDNA Sequences Across the Genome of Lebiasinidae and Ctenoluciidae Species Understudy
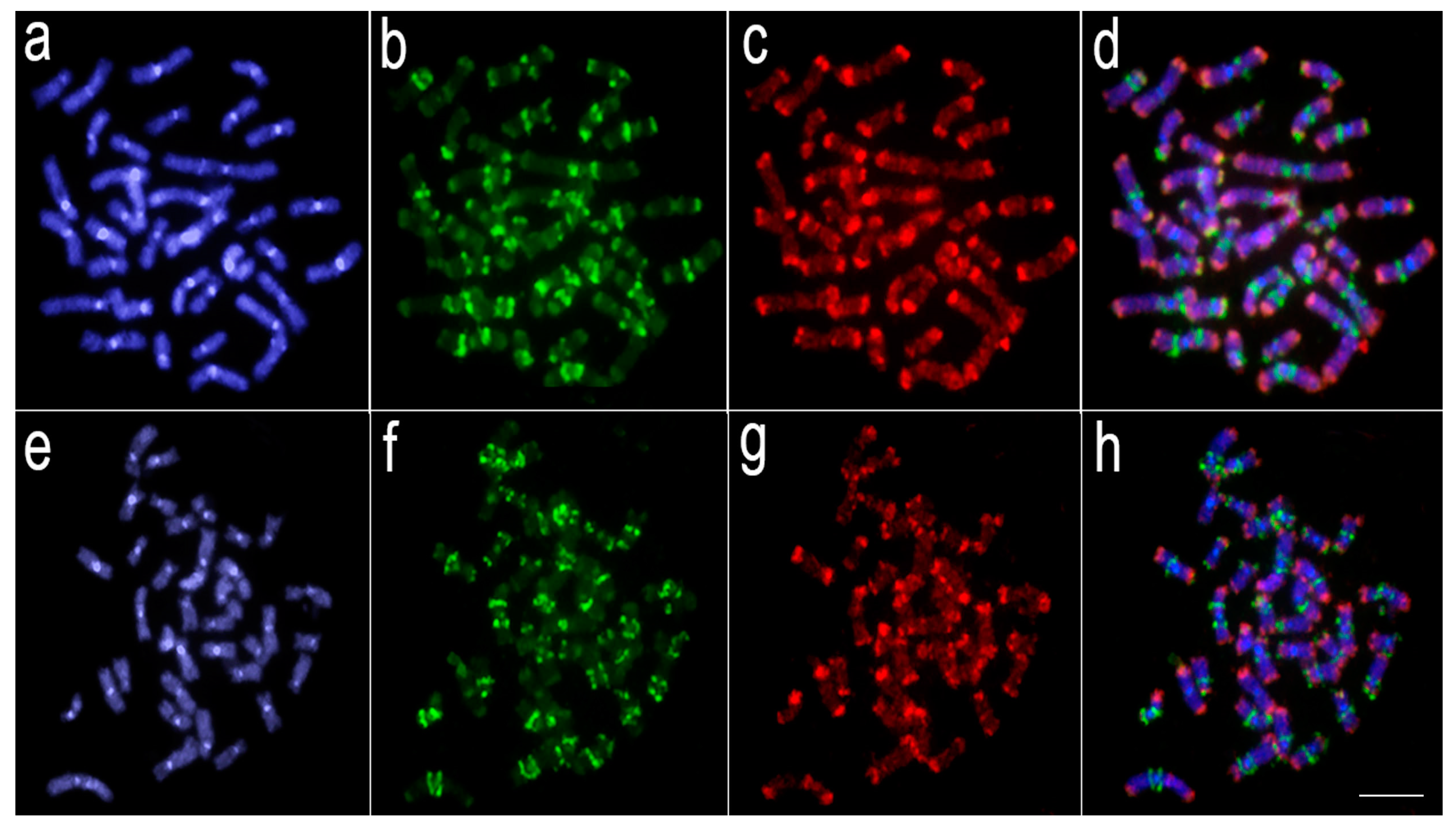
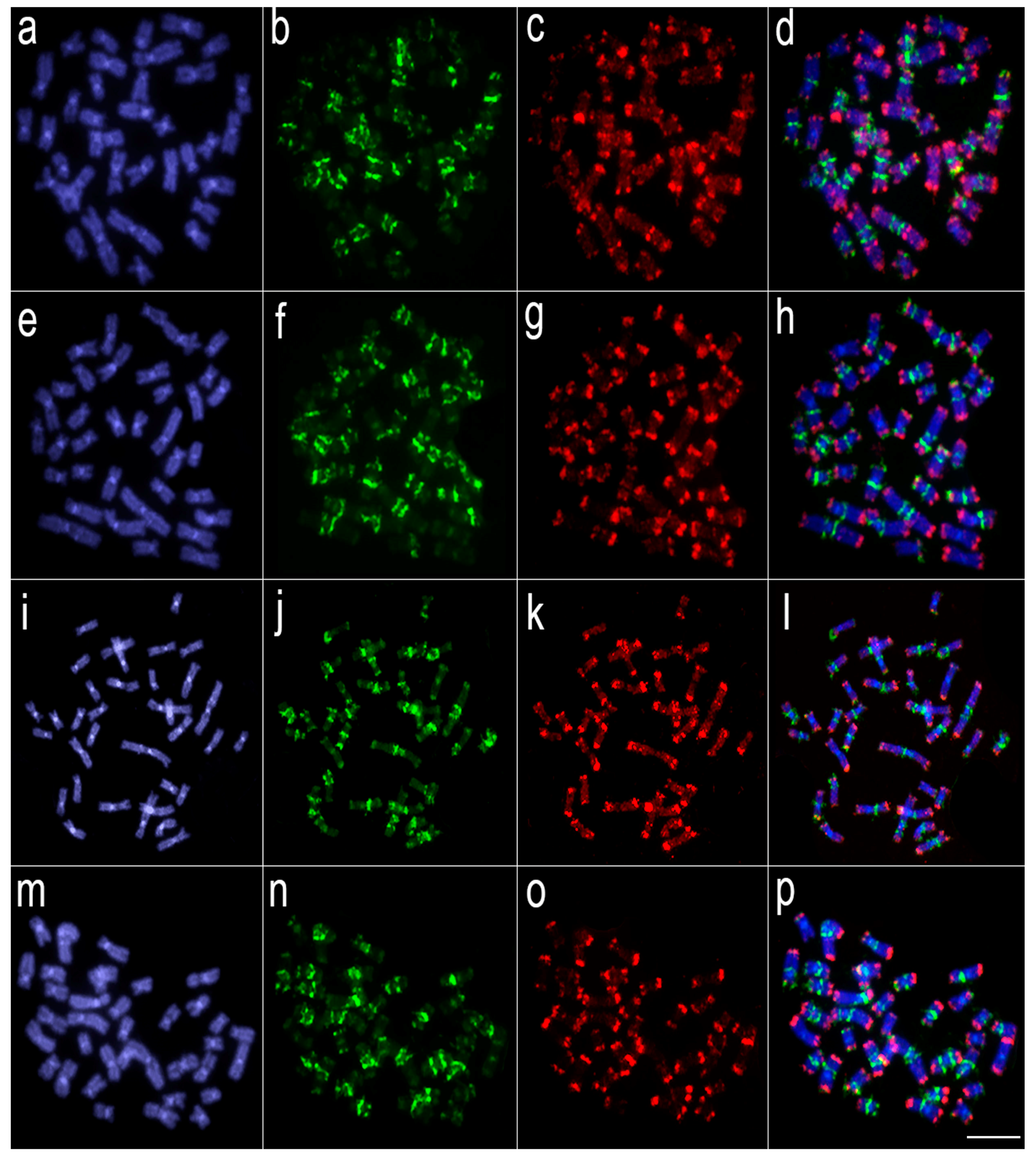
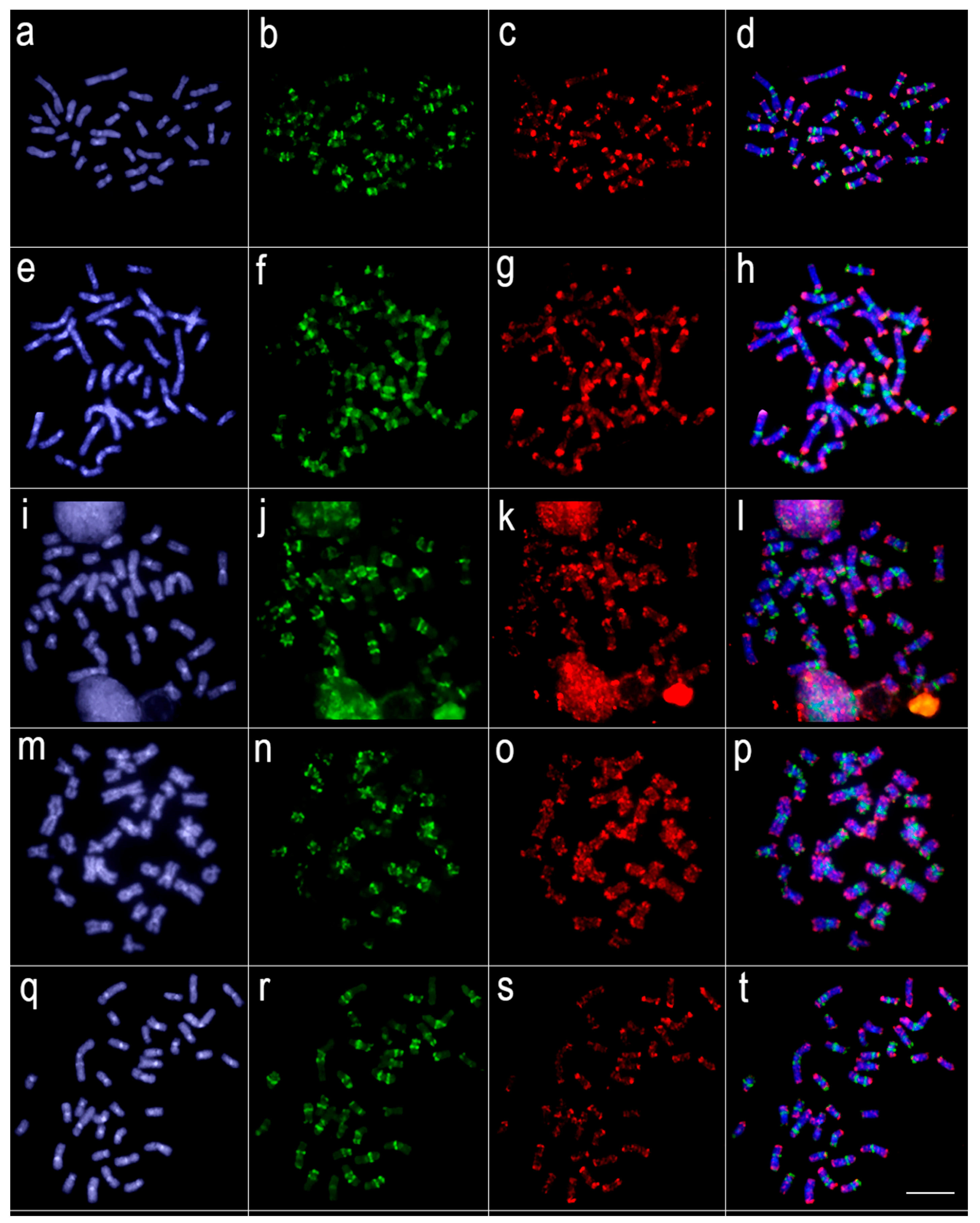
3.2. Comparative Genomic Hybridization (CGH)
4. Discussion
Karyotype and Chromosomal Differentiation in Lebiasinidae
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sember, A.; Bohlen, J.; Šlechtová, V.; Altmanová, M.; Symonová, R.; Ráb, P. Karyotype differentiation in 19 species of river loach fishes (Nemacheilidae, Teleostei): Extensive variability associated with rDNA and heterochromatin distribution and its phylogenetic and ecological interpretation. BMC Evol. Biol. 2015, 15, 251. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, S.H.; Vari, R.P. Miniaturization South American Fishes; an Overview and Discussion. Proc. Biol. Soc. Washingt. 1988, 2, 444–465. [Google Scholar]
- de Moraes, R.L.R.; Bertollo, L.A.C.; Marinho, M.M.F.; Yano, C.F.; Hatanaka, T.; Barby, F.F.; Troy, W.P.; Cioffi, M.d.B. Evolutionary Relationships and Cytotaxonomy Considerations in the Genus Pyrrhulina (Characiformes, Lebiasinidae). Zebrafish 2017, 14, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Souza Sousa, J.F.; Viana, P.F.; Bertollo, L.A.C.; Cioffi, M.d.B.; Feldberg, E. Evolutionary Relationships among Boulengerella Species (Ctenoluciidae, Characiformes): Genomic Organization of Repetitive DNAs and Highly Conserved Karyotypes. Cytogenet. Genome Res. 2017, 152, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Moraes, R.L.R.d.; Sember, A.; Bertollo, L.A.; De Oliveira, E.A.; Rab, P.; Hatanaka, T.; Marinho, M.M.; Liehr, T.; Al-Rikabi, A.B.; Feldberg, E.; et al. Comparative cytogenetics and neo-Y formation in small-sized fish species of the genus Pyrrhulina (Characiformes, Lebiasinidae). Front. Genet. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sassi, F.d.M.C.; Oliveira, E.A.D.; Bertollo, L.A.C.; Nirchio, M.; Hatanaka, T.; Marinho, M.M.F.; Moreira-Filho, O.; Aroutiounian, R.; Liehr, T.; Al-rikabi, A.B.H.; et al. Chromosomal Evolution and Evolutionary Relationships of Lebiasina Species (Characiformes, Lebiasinidae ). Int. J. Mol. Sci. 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Toma, G.A.; De Moraes, R.L.R.; Sassi, F.D.M.C.; Bertollo, L.A.C.; De Oliveira, E.A.; Rab, P.; Sember, A.; Liehr, T.; Hatanaka, T.; Viana, P.F.; et al. Cytogenetics of the small-sized fish, Copeina guttata (Characiformes, Lebiasinidae): Novel insights into the karyotype differentiation of the family. PLoS ONE 2019, 14, 1–13. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 12 February 2020).
- Netto-Ferreira, A.L. Revisão Taxonômica e Relações Interespecíficas de Lebiasinidae (Ostariophysi: Characiformes: Lebiasinidae). Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brazil, 2010. [Google Scholar]
- Ortí, G.; Meyer, A. The radiation of characiform fishes and the limits of resolution of mitochondrial ribosomal DNA sequences. Syst. Biol. 1997, 46, 75–100. [Google Scholar] [CrossRef]
- Buckup, P.A. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleostei: Ostariophysi). Phylogeny Classif. Neotrop. Fishes 1998, 1, 123–144. [Google Scholar]
- Oyakawa, O.T. Relações Filogenéticas das Famílias Pyrrhulinidae, Lebiasinidae e Erythrinidae (Osteichthyes: Characiformes). Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brazil, 1998. [Google Scholar]
- Calcagnotto, D.; Schaefer, S.A.; DeSalle, R. Relationships among characiform fishes inferred from analysis of nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 2005, 36, 135–153. [Google Scholar] [CrossRef]
- De Pinna, M.; Zuanon, J.; Rapp Py-Daniel, L.; Petry, P. A new family of neotropical freshwater fishes from deep fossorial amazonian habitat, with a reappraisal of morphological characiform phylogeny (Teleostei: Ostariophysi). Zool. J. Linn. Soc. 2018, 182, 76–106. [Google Scholar] [CrossRef]
- Oliveira, C.; Avelino, G.S.; Abe, K.T.; Mariguela, T.C.; Benine, R.C.; Ortí, G.; Vari, R.P.; Corrêa Castro, R.M. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 2011, 11, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Arcila, D.; Ortí, G.; Vari, R.; Armbruster, J.W.; Stiassny, M.L.J.; Ko, K.D.; Sabaj, M.H.; Lundberg, J.; Revell, L.J.; Betancur, R.R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nat. Ecol. Evol. 2017, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Betancur, R.; Arcila, D.; Vari, R.P.; Hughes, L.C.; Oliveira, C.; Sabaj, M.H.; Ortí, G. Phylogenomic incongruence, hypothesis testing, and taxonomic sampling: The monophyly of characiform fishes*. Evolution 2019, 73, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Marinho, M.M.F. Relações filogenéticas e revisão taxonômica das espécies do gênero Copella Myers, 1956 (Characiformes: Lebiasinidae). Ph.D. Thesis, Universidade Estadual Paulista “Júlio de Mesquita Filho”, Sao Paulo, Brazil, 27 February 2014. [Google Scholar]
- Kallioniemi, A.; Kallioniemi, O.P.; Sudar, D.; Rutovitz, D.; Gray, J.W.; Waldman, F.; Pinkel, D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef]
- Symonová, R.; Flajšhans, M.; Sember, A.; Havelka, M.; Gela, D.; Kořínková, T.; Rodina, M.; Rábová, M.; Ráb, P.; Flajhans, M.; et al. Molecular cytogenetics in artificial hybrid and highly polyploid sturgeons: An evolutionary story narrated by repetitive sequences. Cytogenet. Genome Res. 2013, 141, 153–162. [Google Scholar] [CrossRef]
- Yano, C.F.; Bertollo, L.A.C.; Liehr, T.; Troy, W.P.; Cioffi, M.D.B. W Chromosome Dynamics in Triportheus Species (Characiformes, Triportheidae): An Ongoing Process Narrated by Repetitive Sequences. J. Hered. 2016, 107, 342–348. [Google Scholar] [CrossRef]
- Oliveira, E.A.d.; Sember, A.; Bertollo, L.A.C.; Yano, C.F.; Ezaz, T.; Moreira-Filho, O.; Hatanaka, T.; Trifonov, V.; Liehr, T.; Al-Rikabi, A.B.H.; et al. Tracking the evolutionary pathway of sex chromosomes among fishes: Characterizing the unique XX/XY1Y2 system in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 2018, 127, 115–128. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; Cioffi, M.d.B.; Moreira-Filho, O. Direct chromosome preparation from Freshwater Teleost Fishes. In Fish Cytogenetic Techniques (Chondrichthyans and Teleosts); Ozouf-Costaz, C., Pisano, E., Eds.; CRC Press: Enfield, CT, USA, 2015; pp. 21–26. [Google Scholar]
- Sember, A.; Oliveira, E.A.d.; Ráb, P.; Bertollo, L.A.C.; Freitas, N.L.d.; Viana, P.F.; Yano, C.F.; Hatanaka, T.; Marinho, M.M.F.; de Moraes, R.L.R.; et al. Centric Fusions behind the Karyotype Evolution of Neotropical Nannostomus Pencilfishes (Characiforme, Lebiasinidae): First Insights from a Molecular Cytogenetic Perspective. Genes 2020, 11, 91. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning, a Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Zwick, M.S.; Hanson, R.E.; Mcknight, T.D.; Islam-Faridi, M.H.; Stelly, D.M.; Wing, R.A.; Price, H.J. A rapid procedure for the isolation of C 0 t-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef]
- Symonová, R.; Sember, A.; Majtánová, Z.; Ráb, P. Characterization of fish genome by GISH and CGH. In Fish Cytogenetic Techniques. Ray-Fin Fishes and Chondrichthyans; Ozouf-Costaz, C., Pisano, E., Eds.; CCR Press: Boca Raton, FL, USA, 2015; pp. 118–131. [Google Scholar]
- Nirchio, M.; Rossi, A.R.; Foresti, F.; Oliveira, C. Chromosome evolution in fishes: A new challenging proposal from Neotropical species. Neotrop. Ichthyol. 2014, 12, 761–770. [Google Scholar] [CrossRef]
- Arai, R. Fish Karyotypes: A Check List; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; Cambridge University Press: Cambridge, UK, 1973. [Google Scholar]
- Sola, L.; Cataudella, S.; Capanna, E. New developments in vertebrate cytotaxonomy III. Karyology of bony fishes: A review. Genetica 1981, 54, 285–328. [Google Scholar] [CrossRef]
- Bertollo, L.A.C. Chromosome Evolution in the Neotropical Erythrinidae Fish Family: An Overview. In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Eds.; CRC Press: Enfield, NH, USA, 2007; pp. 195–211. ISBN 9781578083305. [Google Scholar]
- Cioffi, M.B.; Franco, W.; Ferreira, R.; Bertollo, L.A.C. Chromosomes as tools for discovering Biodiversity—The case of Erythrinidae fish family. In Recent Trends Cytogenet Studies Methodol Appl; Tirunilai, P., Ed.; InTech: Rijeka, Croatia, 2012; pp. 125–146. ISBN 978-953-51-0178-9. [Google Scholar]
- Artoni, R.F.; Bertollo, L.A.C. Cytogenetic studies on Hypostominae (Pisces, Siluriformes, Loricariidae). Considerations on karyotype evolution in the genus Hypostomus. Caryologia 1996, 49, 81–90. [Google Scholar] [CrossRef][Green Version]
- Giuliano-Caetano, L. Polimorfismo cromossômico Robertsoniano em populações de Rineloricaria latirostris (Pisces, Loricariinae). Ph.D. Thesis, Universidade Federal de Sao Carlos-SP, Sao Paulo, Brazil.
- Kavalco, K.F.; Pazza, R.; Bertollo, L.A.C.; Moreira-Filho, O. Karyotypic diversity and evolution of Loricariidae (Pisces, Siluriformes). Heredity 2005, 94, 180–186. [Google Scholar] [CrossRef]
- Goulding, M.; Barthem, R.; Ferreira, E. The Smithsonian Atlas of the Amazon; Smithsonian Books: New York, NY, USA, 2003. [Google Scholar]
- Netto-Ferreira, A.L. Three new species of Lebiasina (Characiformes: Lebiasinidae) from the Brazilian shield border at Serra do Cachimbo, Pará, Brazil. Neotrop. Ichthyol. 2012, 10, 487–498. [Google Scholar] [CrossRef]
- Zimmer, E.A.; Martins, S.L.; Beverly, S.M.; Kan, Y.W.; Wilson, A.C. Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc. Natl. Acad. Sci. USA 1980, 77, 2158–2162. [Google Scholar] [CrossRef]
- Dover, G.A. Molecular drive: A cohesive model of species evolution. Nature 1982, 199, 111–117. [Google Scholar] [CrossRef]
- Roy, V.; Monti-Dedieu, L.; Chaminade, N.; Siljak-Yakovlev, S.; Aulard, S.; Lemeunier, F.; Montchamp-Moreau, C. Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: Ananassae and melanogaster. Heredity 2005, 94, 388–395. [Google Scholar] [CrossRef]
- Symonová, R.; Howell, W. Vertebrate Genome Evolution in the Light of Fish Cytogenomics and rDNAomics. Genes 2018, 9, 96. [Google Scholar] [CrossRef]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Huang, W.; Wang, Y.; Zhou, J. Bacterial community structure in simultaneous nitrification, denitrification and organic matter removal process treating saline mustard tuber wastewater as revealed by 16S rRNA sequencing. Bioresour. Technol. 2017, 228, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad. Ser. B 2014, 90, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Arefjev, V.A. Problems of karyotypic variability in the family Characidae (Pisces, Characiformes) with the description of somatic karyotypes for six species of tetras. Caryologia 1990, 43, 305–319. [Google Scholar] [CrossRef]
- Salvadori, S.; Deiana, A.; Elisabetta, C.; Floridia, G.; Rossi, E.; Zuffardi, O. Colocalization of (TTAGGG)n telomeric sequences and ribosomal genes in Atlantic eels. Chromosome Res. 1995, 3, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Rossi, A.R.; Annesi, F.; Gornung, E. Cytogenetic studies in Sparus auratus (Pisces, Perciformes): Molecular organization of 5S rDNA and chromosomal mapping of 5S and 45S ribosomal genes and of telomeric repeats. Hereditas 2003, 139, 232–236. [Google Scholar] [CrossRef]
- Gornung, E. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet. Genome Res. 2013, 141, 90–102. [Google Scholar] [CrossRef]
- Guerrero, R.F.; Kirkpatrick, M. Local adaptation and the evolution of chromosome fusions. Evolution 2014, 68, 2747–2756. [Google Scholar] [CrossRef]
- Ortiz-Barrientos, D.; Engelstädter, J.; Rieseberg, L.H. Recombination rate evolution and the origin of species. Trends Ecol. Evol. 2016, 31, 226–236. [Google Scholar] [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546–559. [Google Scholar] [CrossRef]
- Ferreira, M.; Kavalco, K.F.; de Almeida-Toledo, L.F.; Garcia, C. Cryptic diversity between two Imparfinis species (Siluriformes, Heptapteridae) by cytogenetic analysis and DNA barcoding. Zebrafish 2014, 11, 306–317. [Google Scholar] [CrossRef]
- Ferreira, M.; Garcia, C.; Matoso, D.A.; de Jesus, I.S.; Cioffi, M.d.B.; Bertollo, L.A.C.; Zuanon, J.; Feldberg, E. The Bunocephalus coracoideus species complex (Siluriformes, Aspredinidae). Signs of a speciation process through chromosomal, genetic and ecological diversity. Front. Genet. 2017, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.P.; Ma, D.M.; Gui, J.F. Triploid origin of the gibel carp as revealed by 5S rDNA localization and chromosome painting. Chromosome Res. 2006, 14, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ye, L.; Chen, Y.; Xiao, J.; Wu, Y.; Tao, M.; Xiao, Y.; Liu, S. The chromosomal constitution of fish hybrid lineage revealed by 5S rDNA FISH. BMC Genet. 2015, 16, 140. [Google Scholar] [CrossRef] [PubMed]
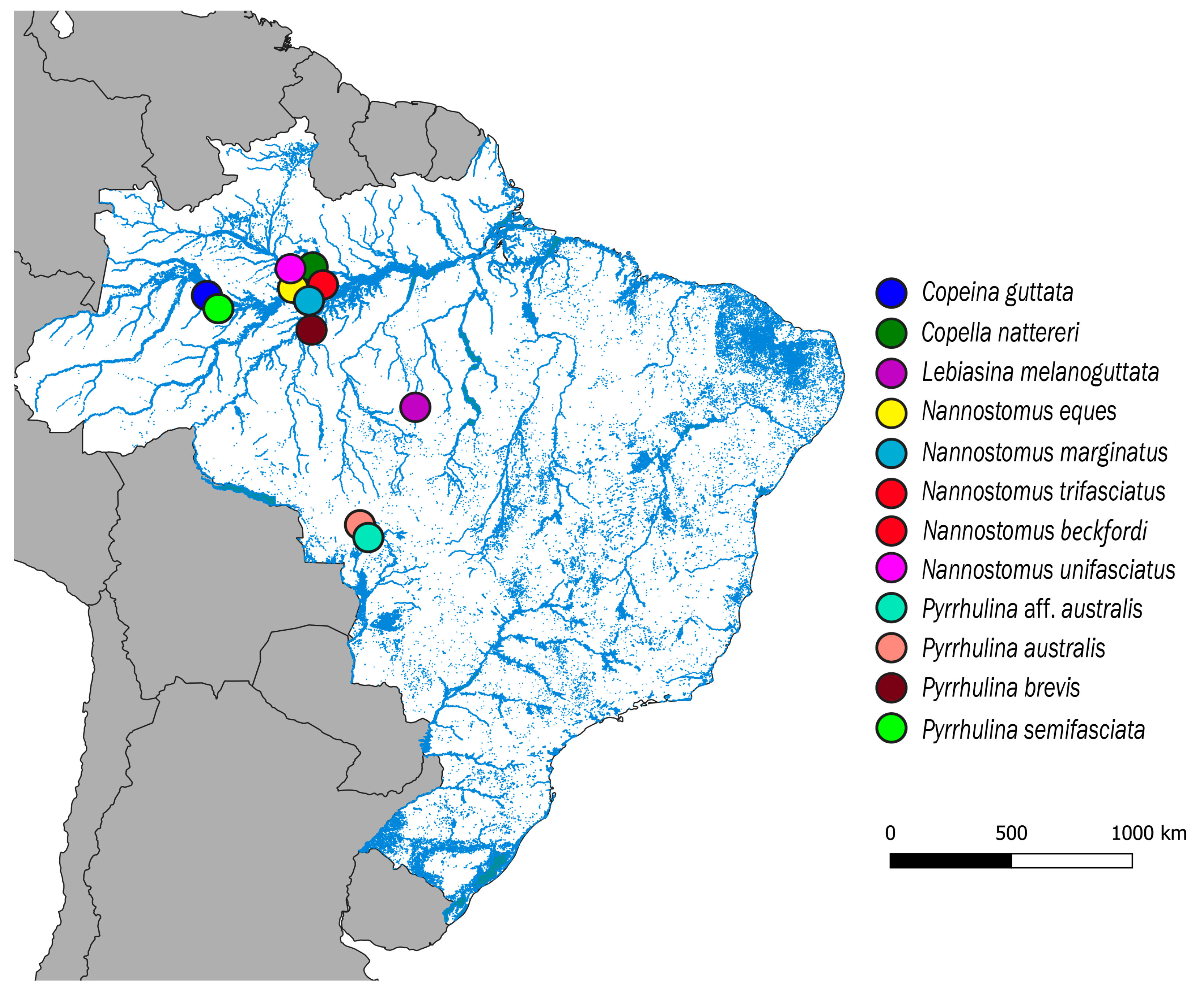

| Species | Locality | N | Deposit Number |
|---|---|---|---|
| Copeina guttata Steindachner, 1876 | Tefé, Amazonas (S03°23′07.7′′, W64°46′43.7′′) | 11♀; 06♂ | MZUSP 124915 |
| Copella nattereri Steindachner, 1876 | Manaus, Amazonas(S02°35′42.9′′, W60°02′23.8′′) | 04♀; 06♂ | MZUSP 124923 |
| Lebiasina melanoguttata Netto-Ferreira, 2012 | Cachoeira da Serra, Pará (S08°58′18,7′′, W54°58′18,7′′) | 22♀; 14♂ | MZUSP 124457 |
| Nannostomus eques Steindachner,1876 | Manaus, Amazonas (S02°47′58.1′′, W60°29′19.8′′) | 02♀; 02♂ | MZUSP 123084 |
| Nannostomus marginatus Eigenmann, 1909 | Manaus, Amazonas (S02°55′53.9′′, W59°58′30.7′′) | 03♀; 05♂ | MZUSP 123079 |
| Nannostomus beckfordi Günther, 1872 | Manaus, Amazonas (S02°55′53.9′′, W59°58′30.7′′) | 09♀; 17♂ | MZUSP 123071 |
| Nannostomus trifasciatus Steindachner, 1876 | Manaus, Amazonas (S02°44′59.6′′, W60°01′37.9′′) | 07♀; 12♂ | MZUSP 123071 |
| Nannostomus unifasciatus Steindachner, 1876 | Manaus, Amazonas (S02°47′58.1′′, W60°29′19.8′′) | 05♀; 07♂ | MZUSP 123083 |
| Pyrrhulina australis Eigenmann & Kennedy, 1903 | Santo Afonso, Mato Grosso (S14°27′25.2′′, W57°34′35.2′′) | 30♀; 18♂ | MZUSP 119079 |
| Pyrrulina aff. australis | Barra do Bugres, Mato Grosso (S15°04′27.5′′, W57°11′05.4′′) | 22♀; 16♂ | MZUSP 119077 |
| Pyrrulina brevis Steindachner, 1876 | Manaus, Amazonas (S02°55′53.9′′, W59°58′30.7′′) | 13♀; 17♂ | MZUSP 124916 |
| Pyrrulina semifasciata Steindachner, 1876 | Tefé, Amazonas (S3°39′45.8′′, W64°35′33.3′′) | 07♀; 12♂ | MZUSP 123073 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sassi, F.d.M.C.; Hatanaka, T.; Moraes, R.L.R.d.; Toma, G.A.; Oliveira, E.A.d.; Liehr, T.; Rab, P.; Bertollo, L.A.C.; Viana, P.F.; Feldberg, E.; et al. An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes). Genes 2020, 11, 365. https://doi.org/10.3390/genes11040365
Sassi FdMC, Hatanaka T, Moraes RLRd, Toma GA, Oliveira EAd, Liehr T, Rab P, Bertollo LAC, Viana PF, Feldberg E, et al. An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes). Genes. 2020; 11(4):365. https://doi.org/10.3390/genes11040365
Chicago/Turabian StyleSassi, Francisco de M. C., Terumi Hatanaka, Renata Luiza R. de Moraes, Gustavo A. Toma, Ezequiel A. de Oliveira, Thomas Liehr, Petr Rab, Luiz A. C. Bertollo, Patrik F. Viana, Eliana Feldberg, and et al. 2020. "An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes)" Genes 11, no. 4: 365. https://doi.org/10.3390/genes11040365
APA StyleSassi, F. d. M. C., Hatanaka, T., Moraes, R. L. R. d., Toma, G. A., Oliveira, E. A. d., Liehr, T., Rab, P., Bertollo, L. A. C., Viana, P. F., Feldberg, E., Nirchio, M., Marinho, M. M. F., Souza, J. F. d. S. e., & Cioffi, M. d. B. (2020). An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes). Genes, 11(4), 365. https://doi.org/10.3390/genes11040365









