Abstract
The plant specific LATERAL ORGAN BOUNDARIES (LOB)-domain (LBD) proteins belong to a family of transcription factors that play important roles in plant growth and development, as well as in responses to various stresses. However, a comprehensive study of LBDs in Brassica napus has not yet been reported. In the present study, 126 BnLBD genes were identified in B. napus genome using bioinformatics analyses. The 126 BnLBDs were phylogenetically classified into two groups and nine subgroups. Evolutionary analysis indicated that whole genome duplication (WGD) and segmental duplication played important roles in the expansion of the BnLBD gene family. On the basis of the RNA-seq analyses, we identified BnLBD genes with tissue or developmental specific expression patterns. Through cis-acting element analysis and hormone treatment, we identified 19 BnLBD genes with putative functions in plant response to abscisic acid (ABA) treatment. This study provides a comprehensive understanding on the origin and evolutionary history of LBDs in B. napus, and will be helpful in further functional characterisation of BnLBDs.
1. Introduction
The LATERAL ORGAN BOUNDARIES (LOB)-domain (LBD)/ASYMMETRIC LEAVES2-LIKE (ASL) gene is a newly discovered transcription factor gene family, playing important roles in various aspects of plant growth and development [1,2,3]. The first LBD gene, named LOB, was found based on the expression analysis of an enhancer trap line [1]. Hitherto, a total of 43 LBD genes have been identified in Arabidopsis, which were divided into two classes. The LBD genes in Class I contain a highly conserved CX2CX6CX3C (X are residues not conserved) zinc-finger-like motif (C block), a GAS (Gly-Ala-Ser) block, and a LX6LX3LX6L leucine zipper-like coiled-coil motif, whereas those in Class II only contain a C block [1]. The C block is approximately 22 amino acids in length, and is associated with DNA-binding. The GAS block is a conserved motif starting with a FX2VH motif and ending with a DP (V/I)G motif. The GAS motif and leucine zipper-like motif are required for protein–protein interaction of LBDs [1,3].
Following the identification of LBD genes in Arabidopsis, LBDs have been found in other plant species, such as Oryza sativa, Malus domestica, Zea mays, Vitis vinifera, Morus notabilis, Lotus japonicas, Medicago truncatula, Brachypodium distachyon, Glycine max, and Eucalyptus grandis. The number of LBD members ranged from 28 to 90 in different plants [4,5,6,7,8,9,10,11,12,13]. However, LBDs have never been identified in any species other than plants, indicating that LBD is a plant-specific gene family [1].
LBDs play important roles in the development of lateral organs, including leaf, root, flower, and embryo development [3]. ASYMMETRIC LEAVES2 (AS2) in Arabidopsis thaliana is involved in the formation of symmetric flat leaf lamina. Overexpression of AtAS2 resulted in upwardly curled leaves that fail to expand [14,15,16], and led to an abnormal adaxial–abaxial pattern via repression of the expression of polarity genes [17]. Overexpression of LBD3 in rice also resulted in narrow and adaxially rolled leaves [18,19]. INDETERMINATE GAMETOPHYTE1 (IG1) in maize was shown to be required for leaf and embryo sac development by affecting the KNOTTED-LIKE HOMEOBOX (KNOX) gene expression [18]. In addition, ADVENTITIOUS ROOTLESS1 (ARL1), LBD16, LBD29, LBD18, and LBD14 have been reported to regulate lateral root formation in Arabidopsis or rice [20,21,22,23,24]. Repression of OsIG1 affects the number of female gametophyte and flower organs [25].
In addition to the functions in lateral organ development, LBDs are also involved in the regulation of anthocyanin and nitrogen metabolism, phytohormone accumulation (e.g., auxin, cytokinin, gibberellin (GA), and brassinosteroid), as well as response to environmental stimuli [3,26]. Mutation of AtLBD25 reduced auxin sensitivity and lateral roots, and resulted in abnormal hypocotyl elongation under dark conditions [27]. Thatcher et al. first revealed that AtLBD20 is regulated by jasmonate acid (JA) signaling, and lbd20 mutant lines showed increased survival after Fusarium oxysporum infection [28]. In banana fruit, MaLBD5 was induced by cold temperature and methyl jasmonate (MeJA) treatment, and was involved in the transcriptional regulation of MeJA-mediated cold tolerance [29].
The conserved domain and gene structure of LBDs in Class II are different from those of Class I, thus the functions of LBDs in different classes are also different. In Arabidopsis, LBD37, LBD38, and LBD39 repressed the anthocyanin synthesis by suppressing the key transcriptional factors (PAP1 and PAP2) of the flavonoid biosynthesis pathway, and affected the nitrogen metabolism by repressing many N-responsive genes [30,31]. Overexpression of OsLBD37 and OsLBD38 delayed the heading date and improved the crop yield in rice [32].
B. napus (AACC, 2n = 38) is a natural polyploid formed ~7500 years ago, which was derived from natural hybridization between B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18) [33]. Gene duplication plays an important role in plant evolution, which greatly contributed to the diversification of gene families, enlarged the genome size, and caused the genome complexity, as well as facilitated the formation of new species with better adaptation, such as species with improved disease resistance and tolerances to complex abiotic and biological stresses [34,35]. As reported, each Brassicaceae species has undergone gene duplication events during evolution [33,36,37]. Chalhoub et al. completed the whole genome sequencing and assembly of B. napus, and 101,040 gene models were generated from 35.5 Gb of seqencing data [33]. The assembled C subgenome (525.8 Mb) is bigger than A subgenome (314.2 Mb) [33]. The duplication history makes B. napus a model species for plant polyploidization research, and the gene expansion and contraction during rapeseed polyploidization would be interesting for gene diversification. How the LBDs are involved in the polyploidization of B. napus would also be of interest. The completion of the whole genome sequence of B. napus greatly facilitated the systematic analyses on the formation, evolution, and function of LBDs in B. napus. In this study, we comprehensively analyzed the structure, conserved motifs, chromosomal location, subcellular localization, and expression pattern of 126 LBDs in B. napus, as well as their synteny during genome duplication. This study provides an important basis for future functional studies of LBDs and may give new insights into improving the disease resistance and stress tolerance of B. napus.
2. Materials and Methods
2.1. Retrieval of LBDs in B. napus
The peptide sequence of B. napus was downloaded from the Genoscope database (http://www.genoscope.cns.fr/brassicanapus) [33]. The 43 Arabidopsis LBDs were acquired from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) [38]. We applied two methods to find all the LBDs in B. napus; firstly, the AtLBDs were used for protein-blast with an e-value of 1×e−10 against the peptide sequences of B. napus. Secondly, the conserved domain of LBDs (LOB domain, DUF260, pfam number: pfam03195) acquired from Pfam (http://pfam.xfam.org/) was used for blast to identify the BnLBDs with DUF260 as a query [39]. Then, the LOB domain in the predicted BnLBDs was screened again with Batch CD-Search (https://www.ncbi.nlm.nih.gov/cdd/) and HMMER (https://www.ebi.ac.uk/Tools/hmmer/) [2,40], and only the sequences with an LOB domain and complete C motif in the N-terminus were used for further analysis. The molecular weight and theoretical pI of the BnLBDs were calculated using the protein isoelectric point calculator (http://isoelectric.org/).
2.2. Phylogenetic Analysis and Characterization of LBD Proteins
The full length protein sequences of LBDs were aligned using the clustalX program (http://www.clustal.org/clustal2/) and edited by Jalview (http://www.jalview.org/). The sequence logos were drawn with WebLogo 3 (http://weblogo.threeplusone.com/). Phylogenetic trees were constructed with the neighbor-joining (NJ) method of MEGA 6.0 (https://www.megasoftware.net/) using the p-distance and pairwise deletion option. The tree reliability was assessed using 1000 bootstrap replications. In the present study, a phylogenetic tree of LBD proteins in B. napus and A. thaliana and a tree of BnLBDs, respectively, were generated.
The exon–intron structures of the BnLBD genes were extracted from the B. napus genome (http://www.genoscope.cns.fr/blat-server/cgi-bin/colza/webBlat). Multiple Em for Motif Elicitation (MEME, http://meme-suite.org/tools/meme) was used to analyze the conserved motifs in BnLBD proteins. The gene structure and the conserved motifs were visualized using the Amazing Optional Gene Viewer in TBtools [41].
2.3. Cis-Acting Element Analysis
To analyze the cis-acting elements of BnLBDs, TBtools was used to obtain the 2000 bp of genomic DNA sequence upstream of coding sequences (CDS). The PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Gent, Belgium) was used to analyze the presence of different cis-acting elements.
2.4. Chromosomal Location, Identification of Orthologous and Paralogous LBD Genes
The length of each chromosome and the location of each BnLBD gene were retrieved from the Genoscope database. BnLBDs were localized on the chromosomes using Map Gene 2 Chromosome v2 (MG2C) (http://mg2c.iask.in/mg2c_v2.0/). Multiple collinear scanning toolkits (MCScanX) were used to analyze gene replication events and synteny relationships among B. napus, A. thaliana, B. rapa, and B. oleracea. To acknowledge the relationship of paralogous LBDs in B. napus, and the orthologous of LBDs among B. napus and other species, the syntenic maps were constructed with the Amazing Super Circos software and the Dual Systeny Plotter in TBtools, respectively [41]. The synonymous rate (Ks), non-synonymous rate (Ka), and Ka/Ks ratio of each gene pair were calculated using KaKs Calculator 2.0 [42].
2.5. Subcellular Localization Analysis of BnLBD Proteins
The subcellular localization of all the BnLBD proteins was predicted with Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (San Diego, CA, USA) and ProtComp v.9.0 in softberry (http://linux1.softberry.com/) (Stockholm, Sweden). In order to verify the predicted results, the coding sequence of BnLBD46, BnLBD104, and BnLBD105 was amplified from the cDNA of B. napus cv. Damor-bzh, and cloned into a green fluorescent protein (GFP) fusion expression vector pMDC83 via an enzyme (SpeI/AscI) digestion-ligation method. All the primers were synthesized by TSINKE Biotech and are listed in Table S1. The Agrobacterium tumefaciens strain GV3101 harboring the 35S:BnLBD-GFP constructs were injected into the abaxial epidermis of Nicotiana benthamiana leaves for transient expression [43]. The florescence images were captured using a confocal laser-scanning microscope (TCS SP8 STED, Leica, Germany).
2.6. Plant Material and Stress Treatment
To investigate the expression pattern of BnLBDs, three replicates of fifteen samples representing the major developmental tissues and organs of B. napus line ‘J9712′ were collected for RNA-seq analysis, including leaf near bolting stage; cotyledon; hypocotyl; root; stem; shoot apical meristem (SAM); 3 mm bud; 6 mm bud; endosperml silique at 14 days after pollination (DAP); and five seed samples at 21, 28, 35, 42, and 50 DAP. Using the RNA-seq data, a heat map of BnLBDs in different developmental stages was generated based on the log10 transformed values of FPKM (fragments per kilobase of transcript per million fragments mapped) values, and if FPKM = 0, then log10FPKM = −3. To investigate the response of BnLBD gene family under abscisic acid (ABA) treatment, the five-week-old seedlings of B. napus line ‘J9712′ were treated with 100 μM ABA. Samples were collected at 0 h, 1 h, 3 h, 6 h, and 12 h after treatment. Six leaves were pooled from three seedlings, and stored at −80 °C for further gene expression analysis.
2.7. RNA Extraction, cDNA Synthesis, and Quantitative Real Time PCR Analysis
Total RNA of leaf samples under ABA treatment was extracted according to the protocal of RNA isolater Total RNA Extraction Reagent (Vazyme, China). The total RNA was reverse transcribed into cDNA using HiScript III RT SuperMix for qPCR (Vazyme, China). qRT-PCR was performed on StepOnePlus Real-Time PCR Syetem (Thermo, Waltham, MA, USA) using PowerUp SYBR Green Master Mixes (Thermo, Waltham, MA, USA). The 2-△△Ct method was used to calculate the relative gene expression levels of BnLBD genes. All the primers were synthesized by TSINKE Biotech and are listed in Table S1.
3. Results
3.1. Identification of BnLBD Proteins
A total of 139 BnLBD proteins were identified in B. napus, including 135 BnLBDs identified using AtLBDs as queries and 136 BnLBDs screened with the LOB domain as query. However, six BnLBDs were excluded because of the absence of an LOB domain and seven BnLBDs were removed for lack of a complete C block in the N-terminus. Thus, we finally identified 126 BnLBD proteins, and these proteins were named with serial numbers from BnLBD1 to BnLBD126 depending on their location on the chromosomes. As B. napus was derived from natural hybridization between two diploids (B. rapa and B. oleracea), while the diploids in Brassica underwent polyploidization events based on A. thaliana, the number of LBDs in B. napus is much less than the theoretical value after the triplication and duplication events, and ~51.16% LBDs might be lost during B. napus evolution. The detailed information of each BnLBD protein is listed in Table S2, including gene identifier (gene ID), genomic location, protein length, isoelectric point (pI), and molecular weight (Mw). The BnLBD genes consisted of 104–358 amino acids (AA) with an average length of 214 AA, while the molecular weight ranged from 11.15 to 40.60 kDa and the pI of BnLBD proteins ranged from 4.73 to 9.08.
Using Plant-mPLoc and ProtComp to predict the subcellular location of 126 BnLBD genes, we found most of them were located in the nucleus. To validate the predicted results, transient expression of BnLBD46, BnLBD104, and BnLBD105 fused with GFP was performed in tobacco, and the subcellular localization of these proteins was consistent with the predicted results (Figure 1).
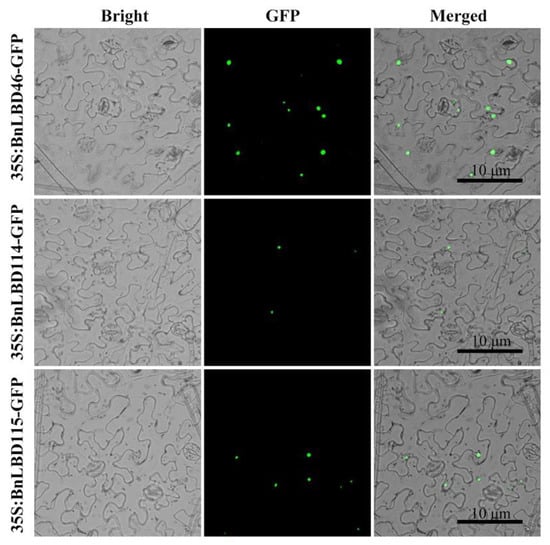
Figure 1.
Subcellular localization of BnLBD46/114/115-GFP. GFP fluorescence is shown in green. Bars = 10 μm. LBD, LATERAL ORGAN BOUNDARIES (LOB)-domain; GFP, green fluorescent protein.
3.2. Sequence Alignment and Phylogenetic Analysis of LBD Proteins
A protein sequence alignment was performed to identify the conserved amino acids and classify the BnLBD proteins. As shown in Figure 2 and Figure S1, a completely conserved CX2CX6CX3C zinc-finger-like motif (C block) existed in all BnLBDs, as reported in other plant species. A further two conserved motifs, a LX6LX3LX6L leucine zipper-like coiled-coil motif and a GAS (Gly-Ala-Ser) block between the C block and the leucine zipper-like motif, were identified in most BnLBDs. Generally, 104 BnLBD proteins with C block, GAS block, and the leucine zipper-like motif were classified into Class I, while 22 BnLBDs with only a C block were classified into Class II of BnLBDs.
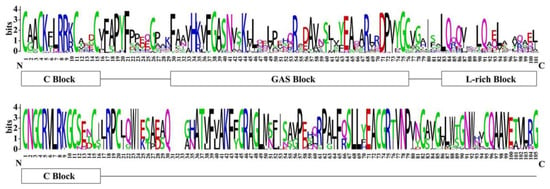
Figure 2.
Sequence logos of LOB domains in BnLBD proteins. Multiple alignment analysis of LOB domains was performed with Clustal X. The sequence logos were drawn by WebLogo 3. The bit score exhibits the information content for each position in the sequence. GAS, Gly-Ala-Ser.
To understand the evolutionary relationship of BnLBDs, a phylogenetic tree was constructed based on the alignment of the BnLBD and AtLBD protein sequences. As shown in Figure 3, the LBD proteins were classified into two groups (Class I and Class II). The larger group (Class I) was further divided into eight subgroups (Class Ia–Class Ih) and Class II was divided into two subgroups (Class IIa and Class IIb). Among the 126 BnLBDs and 43 AtLBDs, 104 BnLBDs and 37 AtLBDs with a complete C block, GAS block, and a leucine zipper-like motif were clustered in Class I, while 22 BnLBDs and 6 AtLBDs with a C block were clustered in Class II. Thirteen BnLBDs clustered in Class IIa were orthologs of AtLBD37, AtLBD38, and AtLBD39, which have been reported as negative regulators of anthocyanin biosynthesis and affect additional nitrogen responses in Arabidopsis [30].
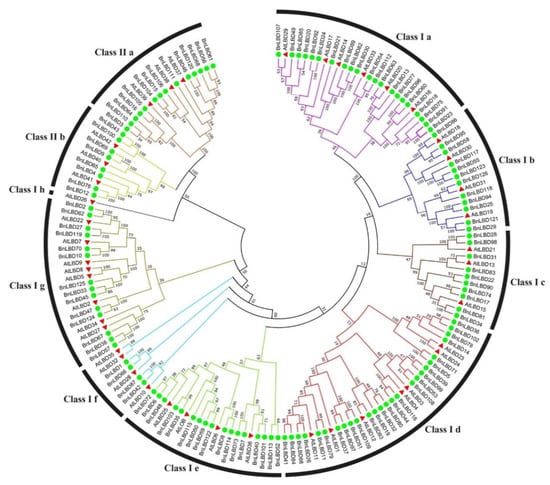
Figure 3.
Phylogenetic relationship of LBD proteins in B. napus and A. thaliana. The different colors indicate different subgroups of the LBD family. The green circles and red triangle represent LBDs from B. napus and A. thaliana, respectively. The numbers on the branches indicate the reliability percent of bootstraps value based on 1000 replications.
3.3. Structural and Conserved Motif Analysis of BnLBD Genes
The alterations in exon–intron structure play an important role in gene function divergence, especially for the duplicate genes [44]. In this study, the exon–intron structure of 126 BnLBD genes was investigated (Figure 4b). Statistical analyses revealed that the exon number of BnLBDs ranged from one to four, of which 69 BnLBDs were identified with two exons, only one gene (BnLBD100) was found with four exons, and 24 BnLBDs contain three exons. The remaining 32 BnLBDs contain one exon, without any intron disrupting the coding sequence. In addition, the genes from the same phylogenetic groups from the same subfamily were more similar in exon–intron structure. For example, most genes in Class If and Class Ie had no intron in the coding sequence region, while a clade in Class Id and one clade in Class IIa contained three exons (Figure 4a, Figure 4b). We also found the length of exons is similar in genes from the same subfamily, but the length of introns varied a lot in some subfamilies, such as BnLBD39/99, BnLBD53/108, and BnLBD95/58, which may contribute to the functional divergence of the duplicated genes.
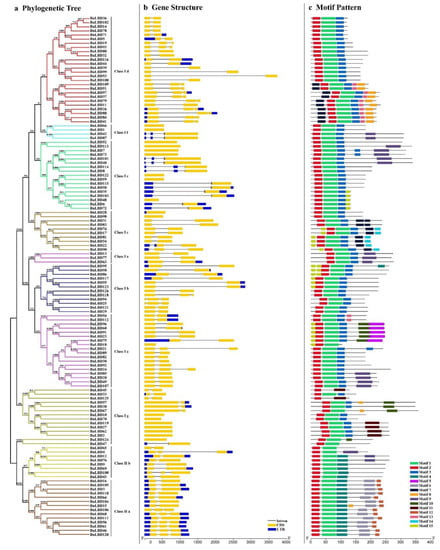
Figure 4.
Phylogenetic relationships, gene structure, and motif composition analysis of the LBD family in B. napus. (a). The phylogenetic tree based on the full-length sequences of BnLBD proteins using MEGA 5 software. The different colors indicate different subgroups of BnLBDs. The number on the branches indicates the reliability percent of bootstraps value based on 1000 replications. (b). Exon/intron structure of BnLBDs. Blue boxes indicate 5′- and 3′-untranslated regions, yellow boxes indicate exons, and black lines indicate introns. The scale bar represents 0.5 kb. (c). The architecture of conserved protein motifs of LBD proteins. Motifs 1–15 are displayed in different colored boxes, and the motif structure is provided in Additional File 3. The scale bar represents 50 amino acids.
We used MEME to analyze the conserved motif of the 126 BnLBD proteins, and a total of 15 conserved motifs in BnLBDs were identified with a length ranging from 14 to 50 amino acids (Figure 4c, Figure S2). We found the structure of BnLBDs was similar in the same subgroup. The conserved motif 1 and motif 2 existed in most BnLBDs, except BnLDB45 and BnLDB125, which lack motif 1. Motif 3 was identified in BnLBDs clustered in Class I, motif 4 existed in Class II, while other motifs were identified in specific subgroups. For example, motif 5 and motif 12 were identified in Class Ia and Class IIa, respectively. Motif 7 was only identified in Class Ic and Class Id.
3.4. Chromosomal Distribution and Genomic Duplication of BnLBD Genes
As reported, both the A and C subgenomes of B. napus have undergone duplications [33,45]. Here, we mapped the BnLBDs onto the B. napus chromosomes, and found 92 BnLBD genes were unevenly distributed across the 19 chromosomes (Figure 5). The remaining 34 BnLBD genes (14 on Ann random chromosomes and 20 on Cnn random chromosomes) were not assigned to specific chromosomes, owing to the incomplete B. napus genome. A total of 61 and 65 BnLBD genes were located on A and C subgenomes, respectively. Chromosome C04 contained the largest number of BnLBD genes (12 genes), followed by Chromosome A03, A05, and C03, with 8, 7, and 7 BnLBDs, respectively. Only one BnLBD was assigned on Chromosome A08 and C06, and no BnLBD was identified on Chromosome A10. The number of BnLBDs was not positively correlated with the chromosome length. According to previous studies, the LBD genes tended to be located on both ends of chromosomes. However, this was not observed in the chromosomal distribution of BnLBDs.
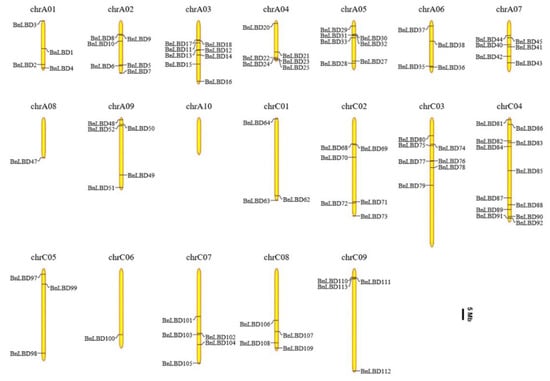
Figure 5.
The distribution of BnLBD genes on the B. napus chromosomes. A total of 92 BnLBD genes are mapped to the 19 chromosomes, and the unmapped 34 BnLBDs are unassembled scaffolds. The chromosomal position of each BnLBD gene was mapped to the B. napus genome. The chromosome number is indicated at the top of each chromosome. The scale is in megabases (Mb).
Gene duplication is universal in all organisms and important in dissecting the novelties in plant evolution [46]. Thus, we investigated the duplication events of the LBD gene family in B. napus (Figure 6, Figure 7), and found that all the 126 BnLBDs were results of duplication events (Table S3), of which 93 BnLBDs were derived from whole-genome duplication (WGD) or segmental duplications, and the other 33 BnLBD genes were results of dispersed duplications. Using MCScanX software, 104 paralogous gene pairs were identified (Table S4). These results indicated that gene duplication largely contributed to the expansion of LBD genes in B. napus genome, and the WGD or segmental duplication events played the main driving role.
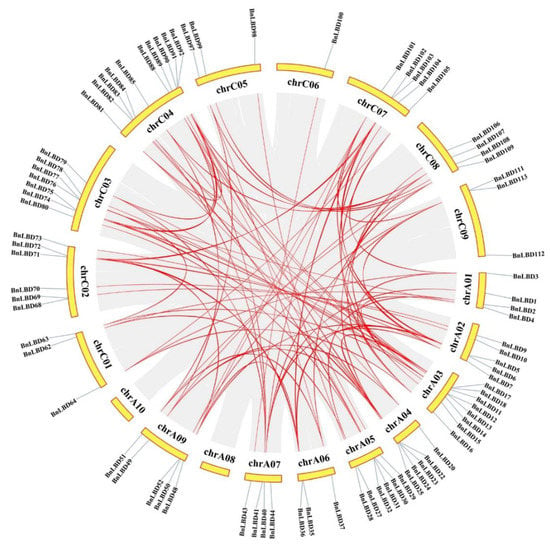
Figure 6.
Chromosomal distribution and interchromosomal relationships of BnLBD genes. Gray lines indicate all synteny blocks in the B. napus genome, and the red lines indicate duplicated BnLBD gene pairs. The chromosome number is indicated on the top of each chromosome.
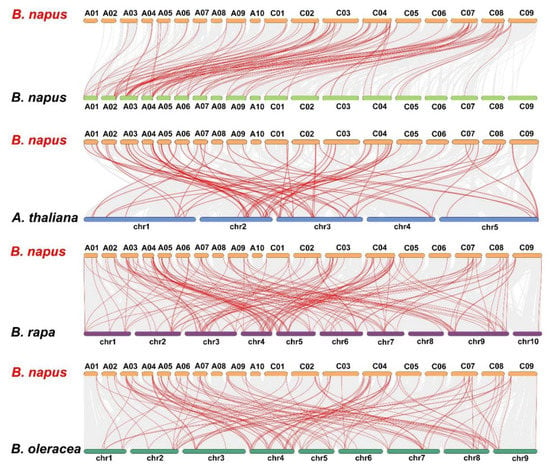
Figure 7.
Synteny analysis of LBD genes in B. napus and three ancestral plant species. Gray lines in the background indicate the collinear blocks within B. napus and other plant genomes, while the red lines highlight the syntenic LBD gene pairs.
To understand the evolution of the BnLBD gene family in the Brassicaceae, we analyzed the syntenic relationships between B. napus and Arabidopsis, B. napus and B. rapa, and B. napus and B. oleracea. Collinearity analysis revealed that a large number of orthologous LBDs existed in B. napus compared with B. rapa, B. oleracea, and Arabidopsis (Figure 7, Table S4). Generally, 100 of the 126 BnLBDs (79.37%) had a syntenic relationship with LBDs in other species, of which 82 BnLBD genes showed a syntenic relationship with Arabidopsis, 95 BnLBDs were syntenic to B. rapa, and 78 BnLBDs had synteny to B. oleracea. We found 86 BnLBD genes were directly inherited from BoLBDs (42 genes) and BrLBDs (44 genes). The synteny analysis indicates that the expansion of the LBD gene family in B. napus was mainly the result of whole-genome duplication.
To characterise the selective pressure on duplicated BnLBD genes during the evolutionary process, Ks, Ka, and Ka/Ks ratios were calculated for the paralogous gene pairs in B. napus, and the orthologous gene pairs among B. napus, B. rapa, B. oleracea, and A. thaliana [47]. We found most of the BnLBD gene pairs with Ka/Ks < 0.5, indicating that the BnLBD gene family might have undergone intense purifying selection in the evolutionary process (Table S3, Figure S3). The divergence time of homologous gene pairs between B. napus–B. napus, B. napus–B. rapa, B.napus–B. oleracea, and B. napus–A. thaliana peaked at 0.11, 0.02, 0.01, and 0.14 (Figure S3), respectively, which means that the divergence of LBD gene pairs in B. napus and A. thaliana was earlier.
3.5. Tissue Expression Analysis of BnLBD Genes
On the basis of the high-throughput RNA sequencing data of different tissues and organs at different development stages of B. napus, we analyzed the tissue specific expression pattern of BnLBD genes, and found 41 BnLBD genes were not or were weakly expressed in all tissues (Figure 8, Table S5). We found all BnLBD genes in Class II and 63 of the 104 BnLBD genes in Class I were expressed in B. napus. Generally, the expression pattern of BnLBD genes could be roughly divided into four types. The first type had low expression in all tissues. The second type was expressed in almost all tissues, such as BnLBD4, BnLBD12, BnLBD76, BnLBD46, and BnLBD120. The third type was expressed in most tissues, but was highly expressed in specific tissues, such as BnLBD28, BnLBD26, BnLBD88, and BnLBD98. The fourth type was only expressed in one or two tissues. For instance, BnLBD38, BnLBD87, BnLBD67, BnLBD42, BnLBD57, BnLBD2, BnLBD27, and BnLBD119 were only expressed in bud, while BnLBD9, BnLBD69, BnLBD63, BnLBD43, BnLBD100, BnLBD86, BnLBD117, BnLBD55, and BnLBD123 were only expressed in the endosperm or seeds at developmental stages. In addition, the expression level of the LBD gene in Class II was generally higher than that in Class I, indicating that the LBD gene in Class II might be more important for the growth and development of rapeseed.
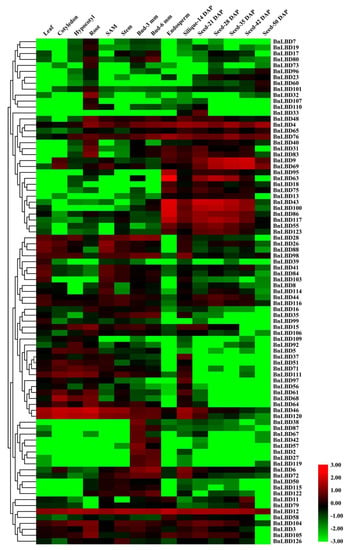
Figure 8.
Expression pattern of BnLBD genes in different tissues and developmental stages. The heatmap is generated based on log10 transformed values of FPKM (the reads per kilobase per million mapped reads) values, and if FPKM = 0, then log10FPKM = −3. BnLBD genes are clustered according to hierarchical clustering. FPKM, fragments per kilobase of transcript per million fragments mapped; SAM, shoot apical meristem; DAP, days after pollination.
3.6. Cis-Acting Element Analysis of BnLBDs
Cis-acting elements play an important role in regulating gene expression, and genes with similar functions may contain the same regulatory elements in their promoters. The cis-elements in 126 BnLBD genes were analyzed using PlantCARE software, and four types of cis-acting elements were found in the promoter region of the BnLBD gene family, including elements related to plant growth and development, abiotic stress responses, hormones responses, and basic promoter elements in eukaryotes (such as CAAT-box and TATA-box) (Figure 9, Table S6). Promoter elements associated with growth and development mainly include GCN4-motif and AACA-motif involved in endosperm expression; CAT-motif related to meristems expression; and a large number of light-response elements such as Box 4, GA-motif, G-box, and TCT-motif. We found the elements related to light response were most common motif in BnLBD promoters, indicating that the BnLBD gene family could be induced by light to regulate plant growth and development. In the second category, mainly cis-elements related to GA, auxin, abscisic acid (ABA), MeJA, and salicylic acid (SA) responses were identified, of which we found the motifs associated with the ABA response were the most abundant, followed by motifs associated with the MeJA response. In addition, we also found motifs related to stresses responses, such as low temperature response (LTR) involved in low temperature response, MYB binding site (MBS) for drought induction, wound-responsive element (WUN-motif) associated with wound response, and anaerobic regulatory element (ARE) essential for anaerobic induction. Overall, these results indicate that BnLBDs might play functions in regulating plant growth and development, response to abiotic stresses, and hormone responses. In addition, we found most BnLBD genes with different types of cis-element in the promoter regions, suggesting that these BnLBDs may be involved in various biological processes and regulatory pathways.

Figure 9.
Cis-element analysis on the promoter regions of BnLBD genes. The presence of different cis-acting elements was determined by the PlantCARE software. Different colored boxes represent different cis-elements.
3.7. The Expression Pattern of BnLBD Genes under ABA Treatment
Phytohormones are essential for plant growth and development, and play important roles in stress response [48]. Several LBD genes have been identified with functions in response to hormone treatments and abiotic stresses. As we found the motifs related to ABA response were the most abundant cis-element in BnLBD promoters, we analyzed the expression pattern of BnLBDs under ABA treatment. A total of thirty BnLBDs with more than five ABA-responsive elements in the promoter regions were analyzed, of which BnLBD37/51/97/109 (orthologous with LBD1 in A. thaliana) and BnLBD9/69 (orthologous with LBD40 in A. thaliana) were significantly up-regulated after ABA treatment. The expressions of BnLBD9 and BnLBD69 were up-regulated under 3 h of ABA treatment, and the expression level under 12 h of ABA treatment was ~25 and 150 times higher than that of the control group, respectively. The expression of BnLBD51 and BnLBD109 was significantly up-regulated under 3 h of ABA treatment. BnLBD37 and BnLBD97 were strongly induced by 1 h of ABA treatment (Figure 10). Besides, we found another 11 BnLBD genes that were not induced by ABA treatment. These results indicate that 19 members of the BnLBD gene family might be involved in the ABA signaling pathway, which is consistent with the result of cis-element analysis in the promoters.
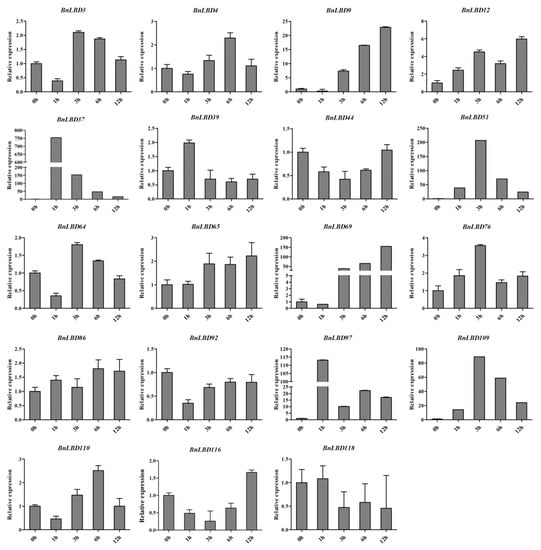
Figure 10.
The expression pattern of BnLBD genes under abscisic acid (ABA) treatment. Data were normalized to BnActin 2 gene and vertical bars indicate standard deviation.
4. Discussion
The LBD genes encode a class of plant-specific transcription factors, which play important roles in regulating the plant growth and development, especially in the development of lateral organs [1,14,15]. The LBD gene family has been extensively studied in different plants species, but the comprehensive analysis of LBDs in the oil crop B. napus has not yet been reported. In this study, we identified 126 BnLBD genes in B. napus. These BnLBD genes were divided into two groups and ten subgroups based on the evolutionary relationships, which is highly consistent with the reports in Arabidopsis, G. max, and Z. mays [1,6,12]. The variations in the structure of BnLBDs might be because of the gain or loss of introns/exons in the long-term evolutionary process. On the basis of the conserved motif analysis of BnLBDs, we suspect that group-specific and subgroup-specific motifs might be responsible for the functional divergence of the BnLBD gene family. In addition, the similar motif structure, exon–intron pattern, and protein sequences in each subgroup of BnLBDs might indicate the gene replication events during the evolutionary process.
B. napus, as an allopolyploid, was formed by natural hybridization between B. rapa and B. oleracea [33]. There are two main mechanisms for gene duplication, WGD /segmental duplication and tandem duplication [49]. WGD/segmental duplication is very common in plant genomes, as most plants have undergone polyploidy events during evolution, and thus retain many duplicated chromosome segments in their genomes [50,51]. Compared with other plant species such as Arabidopsis, O. sativa, Z. mays, G. max, and Populus, which contain 43, 35, 44, 80, and 57 LBD genes [1,6,12,13], respectively, we found B. napus with the largest number of LBD genes, indicating the genome duplication during evolution of B. napus greatly contributed to the diversification of BnLBDs. Further, all 126 BnLBD genes were identified as the result of gene duplication, 93 of them were identified with WGD or segmental duplications history, and the rest resulted from dispersed duplications. This indicated that WGD and segmental duplications are the primary force for the expansion of the BnLBD gene family.
In addition, tandem replication is another type of gene replication characterized by the presence of multiple members of a gene family in the same or adjacent intergenic regions [49]. In this study, we conducted a genome-wide analysis to identify all tandem replication gene pairs, and 3149 gene pairs were identified with tandem duplication, accounting for ~6% of the genes in the B. napus genome. Unfortunately, none of the LBD family members were found in these gene pairs. Additionally, 33 BnLBD genes were identified with dispersed duplication. Hitherto, little research on dispersed gene duplication has been reported in plant species. In the human genome, dispersed duplications such as transposon insertion and copy number variation are very common [52]. In heteroploid plants, the expansion of the genome is usually accompanied by the insertion of transposon factors [53,54]. Transposable elements in Brassica species are also abundant [54,55]. However, whether these transposons are one of the reasons for the expansion of LBD gene family is still unclear.
After gene duplication, not all the duplicated genes are retained or keep the original function. In theory, non-functionalization, neofunctionalization, and subfunctionalization are the three possible evolutionary consequences for duplicated genes [46]. Comparative genomic studies have shown that Brassica species, such as B. rapa and B. oleracea, diverged from a common ancestor A. thaliana via whole-genome triplication (WGT) approximately 20 to 40 million years ago [33,36,56]. Thus, three copies of each A. thaliana gene should exist in B. rapa and B. oleracea. The Arabidopsis genome contains 43 LBD genes, therefore, B. rapa and B. oleracea should contain more than 120 LBD genes after WGT, and ultimately result in even more BnLBD genes in B. napus. However, only 126 BnLBD genes were identified in this study, indicating that more than half of the LBD genes were lost after WGT. As reported, B. napus was derived from natural hybridization between two diploids (B. rapa and B. oleracea), while the diploids in Brassica have undergone polyploidization events based on Arabidopsis thaliana. Thus, we compared B. napus with A. thaliana to analyze the gene loss after WGD, and found 38.02% of genes were lost after WGD in B. napus (27,169 in A. thaliana and 101,040 in B. napus), while 51.16% of LBDs were lost in B. napus (43 in A. thaliana and 126 in B. napus). Compared with the gene loss rate in the whole genome, we may speculate the BnLBDs have undergone strong selection during B. napus evolution, and the LBDs kept should have interesting functions in B. napus development. On the basis of the phylogenetic tree, we can see that most AtLBD genes have more than one orthologous gene in B. napus, indicating that the LBD gene family has been expanded. However, most AtLBD genes have less than six orthologous genes in B. napus, which was less than expected. This agreed with the shrink of gene number in B. napus, indicating the BnLBD gene family was contracted during B. napus diversification. Besides, the orthologs of six AtLBDs (AtLBD8, AtLBD9, AtLBD26, AtLBD32, and AtLBD34) were not found in the B. napus genome, indicating that all the duplicated copies of these LBDs were lost during the evolutionary process. These lost LBDs might be redundant genes that were gradually replaced by other genes with similar functions. Previous studies also showed that all BnLBD genes have undergone intense purification selection, which plays a key role in maintaining the number of genes (Table S2, Figure S3).
According to the tissue expression pattern of BnLBDs, we found that not all the remaining BnLBD genes could be expressed normally, and more than 40 BnLBD genes were not expressed in the 15 tissues we analyzed. We suspect these genes might be expressed at specific developmental stages that were not covered in this study, or they were silenced after WGT. Another possibility is that these genes are pseudogenes. In addition, neofunctionalization and subfunctionalization may also greatly contribute as evolutionary forces in the divergence of the BnLBD gene family. Most of the paralogs in the BnLBD gene family are highly conserved at both the DNA and protein levels, including gene structure, protein sequence, and motif composition. The differences existing in some BnLBD gene pairs would lead to gene functional divergence and help to drive new functions. In Arabidopsis, LBD37/38/39 have been confirmed with functions in regulating anthocyanin biosythesis, and they are transcription factors located in the nucleus. Subcellular localization of BnLBD46/104/105, which are homologous to LBD37/38/39, indicated they are also nucleus anchered proteins, indicating they may with similar functions as LBD37/38/39.
Some of gene pairs were identified with variations in gene structure, such as loss or gain of motif in BnLBD38/57 and BnLBD15/104, as well as segmental loss or gain of intron in BnLBD11/79 and BnLBD118/126. Furthermore, the differential expression pattern of duplicated BnLBD gene pairs also indicated that these genes might have undergone functional differentiations. For example, BnLBD37 and BnLBD51, the orthologs of AtLBD1, were identified with different expression patterns in different tissues and under stress conditions. This indicates that BnLBD37 and BnLBD51 might have undergone functional divergence, and either generated new functions or inherited partial functions of ancestral genes. Besides, we found several couples of BnLBD homologs with different expression patterns during rapeseed development. For instance, we also found BnLBD104 and BnLBD106 (orthologous with LBD38 in A. thaliana) with totally different expression patterns throughout rapeseed development, and BnLBD12 and BnLBD65 (orthologous with AtLBD41 in A. thaliana) with different expression patterns in different developmental stages of B. napus.
Many studies have revealed that the LBD gene family plays significant roles in plant growth and development, as well as in responding to biotic and abiotic stresses [57,58]. On the basis of the tissue expression pattern, we found some valuable BnLBD genes might have functions in specific physiological processes. For instance, BnLBD46/120 and BnLBD15/104 were highly expressed in the root tissues, and their orthologous genes in A. thaliana (AtLBD37 and AtLBD38) were also reported with high expression in root tissues and functioned as the repressors of N-repressed gene and important molecular components in plant NO3−/N signaling. This indicates that BnLBD46/120 and BnLBD15/104 may have the same function in B. napus [30,59].
The functional roles of BnLBD genes related to different stresses were also speculated in this study. On the basis of PlantCARE software, we identified a large amount of cis-acting elements, which were associated with abiotic stress responses and hormones responses. All BnLBD promoters contain at least one cis-element associated with abiotic stress or hormonal response, suggesting that these genes may play functions in plant response to abiotic stress and hormonal stress. To further confirm this speculation, we analyzed the expression pattern of BnLBD genes to ABA treatment, and found the expression level of 19 BnLBDs was significantly induced after ABA treatment. This indicates that these BnLBD genes play functions in plant response to ABA treatment and may participate in plant responses to biotic and abiotic stresses. In general, this study provided general information on the diversification and putative functions of LBD genes in B. napus, and would be helpful in further functional studies of BnLBDs.
5. Conclusions
In this study, we systematically analyzed 126 LBD genes in B. napus, which were further classified into nine subgroups, and each group was clustered with similar gene structure and motif composition. Collinear analysis confirmed the high homology among BnLBDs and LBDs in A. thaliana, B. rapa, and B. oleracea. The evolutionary analysis indicated that whole-genome duplication and segmental duplication are the primary sources for the expansion of LBD genes in the B. napus genome. After polyploidization, gene loss and functional divergence occurred in the BnLBD gene family. On the basis of tissue expression, we identified the BnLBD genes with putative roles in specific plant growth and developmental stages. Through promoter analysis and hormone treatment, we identified 19 BnLBD genes involved in plant response to ABA treatment. Generally, the bioinformatics and expression analysis of BnLBDs provided us with a basic, but comprehensive understanding of the origin and evolutionary history of LBDs in B. napus, and also provided a theoretical basis for the further functional study on BnLBDs.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/3/280/s1, Figure S1. Multiple sequence alignment of LOB domains in BnLBD proteins using ClustalX, Figure S2. Conserved protein motifs BnLBDs family. The bit score exhibits the information content for each position in the sequence, Figure S3. Ka/Ks value and divergence time distribution of LBD genes. (a). B. napus-B. napus. (b) B. napus-B. rapa (c). B. napus-B. oleracea. (d). B. napus-A. thaliana, Table S1. The primers used in this study, Table S2. The information of BnLBD gene family in B. napus, Table S3. Duplication type of BnLBD genes, Table S4. Ka/Ks calculation of the duplicated LBD gene pairs, Table S5. The RNA-seq data of BnLBD genes in different tissues and developmental stages, Table S6. The cis-elements on the promoter regions of BnLBD genes.
Author Contributions
T.X. performed the experiments and drafted the manuscript; L.Z., H.R., and T.X. analyzed the RNA-seq data; X.C. and J.W. sampled the plant materials; J.J., J.B., and Y.W. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFE0108000, 2016YFD0101000), the National Natural Science Foundations (31972963, 31771825, 31771824), the Natural Science Foundation of Jiangsu Province (BK20180101, BE2018356), Jiangsu Province Government Project (BM2018003)/The Open Funds of the Key Laboratory of Plant Functional Genomics of the Ministry of Education (ML201804), the Undergraduate Training Program for Innovation and Entrepreneurship (X20190712), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Liu, C.Y.; Liu, C.X.; Zhao, Y.L.; Xu, R.R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, B.; Zeng, Q.; Xiang, Z.; He, N. Identification and characterization of Lateral Organ. Boundaries Domain genes in mulberry, Morus notabilis. Meta Gene 2016, 8, 44–50. [Google Scholar] [CrossRef]
- Yang, T.; Fang, G.Y.; He, H.; Chen, J. Genome-wide identification, evolutionary analysis and expression profiles of LATERAL ORGAN BOUNDARIES DOMAIN gene family in Lotus japonicus and Medicago truncatula. PLoS ONE 2016, 11, e0161901. [Google Scholar] [CrossRef]
- Gombos, M.; Zombori, Z.; Szecsenyi, M.; Sandor, G.; Kovacs, H.; Gyorgyey, J. Characterization of the LBD gene family in Brachypodium: A phylogenetic and transcriptional study. Plant Cell Rep. 2017, 36, 61–79. [Google Scholar] [CrossRef]
- Grimplet, J.; Pimentel, D.; Agudelo-Romero, P.; Martinez-Zapater, J.M.; Fortes, A.M. The LATERAL ORGAN BOUNDARIES domain gene family in grapevine: Genome-wide characterization and expression analyses during developmental processes and stress responses. Sci. Rep. 2017, 7, 15968. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Shi, G.; Du, H.; Wang, H.; Zhang, Z.; Hu, D.; Wang, J.; Huang, F.; Yu, D. Genome-wide analysis of soybean LATERAL ORGAN BOUNDARIES Domain-containing genes: A functional investigation of GmLBD12. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the LATERAL ORGAN BOUNDARIES DOMAIN gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Some, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef]
- Evans, M.M. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- Li, C.; Zou, X.; Zhang, C.; Shao, Q.; Liu, J.; Liu, B.; Liu, H.; Zhao, T. OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS ONE 2016, 11, e0156413. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S.; Yu, X.; Yu, J.; He, X.; Zhang, S.; Shou, H.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, X.; Wang, G.; Liu, H.; Zhu, J. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot. 2012, 110, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Cho, C.; Kim, J. Lateral Organ. Boundaries Domain16 and 18 act downstream of the AUXIN 1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015, 168, 1792–1806. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.; Yong Kang, N.; Cho, C.; Joon Seo, P.; Chung Suh, M.; Kim, J. LBD14/ASL17 positively regulates lateral root formation and is involved in ABA response for root architecture in Arabidopsis. Plant Cell Physiol. 2017, 58, 2190–2201. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, W.; Huang, Y.; Niu, X.; Zhao, Y.; Han, Y.; Liu, Y. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 2015, 66, 99–112. [Google Scholar] [CrossRef]
- Bell, E.M.; Lin, W.C.; Husbands, A.Y.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef] [PubMed]
- Mangeon, A.; Bell, E.M.; Lin, W.C.; Jablonska, B.; Springer, P.S. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 2011, 62, 221–233. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.; Kazan, K.; Manners, J.M. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef]
- Ba, L.J.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. J. Agric. Food Chem. 2016, 64, 738–745. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; dePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Mandakova, T.; Wu, J.; Xie, Q.; Lysak, M.A.; Wang, X. Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 2013, 25, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Cheung, K.; Kitschke, M.; Bures, P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 2007, 145, 402–410. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Sonnhammer, E.L.L.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Wang, D.P.; Wan, H.L.; Zhang, S.; Yu, J. γ-MYN: A new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct. 2009, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, S.; He, F.; Zhu, J.; Hu, S.; Yu, J. How do variable substitution rates influence Ka and Ks calculations? Genom. Proteom. Bioinf. 2009, 7, 116–127. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Parkin, I.A.; Gulden, S.M.; Sharpe, A.G.; Lukens, L.; Trick, M.; Osborn, T.C.; Lydiate, D.J. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 2005, 171, 765–781. [Google Scholar] [CrossRef]
- Kroon, M.; Lameijer, E.W.; Lakenberg, N.; Hehir-Kwa, J.Y.; Thung, D.T.; Slagboom, P.E.; Kok, J.N.; Ye, K. Detecting dispersed duplications in high-throughput sequencing data using a database-free approach. Bioinformatics 2016, 32, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Sabot, F.; Guyot, R.; Wicker, T.; Chantret, N.; Laubin, B.; Chalhoub, B.; Leroy, P.; Sourdille, P.; Bernard, M. Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol. Genet. Genom. 2005, 274, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Sarilar, V.; Palacios, P.M.; Rousselet, A.; Ridel, C.; Falque, M.; Eber, F.; Chevre, A.M.; Joets, J.; Brabant, P.; Alix, K. Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. New Phytol. 2013, 198, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.X.; Du, J.C.; Lin, F.; Tong, C.B.; Yu, J.Y.; Huang, S.M.; Wang, X.W.; Liu, S.Y.; Ma, J.X. Shifts in the evolutionary rate and intensity of purifying selection between two Brassica genomes revealed by analyses of orthologous transposons and relics of a whole genome triplication. Plant J. 2013, 76, 211–222. [Google Scholar]
- Dun, X.; Shen, W.; Hu, K.; Zhou, Z.; Xia, S.; Wen, J.; Yi, B.; Shen, J.; Ma, C.; Tu, J.; et al. Neofunctionalization of duplicated Tic40 genes caused a gain-of-function variation related to male fertility in Brassica oleracea lineages. Plant Physiol. 2014, 166, 1403–1419. [Google Scholar] [CrossRef]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.M.; Springer, P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).