Model-Based Clustering with Measurement or Estimation Errors
Abstract
1. Introduction
2. Materials and Methods
2.1. Review of MCLUST Model
2.2. MCLUST-ME Model
2.3. Expectation-Maximization (EM) Algorithm
- M step: Given current estimates of , maximize the complete-data log-likelihood with respect to .
- E step: Given estimates from last M step, for all and , compute the membership probabilities
2.4. Initial Values
2.5. Model Selection
2.6. Decision Boundaries for Two-Group Clustering
2.6.1. MCLUST Boundary
2.6.2. MCLUST-ME Boundary
2.7. Related Methods
3. Results
3.1. Simulation 1: Clustering Performance
3.1.1. Data Generation
- (1)
- Generate i.i.d. from Bernoulli. For each i, will serve as indicator for error, and on average, a proportion of data points will be associated with error.
- (2)
- Generate i.i.d. from Bernoulli. Parameter will be the mixing proportion.
- (3)
- For , generate from
3.1.2. Simulation Procedure
- (1)
- Choose a value for from .
- (2)
- Randomly select a random seed.
- (3)
- Generate a random sample following Section 3.1.1.
- (4)
- Run MCLUST and MCLUST-ME, fixing . Initiate with true memberships.
- (5)
- Repeat – for 100 different seeds.
- (6)
- Repeat – for each value of .
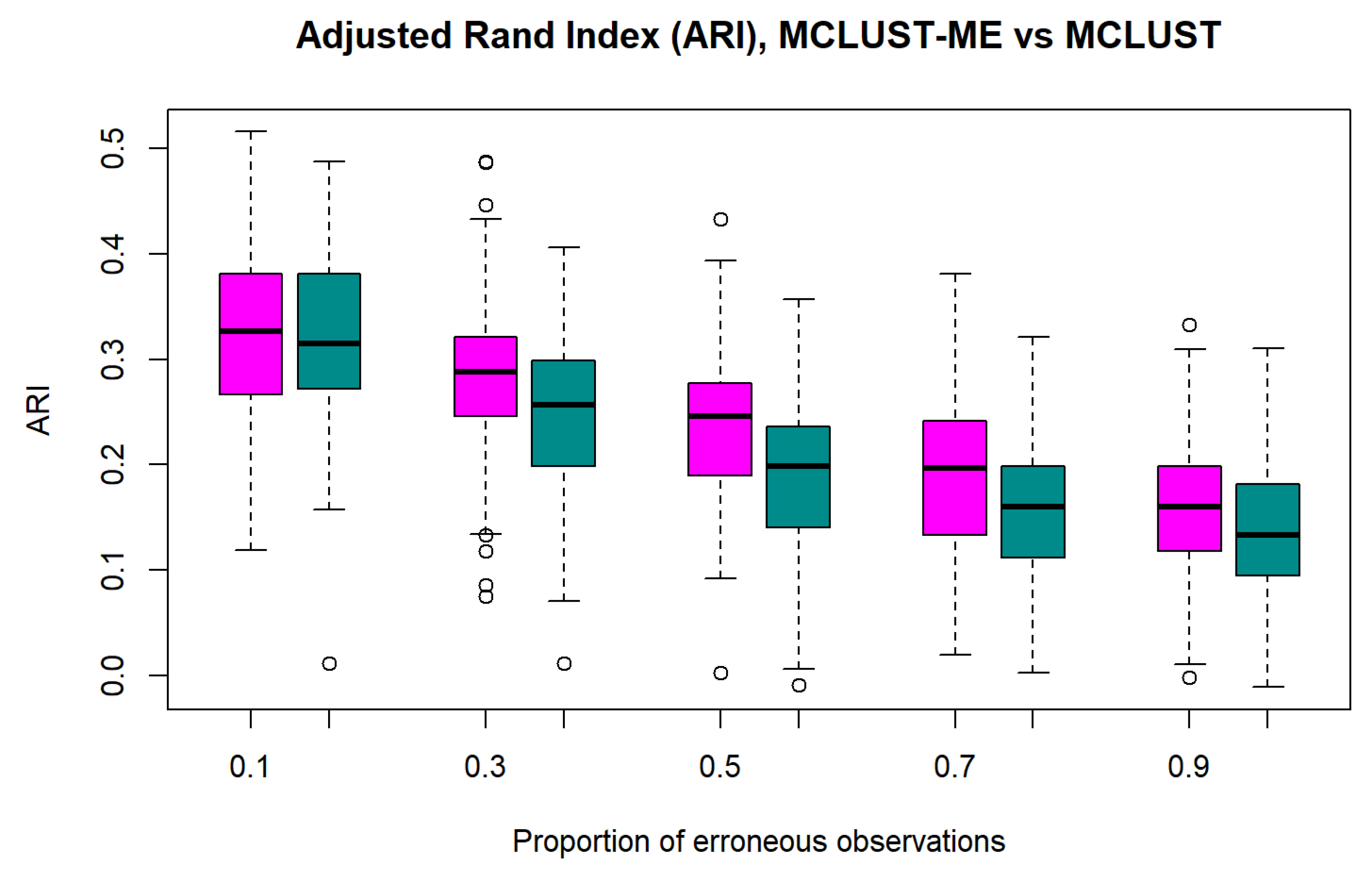
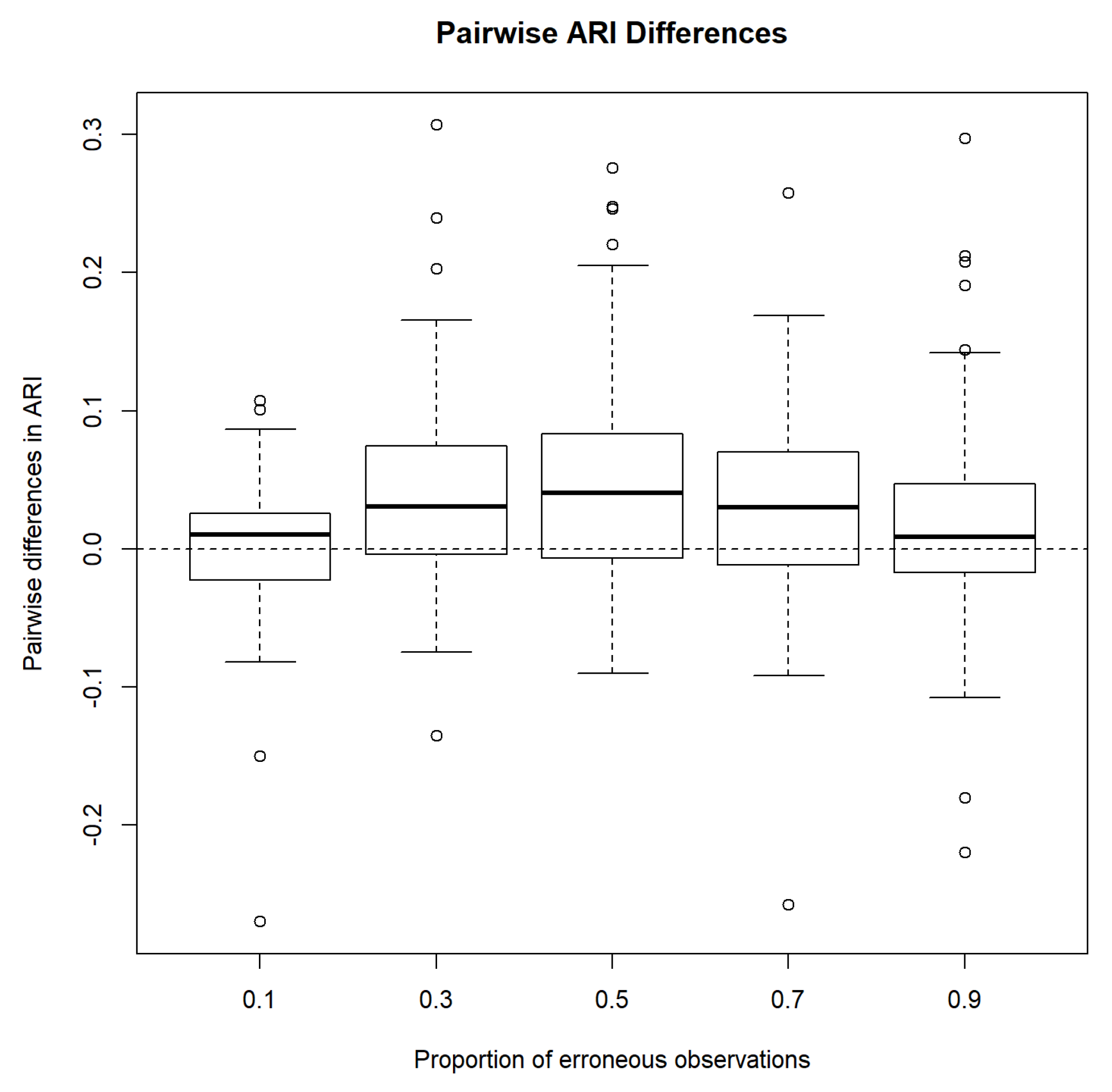
3.1.3. The Adjusted Rand Index
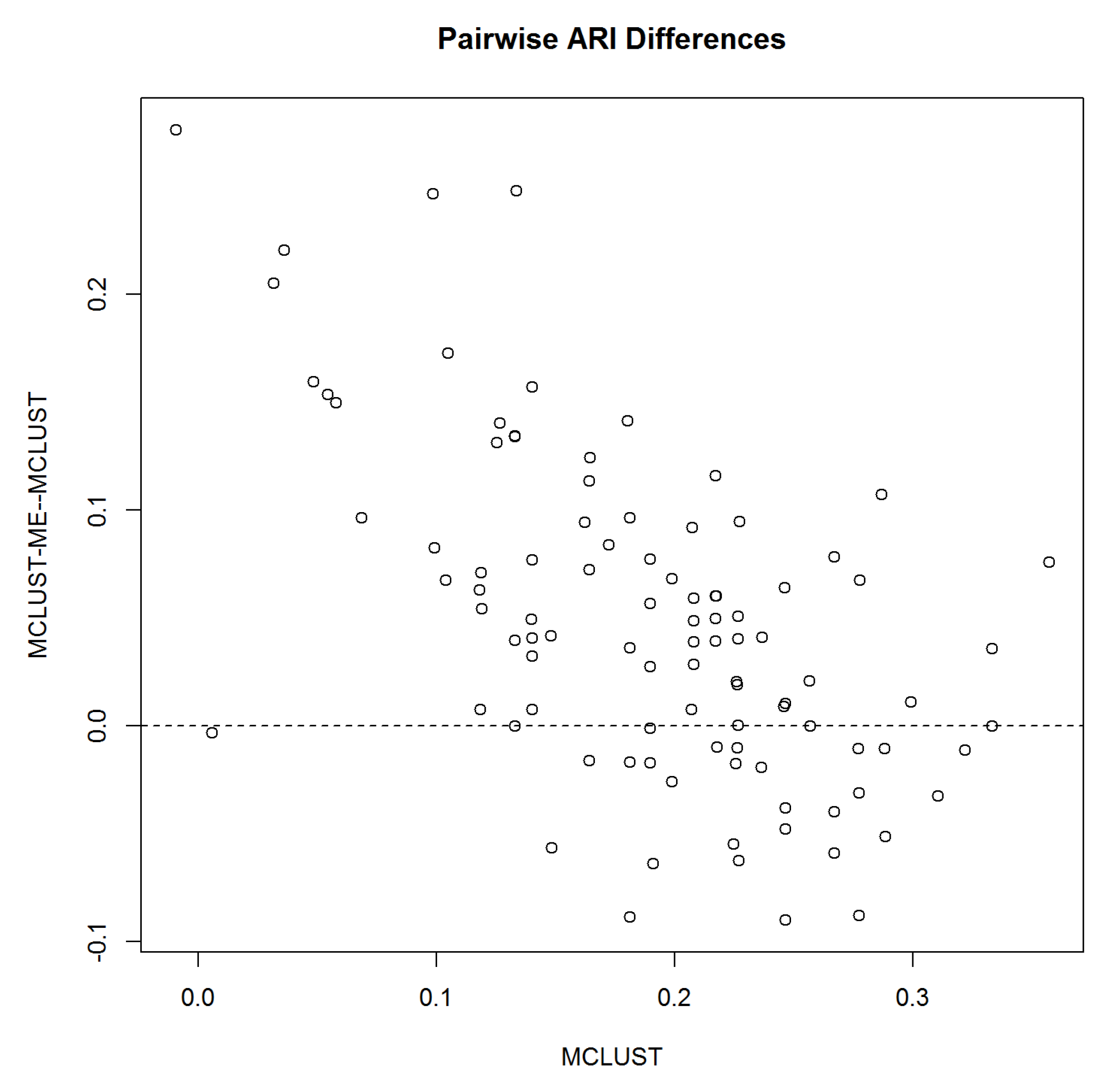
3.1.4. Simulation 1 Results
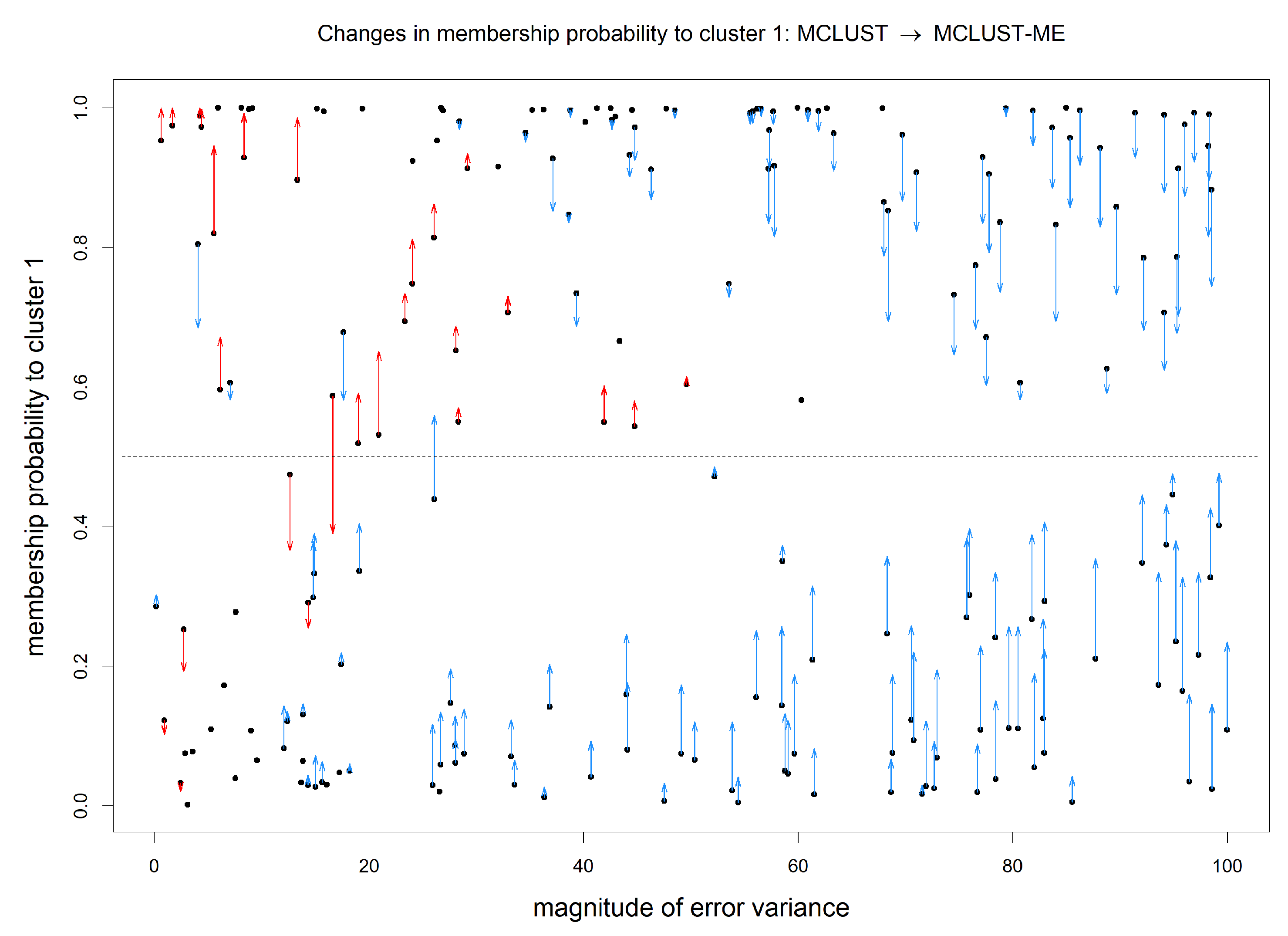
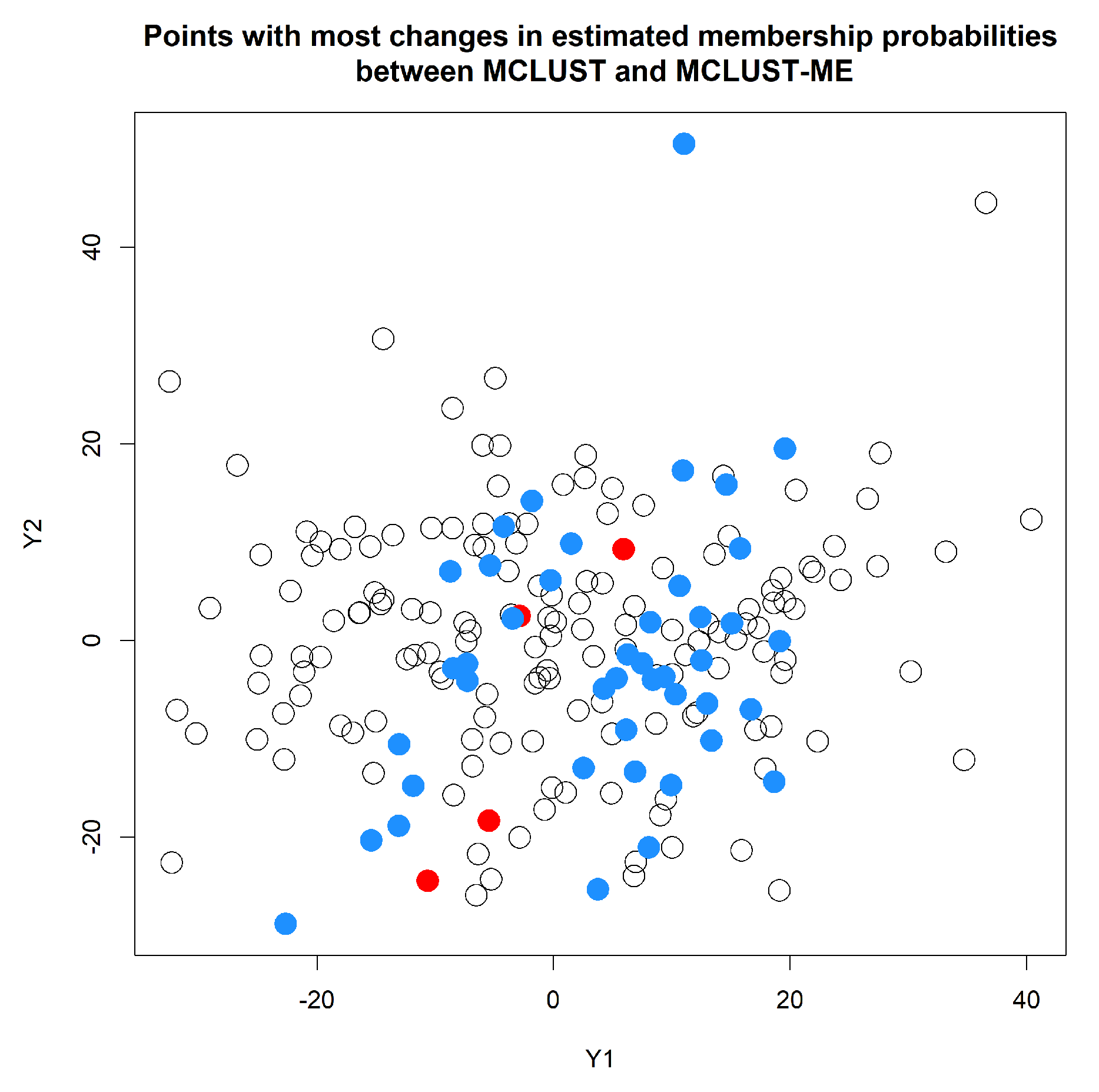
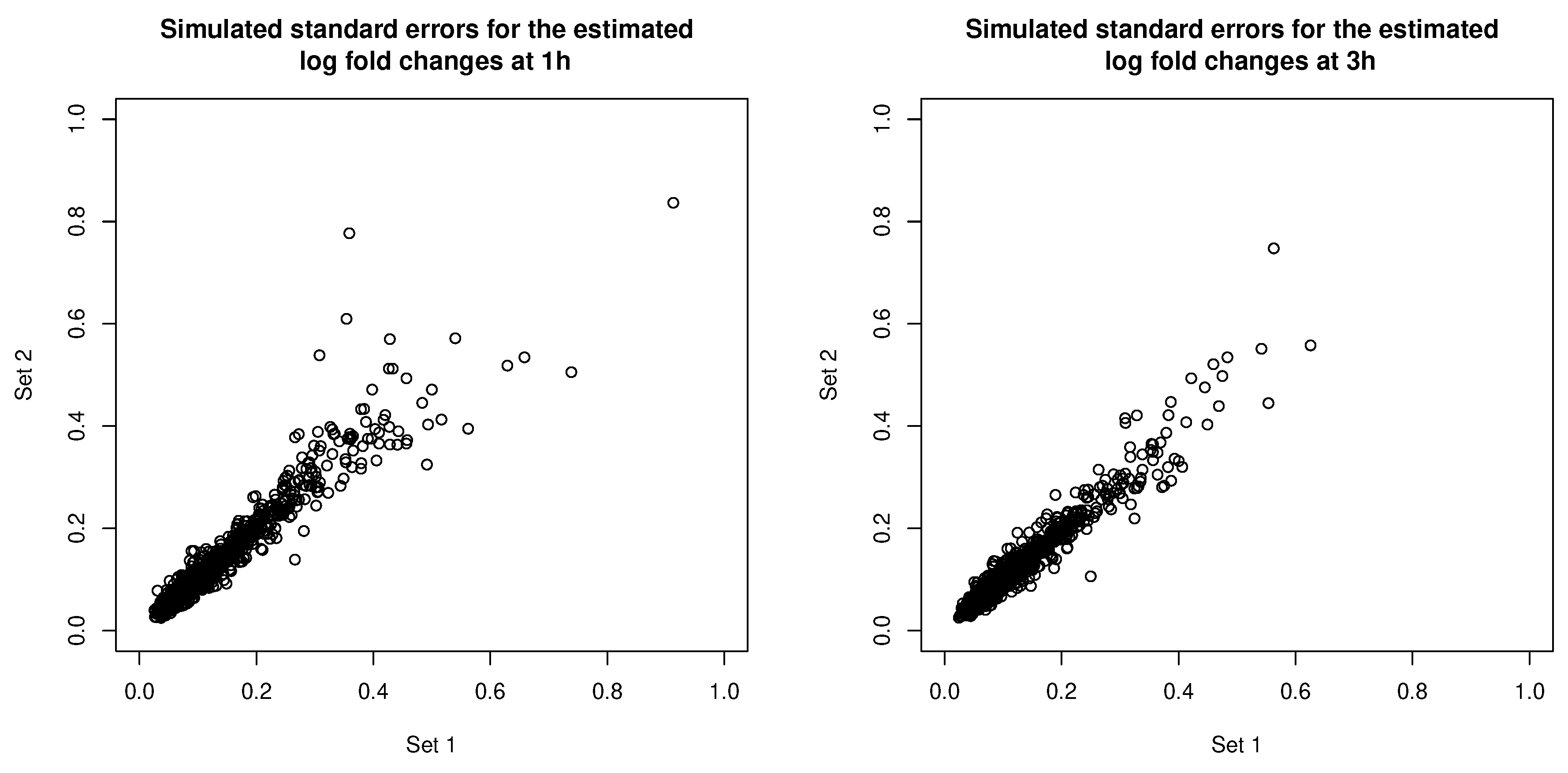
3.2. Simulation 2: Clustering Uncertainties and Magnitudes of Error Covariances
3.2.1. Data Generation
- (1)
- Generate i.i.d. from Uniform, where denotes the magnitude of error covariance for observation i.
- (2)
- Generate i.i.d. from Bernoulli. Parameter will be the mixing proportion.
- (3)
- For , generate fromwhere denotes the 2-dimensional identity matrix.
3.2.2. Simulation Procedure
- (1)
- Generate a random sample following Section 3.2.1.
- (2)
- Run MCLUST and MCLUST-ME, fixing . Initiate with true memberships.
- (3)
- Record cluster membership probabilities and MLEs for model parameters upon convergence.
3.2.3. Simulation 2 Results
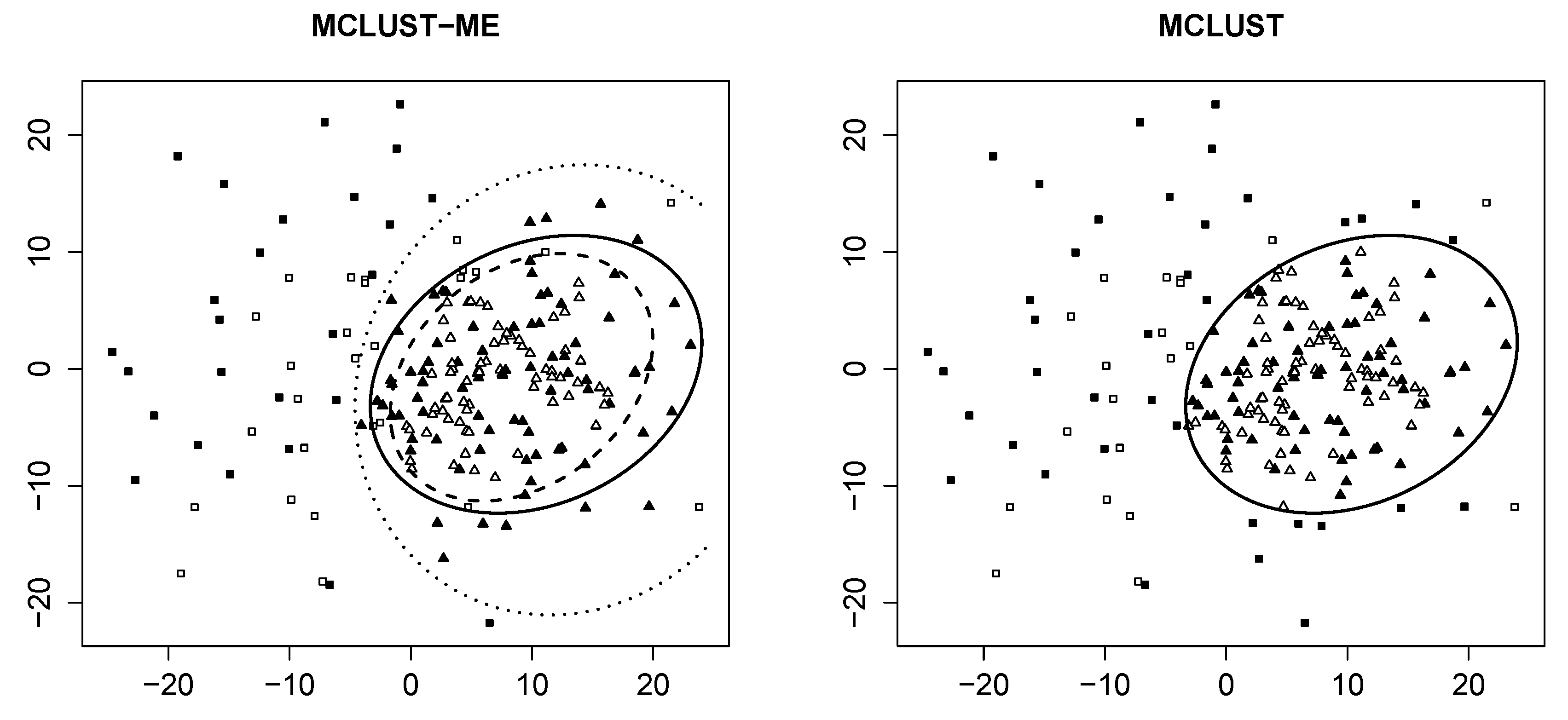
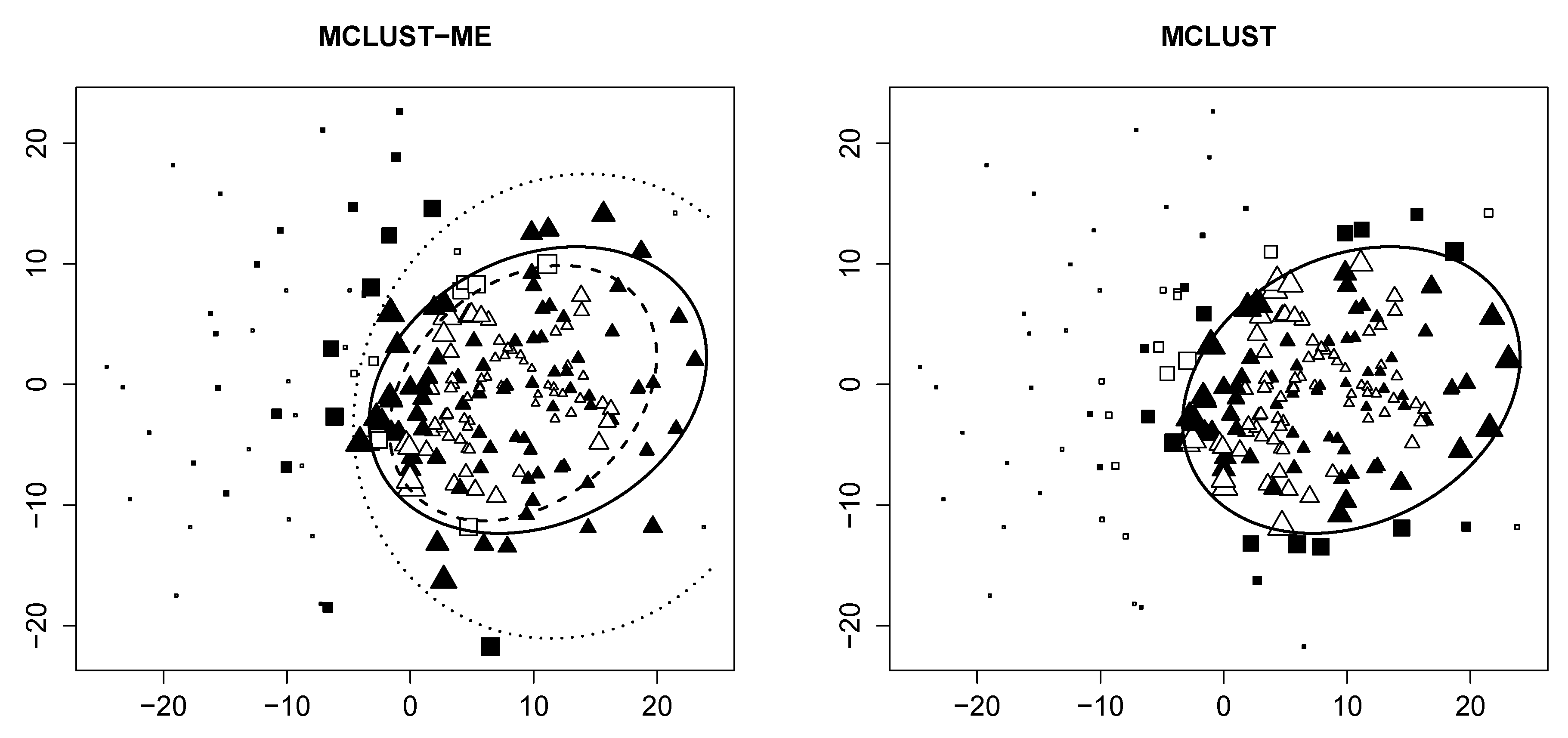
3.3. A Real Data Example
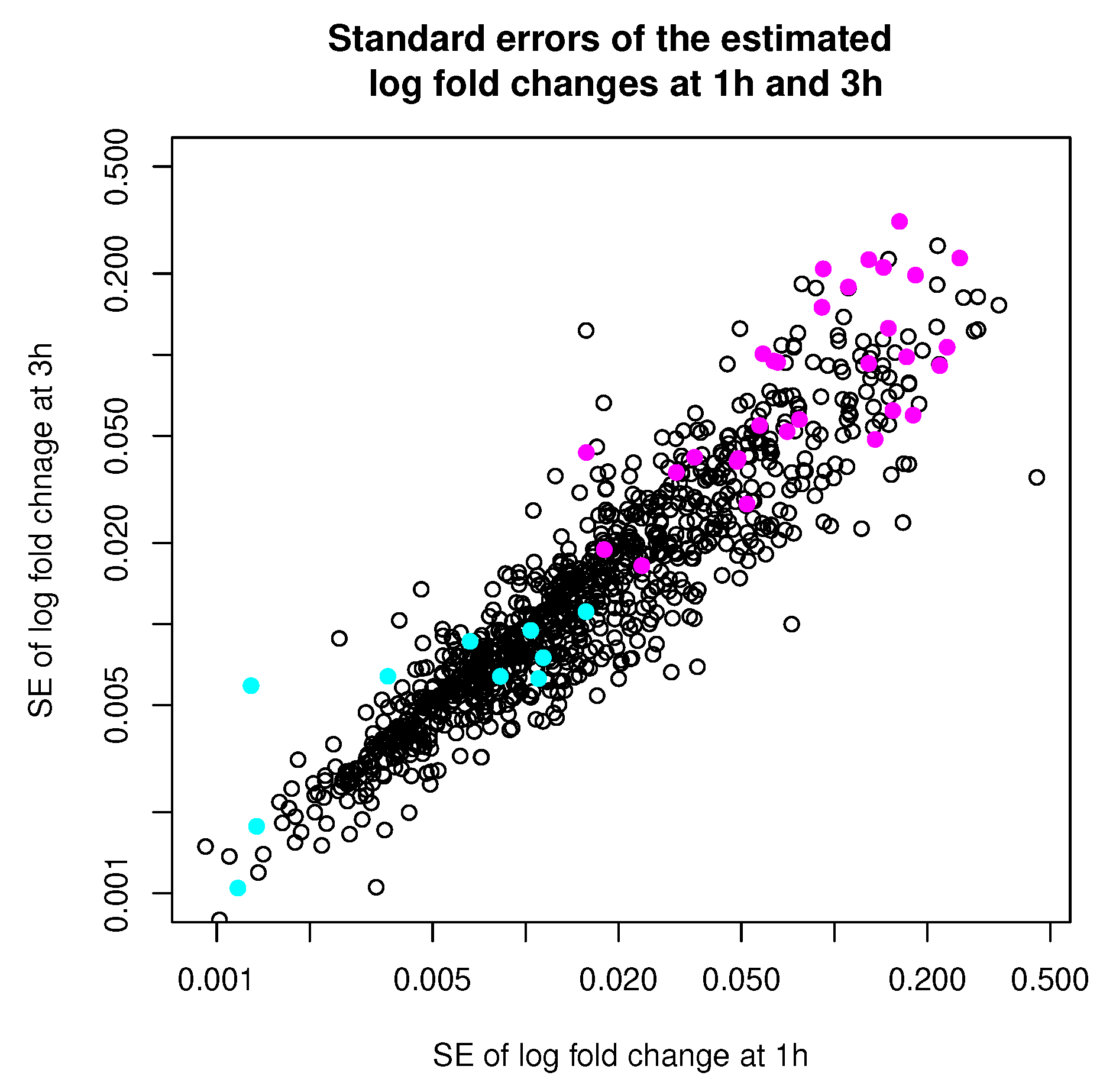
3.3.1. Data Description
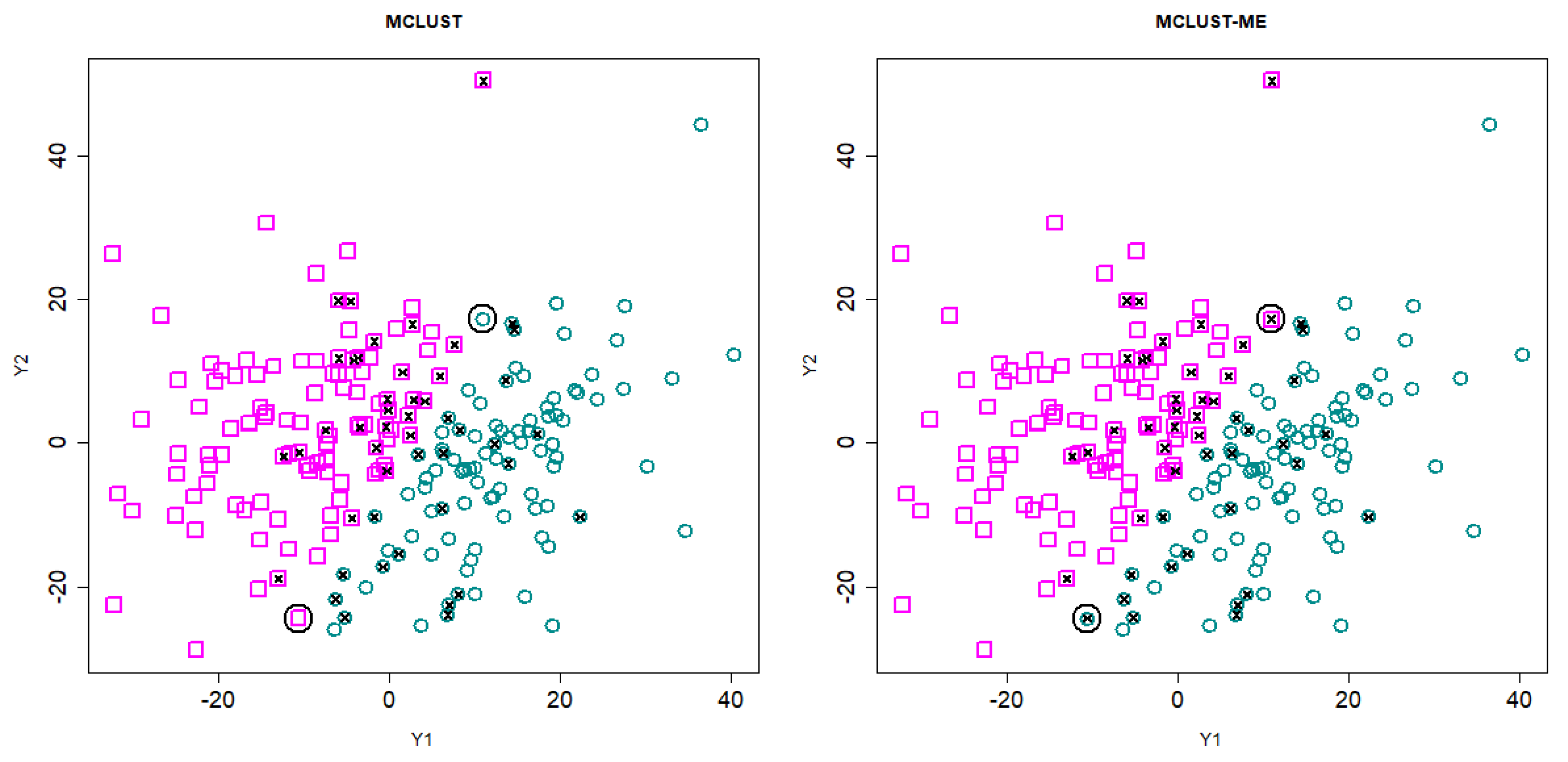
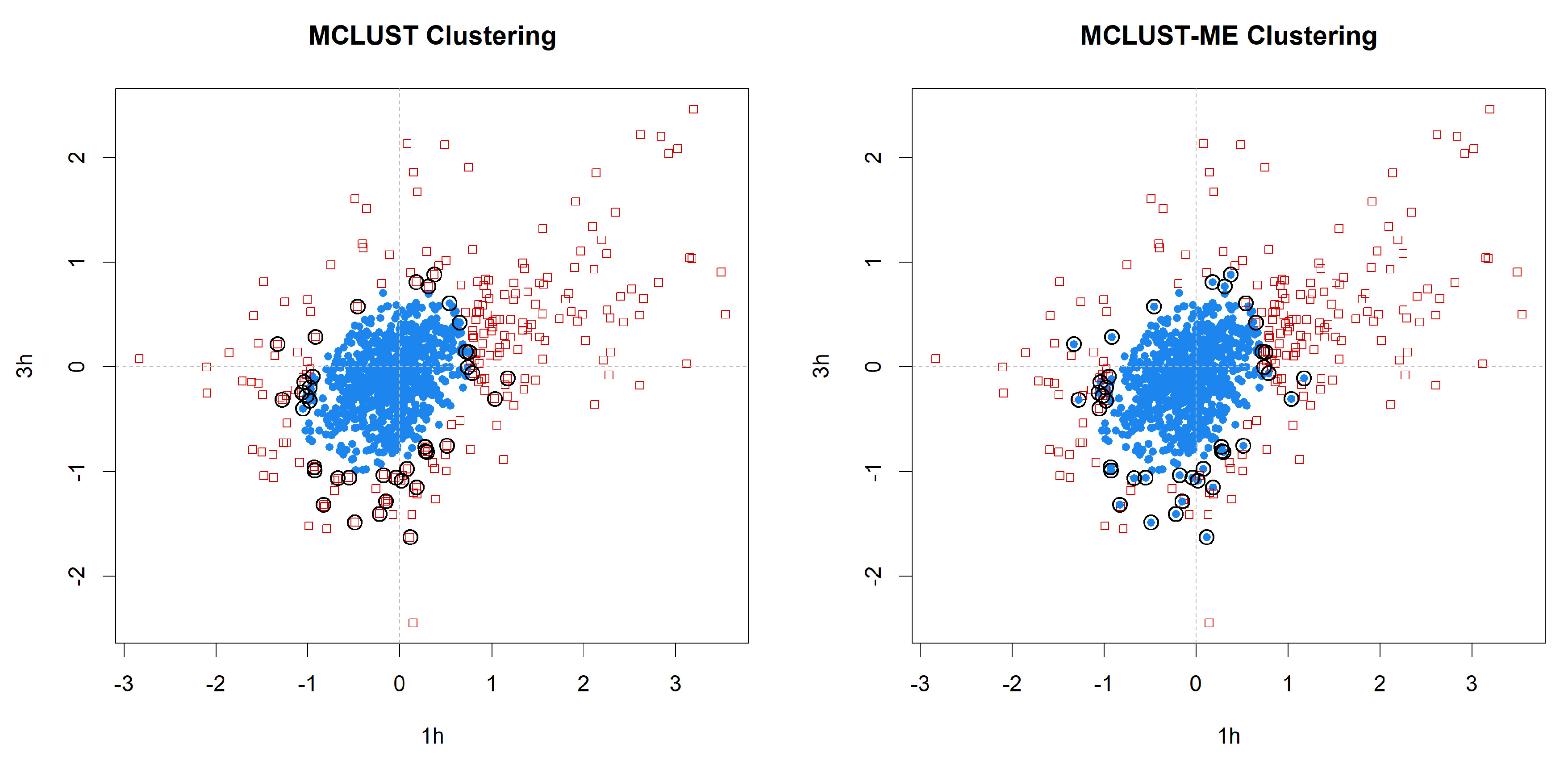
3.3.2. Cluster Analysis
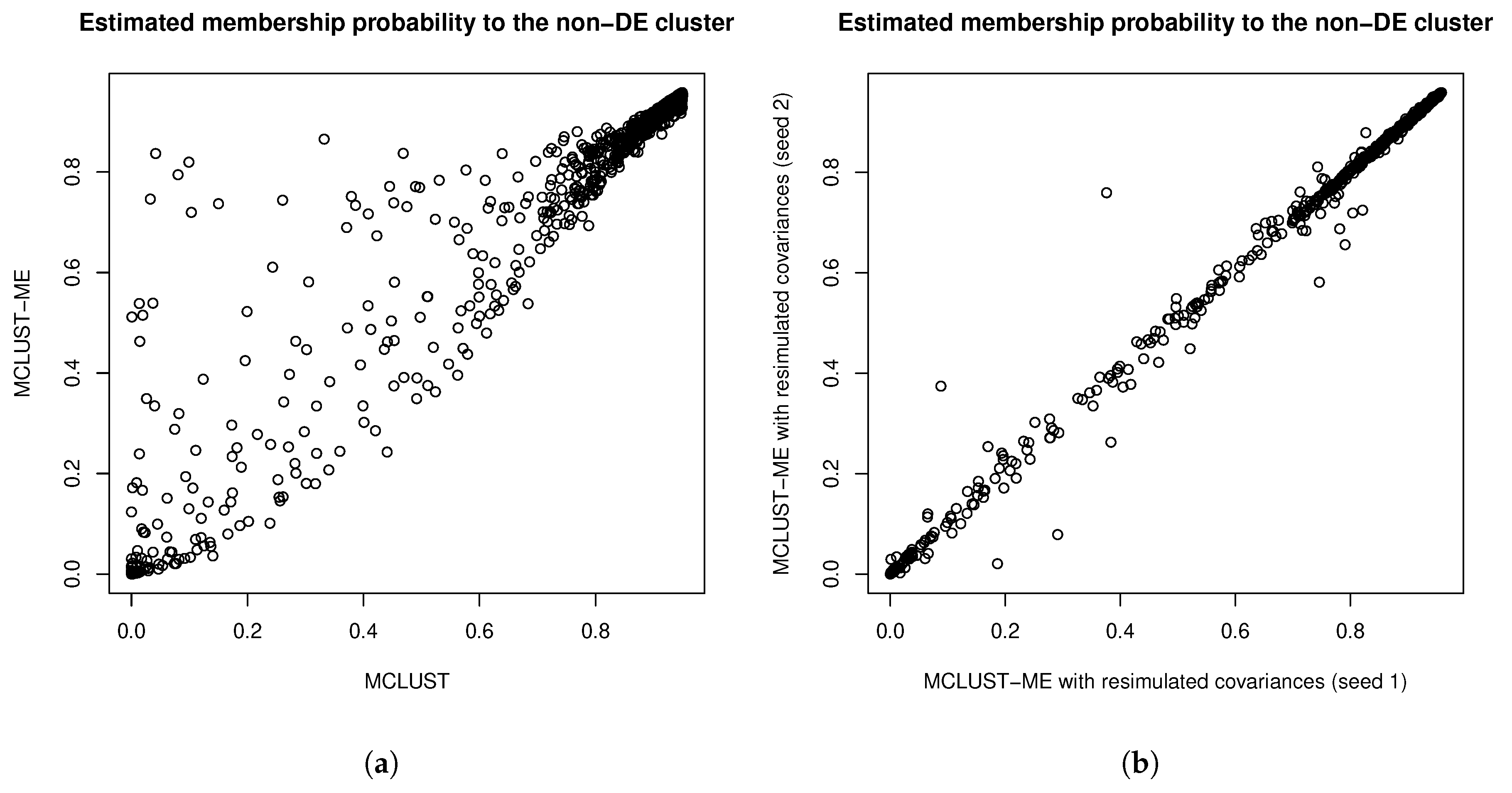
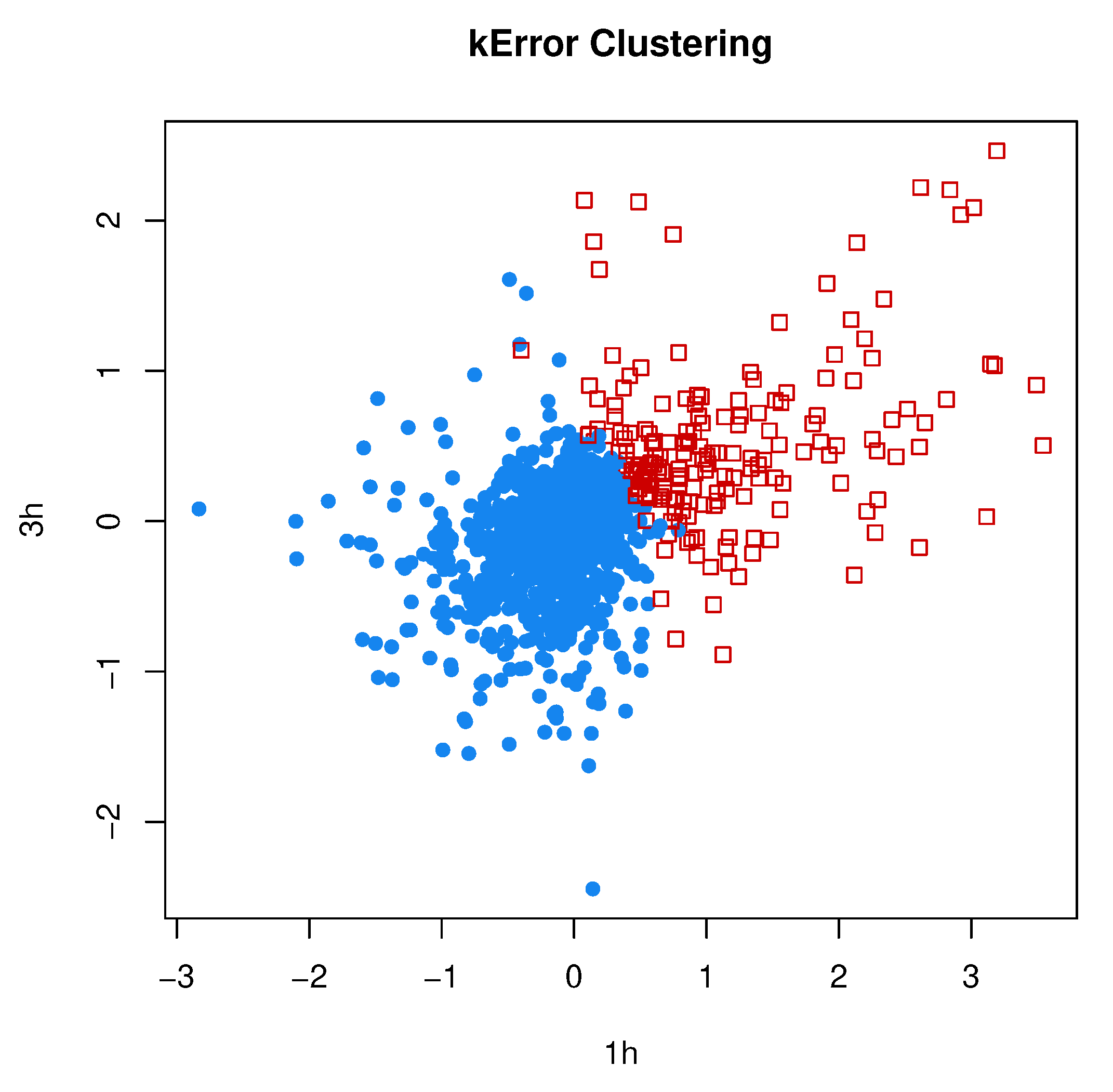
3.3.3. Comparison to kError
4. Conclusions and Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | Adjusted Rand index |
| BIC | Bayesain information criterion |
| DE | Differentially expressed |
| EM | Expectation-maximization |
| MLE | Maximum likelihood estimate(s) |
| iid | independent and identically distributed |
| RI | Rand index |
References
- Fraley, C.; Raftery, A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 2002, 97, 611–631. [Google Scholar] [CrossRef]
- Bouveyron, C.; Celeux, G.; Murphy, T.B.; Raftery, A.E. Model-Based Clustering and Classification for Data Science: With Applications in R; Cambridge University Press: Cambridge, UK, 2019; Volume 50. [Google Scholar]
- Wolfe, J.H. Pattern clustering by multivariate mixture analysis. Multivar. Behav. Res. 1970, 5, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Fraley, C.; Raftery, A.E. Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. J. Classif. 2003, 20, 263–286. [Google Scholar] [CrossRef]
- Fraley, C.; Raftery, A.E.; Murphy, T.B.; Scrucca, L. Mclust Version 4 for R: Normal Mixture Modeling for Model-Based Clustering, Classification, and Density Estimation; Tech. Rep. No. 597; Department of Statistics, University of Washington: Washington, DC, USA, 2012. [Google Scholar]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Banfield, J.D.; Raftery, A.E. Model-based Gaussian and non-Gaussian clustering. Biometrics 1993, 49, 803–821. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Society. Ser. B (Methodological) 1977, 39, 1–38. [Google Scholar]
- Celeux, G.; Govaert, G. Gaussian parsimonious clustering models. Pattern Recognit. 1995, 28, 781–793. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Dasgupta, A.; Raftery, A.E. Detecting features in spatial point processes with clutter via model-based clustering. J. Am. Stat. Assoc. 1998, 93, 294–302. [Google Scholar] [CrossRef]
- Zhang, W. Model-based Clustering Methods in Exploratory Analysis of RNA-Seq Experiments. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, 2017. [Google Scholar]
- Dwyer, P.S. Some applications of matrix derivatives in multivariate analysis. J. Am. Stat. Assoc. 1967, 62, 607–625. [Google Scholar] [CrossRef]
- Byrd, R.H.; Lu, P.; Nocedal, J.; Zhu, C. A limited memory algorithm for bound constrained optimization. SIAM J. Sci. Comput. 1995, 16, 1190–1208. [Google Scholar] [CrossRef]
- Murtagh, F.; Raftery, A.E. Fitting straight lines to point patterns. Pattern Recognit. 1984, 17, 479–483. [Google Scholar] [CrossRef]
- Fraley, C. Algorithms for model-based Gaussian hierarchical clustering. SIAM J. Sci. Comput. 1998, 20, 270–281. [Google Scholar] [CrossRef]
- Karlis, D.; Xekalaki, E. Choosing initial values for the EM algorithm for finite mixtures. Comput. Stat. Data Anal. 2002, 41, 577–900. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, N.R. Clustering data with measurement errors. Comput. Stat. Data Anal. 2007, 51, 6084–6101. [Google Scholar] [CrossRef]
- Tjaden, B. An approach for clustering gene expression data with error information. BMC Bioinform. 2006, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Rand, W.M. Objective Criteria for the Evaluation of Clustering Methods. J. Am. Stat. Assoc. 1971, 66, 846–850. [Google Scholar] [CrossRef]
- Santos, J.M.; Embrechts, M. On the use of the adjusted Rand index as a metric for evaluating supervised classification. In Proceedings of the ICANN (International Conference on Artificial Neural Networks), Limassol, Cyprus, 14–17 September 2009; Springer: Berlin, Germany, 2009; pp. 175–184. [Google Scholar]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Qannari, E.M.; Courcoux, P.; Faye, P. Significance test of the adjusted Rand index. Application to the free sorting task. Food Qual. Prefer. 2014, 32, 93–97. [Google Scholar] [CrossRef]
- Di, Y. Single-gene negative binomial regression models for RNA-Seq data with higher-order asymptotic inference. Stat. Its Interface 2015, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
| Gene ID | Log Fold Change (SE) 1 h | Log Fold Change (SE) 3 h | MCLUST | MCLUST−ME |
|---|---|---|---|---|
| AT2G42230 | −0.277 (0.006) | 0.152 (0.006) | 0.920 | 0.921 |
| AT3G56110 | 0.081 (0.121) | 0.228 (0.099) | 0.919 | 0.895 |
| AT1G23330 | 0.351 (0.018) | −0.209 (0.012) | 0.862 | 0.870 |
| AT5G23060 | −0.243 (0.005) | −0.909 (0.005) | 0.684 | 0.751 |
| AT5G06240 | −0.680 (0.022) | 0.103 (0.012) | 0.774 | 0.764 |
| AT3G20350 | −0.952 (0.007) | −0.090 (0.009) | 0.562 | 0.396 |
| AT1G30440 | −1.056 (0.010) | −0.398 (0.009) | 0.511 | 0.375 |
| AT1G30490 | −0.983 (0.008) | −0.322 (0.006) | 0.612 | 0.480 |
| AT1G23400 | −1.017 (0.011) | −0.275 (0.006) | 0.547 | 0.418 |
| AT1G17980 | 0.734 (0.001) | −0.001 (0.006) | 0.524 | 0.363 |
| AT2G30890 | −1.040 (0.150) | −0.142 (0.125) | 0.445 | 0.771 |
| AT5G15160 | −0.044 (0.129) | −1.059 (0.225) | 0.332 | 0.866 |
| AT5G45310 | −0.221 (0.162) | −1.404 (0.313) | 0.042 | 0.837 |
| AT5G46871 | 0.373 (0.065) | 0.886 (0.094) | 0.305 | 0.581 |
| AT2G22240 | 0.076 (0.016) | −0.975 (0.043) | 0.371 | 0.690 |
| Partition | Q | |
|---|---|---|
| R | Pair in same group | Pair in different groups |
| Pair in Same Group | a | b |
| Pair in Different Groups | c | d |
| 0.1 | 0.3 | 0.5 | 0.7 | 0.9 | |
|---|---|---|---|---|---|
| -value | 0.256 | 0 | 0 | 0 | 0.002 |
| MCLUST-ME | |||
|---|---|---|---|
| MCLUST | Non-DE | DE | |
| Non-DE | 775 | 10 | |
| DE | 30 | 185 | |
| MCLUST-ME Run 1 | |||
|---|---|---|---|
| MCLUST-ME Run 2 | Non-DE | DE | |
| Non-DE | 801 | 2 | |
| DE | 6 | 191 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Di, Y. Model-Based Clustering with Measurement or Estimation Errors. Genes 2020, 11, 185. https://doi.org/10.3390/genes11020185
Zhang W, Di Y. Model-Based Clustering with Measurement or Estimation Errors. Genes. 2020; 11(2):185. https://doi.org/10.3390/genes11020185
Chicago/Turabian StyleZhang, Wanli, and Yanming Di. 2020. "Model-Based Clustering with Measurement or Estimation Errors" Genes 11, no. 2: 185. https://doi.org/10.3390/genes11020185
APA StyleZhang, W., & Di, Y. (2020). Model-Based Clustering with Measurement or Estimation Errors. Genes, 11(2), 185. https://doi.org/10.3390/genes11020185




