Suppression of Notch Signaling Stimulates Progesterone Synthesis by Enhancing the Expression of NR5A2 and NR2F2 in Porcine Granulosa Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. siRNAs and Cell Transfection
2.3. RT-qPCR Assay
2.4. Western Blot Analysis
2.5. ELISA
2.6. Statistical Analysis
3. Results
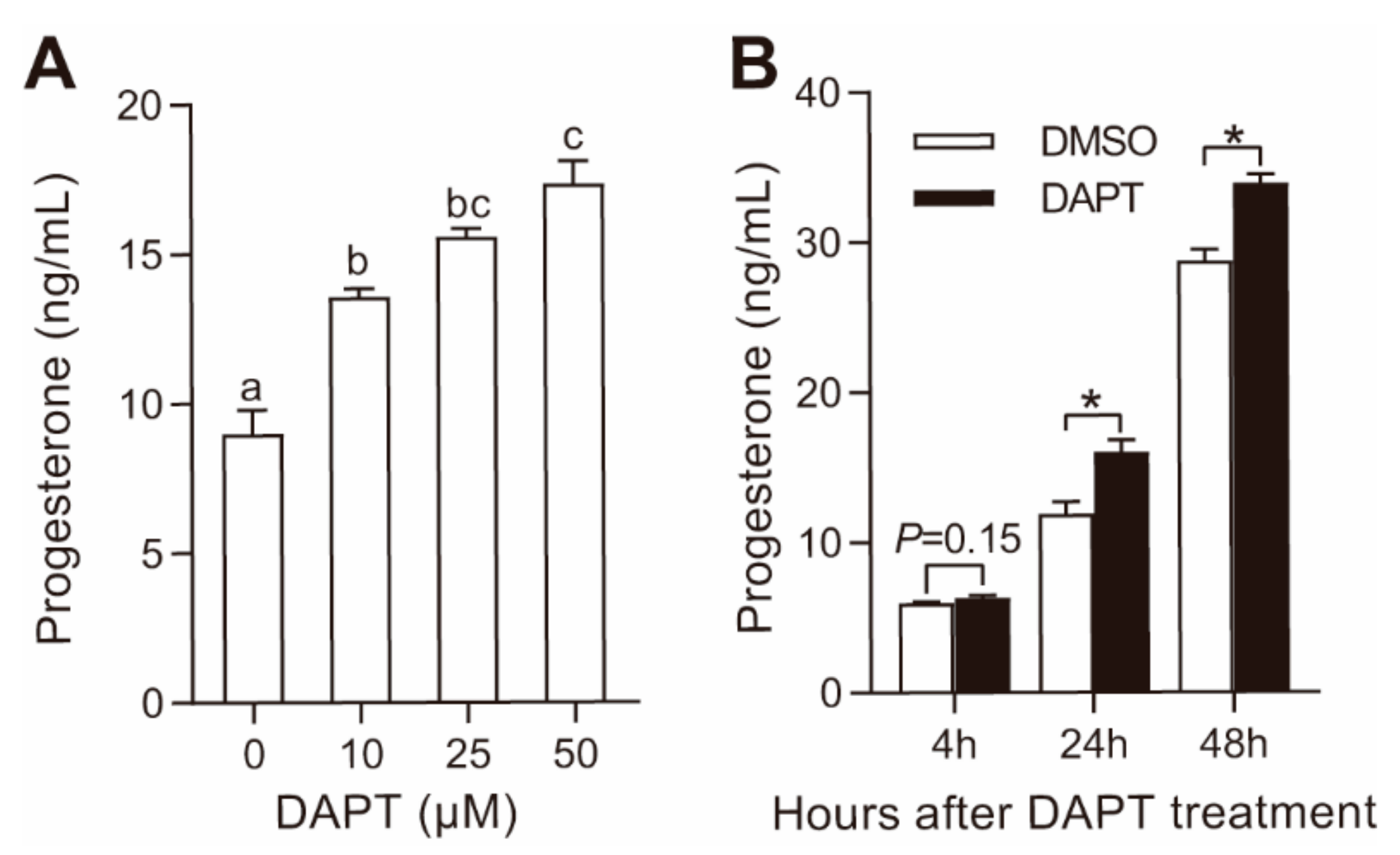
3.1. DAPT Treatment Stimulated Progesterone Secretion in pGCs
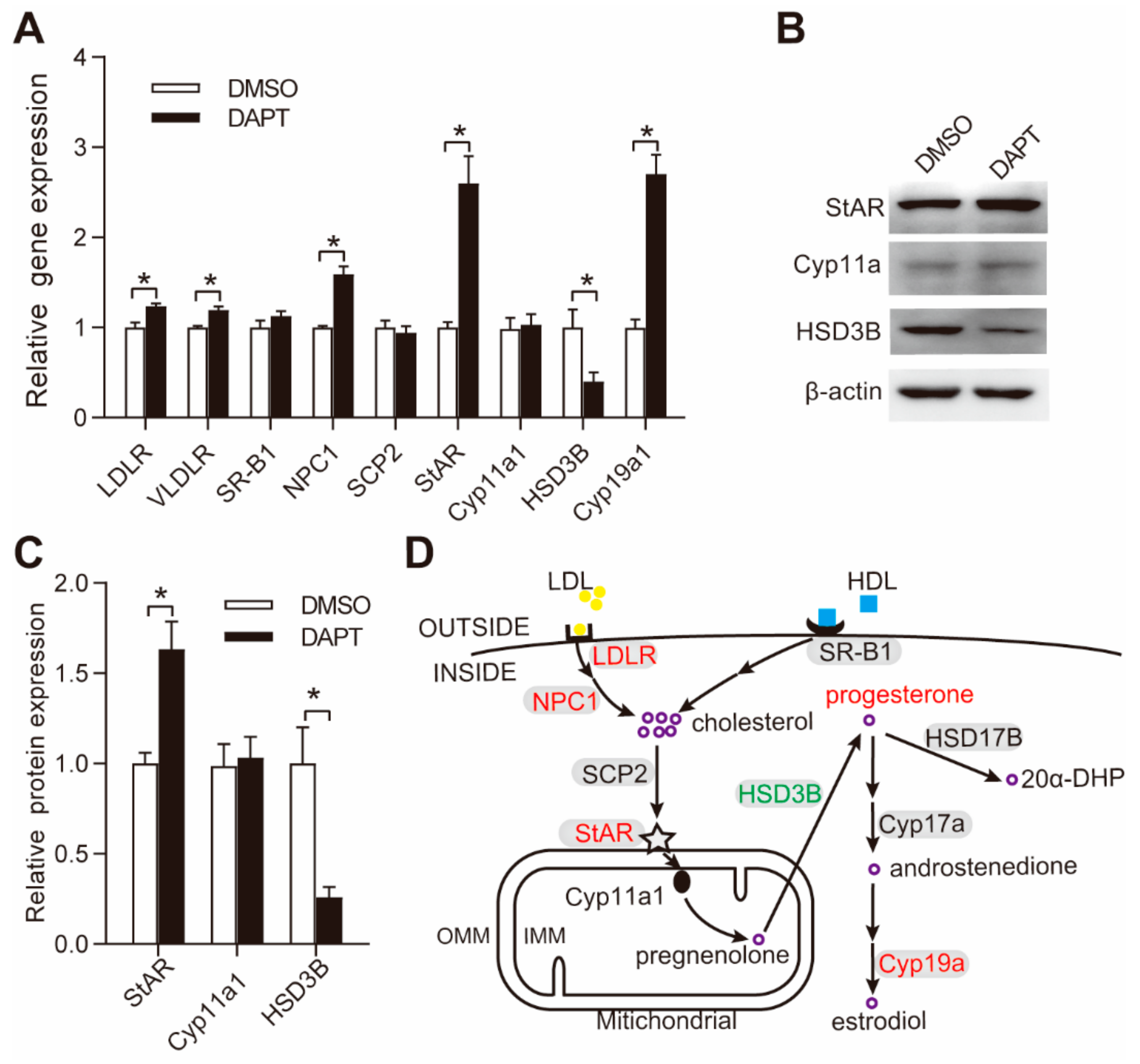
3.2. DAPT Treatment Altered the Expression of Genes Involved in Steroidogenesis
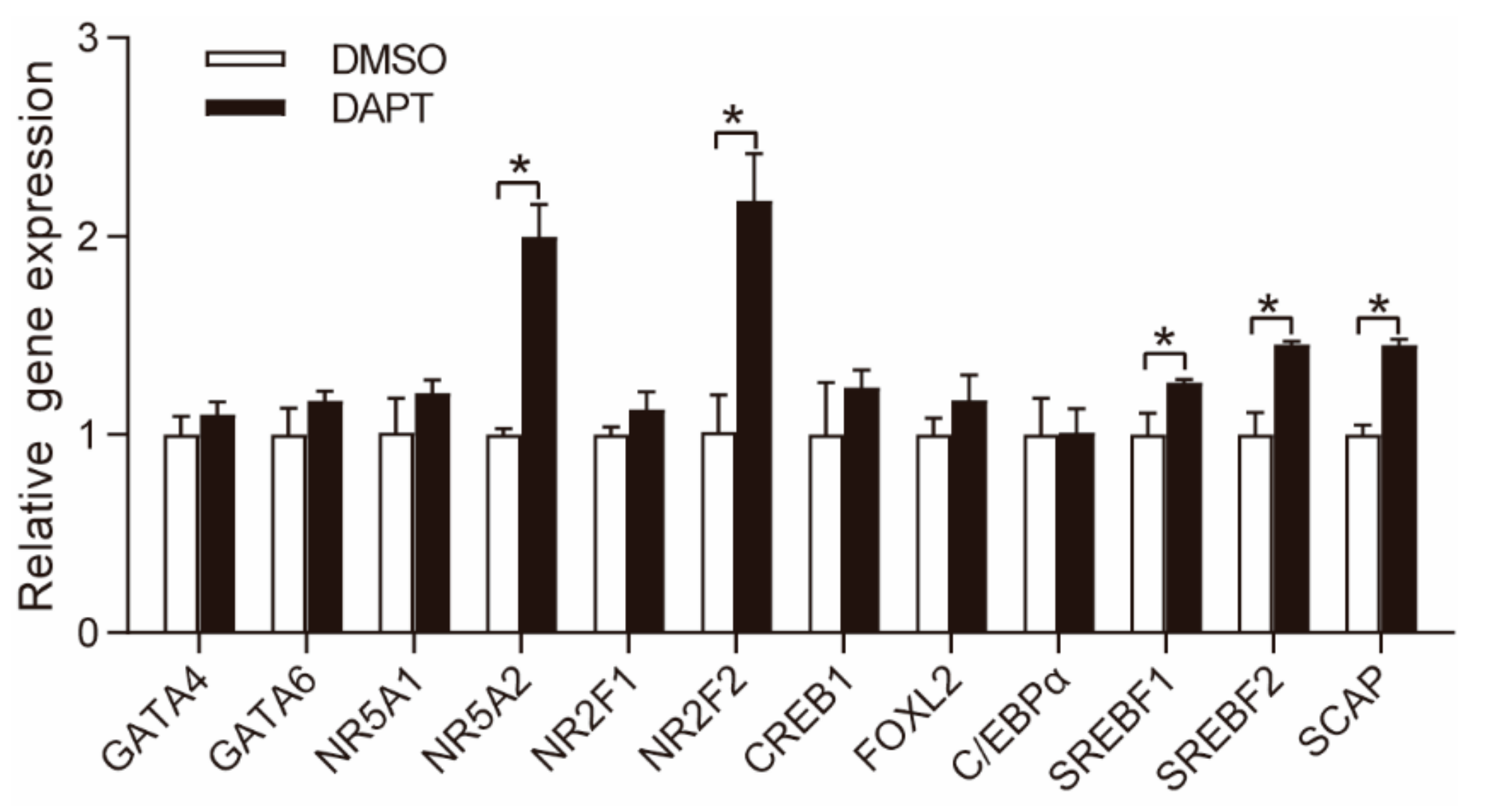
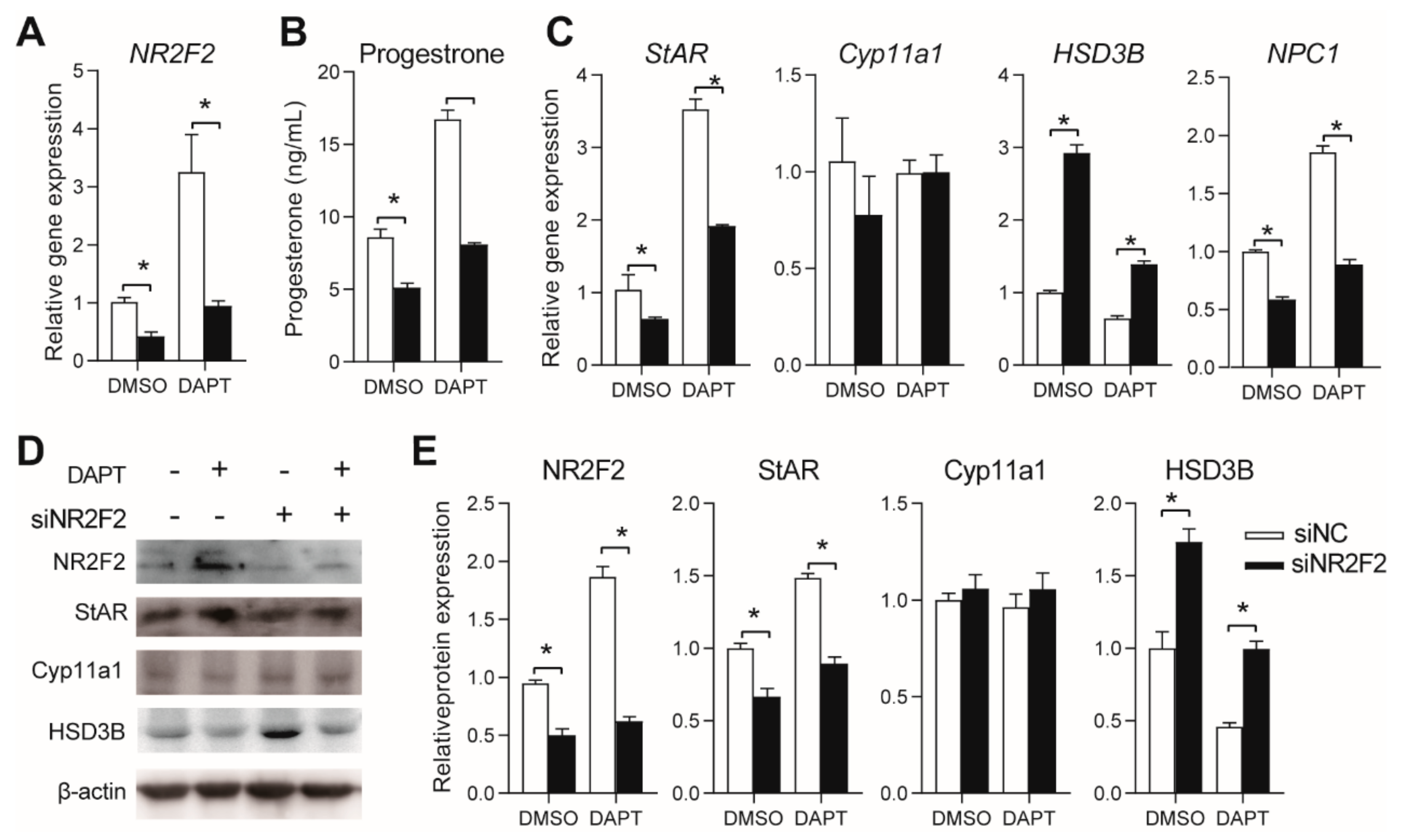
3.3. DAPT Treatment Stimulated the Expression of NR5A2 and NR2F2
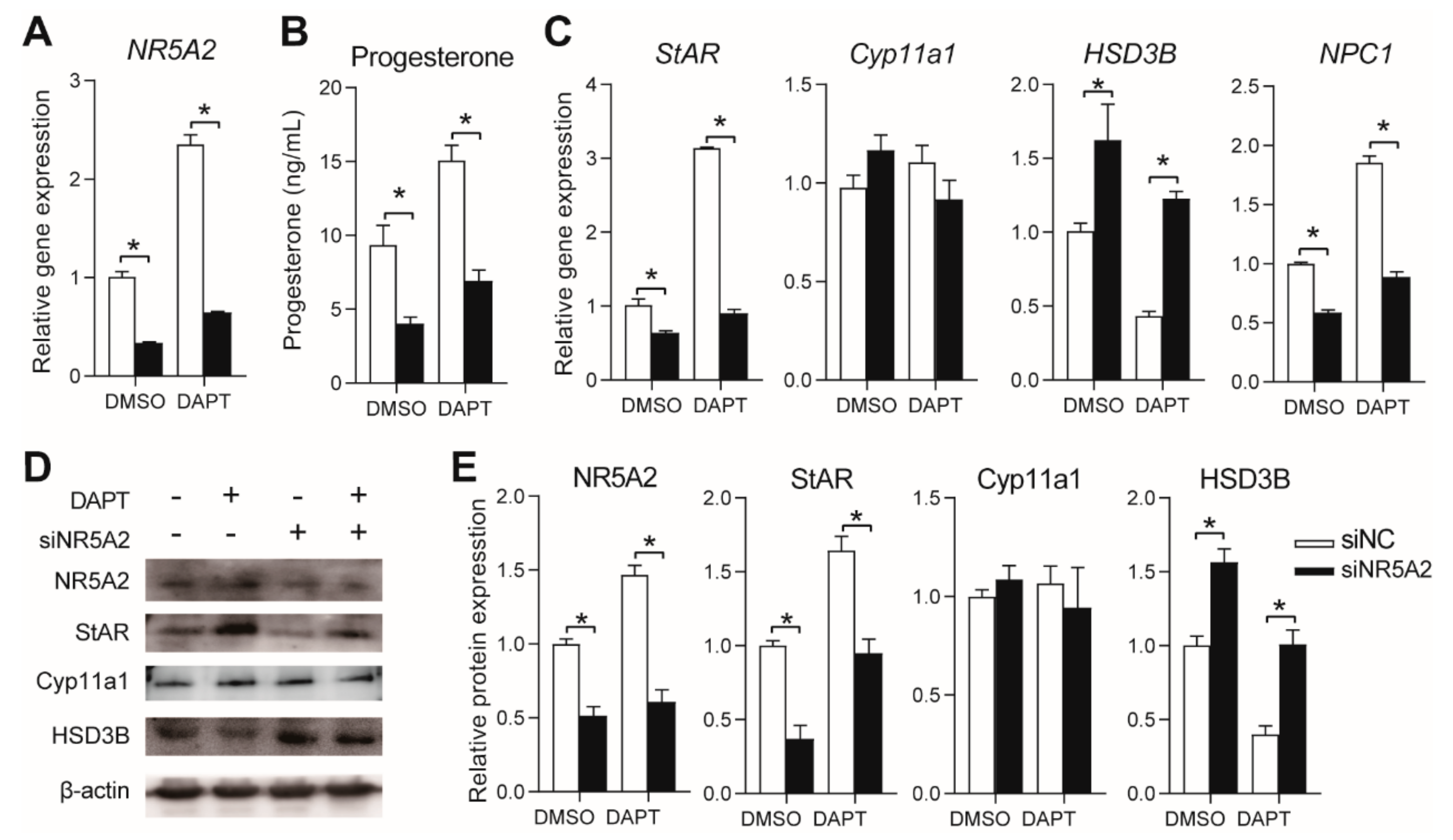
3.4. Effects of NR5A2 Knockdown on Progesterone Secretion in pGCs
3.5. Effects of NR2F2 Knockdown on Progesterone Secretion in pGCs
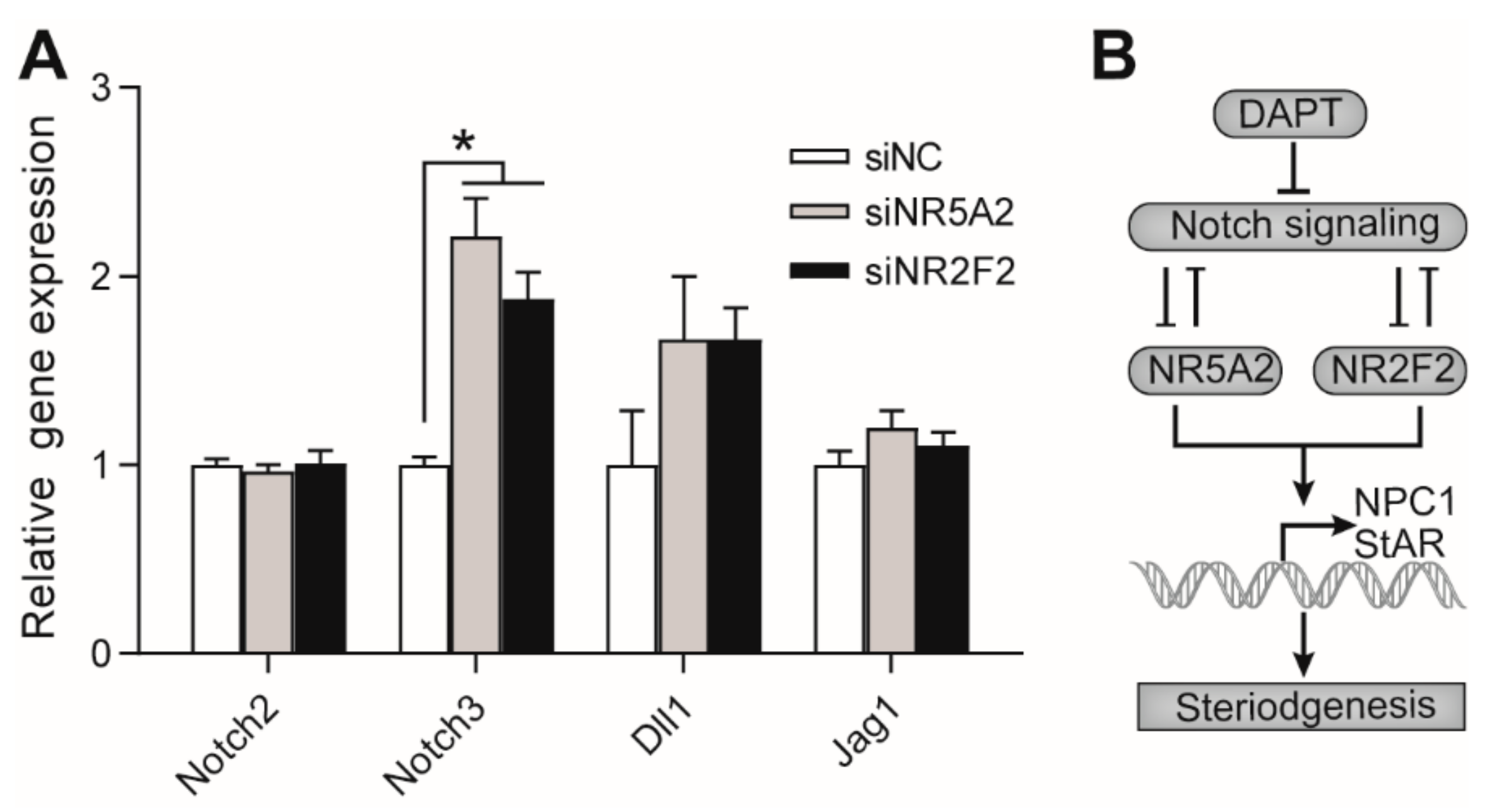
3.6. Effects of NR5A2 and NR2F2 Knockdown on the Gene Expression of Notch Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stocco, C.; Telleria, C.; Gibori, G. The molecular control of corpus luteum formation, function, and regression. Endocr. Rev. 2007, 28, 117–149. [Google Scholar] [CrossRef]
- Miller, W.L.; Auchus, R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011, 32, 81–151. [Google Scholar] [CrossRef]
- Miller, W.L. Steroidogenesis: Unanswered Questions. Trends Endocrinol. Metab. 2017, 28, 771–793. [Google Scholar] [CrossRef]
- Vanorny, D.A.; Mayo, K.E. The role of Notch signaling in the mammalian ovary. Reproduction 2017, 153, R187–R204. [Google Scholar] [CrossRef]
- Knight, P.G.; Glister, C. TGF-β superfamily members and ovarian follicle development. Reproduction 2006, 132, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Xu, J.X.; Gridley, T. Notch2 is required in somatic cells for breakdown of ovarian germ-cell nests and formation of primordial follicles. BMC Biol. 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Vanorny, D.A.; Prasasya, R.D.; Chalpe, A.J.; Kilen, S.M.; Mayo, K.E. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol. Endocrinol. 2014, 28, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Trombly, D.J.; Woodruff, T.K.; Mayo, K.E. Suppression of Notch Signaling in the Neonatal Mouse Ovary Decreases Primordial Follicle Formation. Endocrinology 2009, 150, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, E.; Bao, R.; Xu, P.; Feng, F.; Wen, W.; Dong, Q.; Hu, C.; Xiao, L.; Tang, M.; et al. Notch signalling regulates steroidogenesis in mouse ovarian granulosa cells. Reprod. Fertil. Dev. 2019. [Google Scholar] [CrossRef]
- George, R.M.; Hahn, K.L.; Rawls, A.; Viger, R.S.; Wilson-Rawls, J. Notch signaling represses GATA4-induced expression of genes involved in steroid biosynthesis. Reproduction 2015, 150, 383–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prasasya, R.D.; Mayo, K.E. Notch Signaling Regulates Differentiation and Steroidogenesis in Female Mouse Ovarian Granulosa Cells. Endocrinology 2018, 159, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.; Peluffo, M.C.; Stouffer, R.L.; Irusta, G.; Tesone, M. Role of the DLL4-NOTCH system in PGF2alpha-induced luteolysis in the pregnant rat. Biol. Reprod. 2011, 84, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Accialini, P.; Hernandez, S.F.; Bas, D.; Pazos, M.C.; Irusta, G.; Abramovich, D.; Tesone, M. A link between Notch and progesterone maintains the functionality of the rat corpus luteum. Reproduction 2015, 149, 1–10. [Google Scholar] [CrossRef]
- Qin, L.; Xu, J.; Wu, Z.; Zhang, Z.; Li, J.; Wang, C.; Long, Q. Notch1-mediated signaling regulates proliferation of porcine satellite cells (PSCs). Cell. Signal. 2013, 25, 561–569. [Google Scholar] [CrossRef]
- Jiao, Y.; Huang, B.; Chen, Y.; Hong, G.; Xu, J.; Hu, C.; Wang, C. Integrated Analyses Reveal Overexpressed Notch1 Promoting Porcine Satellite Cells’ Proliferation through Regulating the Cell Cycle. Int. J. Mol. Sci. 2018, 19, 271. [Google Scholar] [CrossRef]
- Lei, T.; Bi, Y.; Gao, M.J.; Gao, S.M.; Zhou, L.L.; Zheng, H.L.; Chen, X.D. HES1 inhibits adipogenesis of porcine mesenchymal stem cells via transcriptional repression of FAD24. Domest. Anim. Endocrinol. 2013, 45, 28–32. [Google Scholar] [CrossRef]
- Frias, A.; Lambies, G.; Vinas-Castells, R.; Martinez-Guillamon, C.; Dave, N.; Garcia de Herreros, A.; Diaz, V.M. A Switch in Akt Isoforms Is Required for Notch-Induced Snail1 Expression and Protection from Cell Death. Mol. Cell. Biol. 2015, 36, 923–940. [Google Scholar] [CrossRef]
- Cai, L.; Sun, A.; Li, H.; Tsinkgou, A.; Yu, J.; Ying, S.; Chen, Z.; Shi, Z. Molecular mechanisms of enhancing porcine granulosa cell proliferation and function by treatment in vitro with anti-inhibin α subunit antibody. Reprod. Biol. Endocrinol. 2015, 13, 26. [Google Scholar] [CrossRef]
- Li, H.; Guo, S.; Cai, L.; Ma, W.; Shi, Z. Lipopolysaccharide and heat stress impair the estradiol biosynthesis in granulosa cells via increase of HSP70 and inhibition of smad3 phosphorylation and nuclear translocation. Cell. Signal. 2017, 30, 130–141. [Google Scholar] [CrossRef]
- Schrade, A.; Kyronlahti, A.; Akinrinade, O.; Pihlajoki, M.; Hakkinen, M.; Fischer, S.; Alastalo, T.P.; Velagapudi, V.; Toppari, J.; Wilson, D.B.; et al. GATA4 Is a Key Regulator of Steroidogenesis and Glycolysis in Mouse Leydig Cells. Endocrinology 2015, 156, 1860–1872. [Google Scholar] [CrossRef]
- Bertolin, K.; Gossen, J.; Schoonjans, K.; Murphy, B.D. The orphan nuclear receptor Nr5a2 is essential for luteinization in the female mouse ovary. Endocrinology 2014, 155, 1931–1943. [Google Scholar] [CrossRef]
- Convissar, S.M.; Bennett, J.; Baumgarten, S.C.; Lydon, J.P.; DeMayo, F.J.; Stocco, C. GATA4 and GATA6 Knockdown During Luteinization Inhibits Progesterone Production and Gonadotropin Responsiveness in the Corpus Luteum of Female Mice. Biol. Reprod. 2015, 93, 133. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Komiyama, J.; Viger, R.S.; Okuda, K. The expression of the nuclear receptors NR5A1 and NR5A2 and transcription factor GATA6 correlates with steroidogenic gene expression in the bovine corpus luteum. Mol. Reprod. Dev. 2009, 76, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Liu, Z.; Shimada, M.; Sterneck, E.; Johnson, P.F.; Hedrick, S.M.; Richards, J.S. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 2009, 324, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Large, M.J.; Duggavathi, R.; DeMayo, F.J.; Lydon, J.P.; Schoonjans, K.; Kovanci, E.; Murphy, B.D. Liver receptor homolog-1 is essential for pregnancy. Nat. Med. 2013, 19, 1061–1066. [Google Scholar] [CrossRef]
- Stergiopoulos, A.; Politis, P.K. Nuclear receptor NR5A2 controls neural stem cell fate decisions during development. Nat. Commun. 2016, 7, 12230. [Google Scholar] [CrossRef]
- Lin, F.J.; Qin, J.; Tang, K.; Tsai, S.Y.; Tsai, M.J. Coup d’Etat: An orphan takes control. Endocr. Rev. 2011, 32, 404–421. [Google Scholar] [CrossRef]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Zhang, C.P.; Yang, J.L.; Zhang, J.; Li, L.; Huang, L.; Ji, S.Y.; Hu, Z.Y.; Gao, F.; Liu, Y.X. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology 2011, 152, 2437–2447. [Google Scholar] [CrossRef]
- Benedito, R.; Roca, C.; Sorensen, I.; Adams, S.; Gossler, A.; Fruttiger, M.; Adams, R.H. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 2009, 137, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Rutz, S.; Mordmuller, B.; Sakano, S.; Scheffold, A. Notch ligands Delta-like1, Delta-like4 and Jagged1 differentially regulate activation of peripheral T helper cells. Eur. J. Immunol. 2005, 35, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Murta, D.; Batista, M.; Silva, E.; Trindade, A.; Mateus, L.; Duarte, A.; Lopes-da-Costa, L. Differential expression of Notch component and effector genes during ovarian follicle and corpus luteum development during the oestrous cycle. Reprod. Fertil. Dev. 2015, 27, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Gevry, N.; Schoonjans, K.; Guay, F.; Murphy, B.D. Cholesterol supply and SREBPs modulate transcription of the Niemann-Pick C-1 gene in steroidogenic tissues. J. Lipid Res. 2008, 49, 1024–1033. [Google Scholar] [CrossRef]
- Kathiriya, I.S.; King, I.N.; Murakami, M.; Nakagawa, M.; Astle, J.M.; Gardner, K.A.; Gerard, R.D.; Olson, E.N.; Srivastava, D.; Nakagawa, O. Hairy-related transcription factors inhibit GATA-dependent cardiac gene expression through a signal-responsive mechanism. J. Biol. Chem. 2004, 279, 54937–54943. [Google Scholar] [CrossRef]
- Xiang, F.; Sakata, Y.; Cui, L.; Youngblood, J.M.; Nakagami, H.; Liao, J.K.; Liao, R.; Chin, M.T. Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H1997–H2006. [Google Scholar] [CrossRef]
- Fischer, A.; Klattig, J.; Kneitz, B.; Diez, H.; Maier, M.; Holtmann, B.; Englert, C.; Gessler, M. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol. Cell. Biol. 2005, 25, 8960–8970. [Google Scholar] [CrossRef]
- Hallaq, R.; Volpicelli, F.; Cuchillo-Ibanez, I.; Hooper, C.; Mizuno, K.; Uwanogho, D.; Causevic, M.; Asuni, A.; To, A.; Soriano, S.; et al. The Notch intracellular domain represses CRE-dependent transcription. Cell. Signal. 2015, 27, 621–629. [Google Scholar] [CrossRef]
- You, L.R.; Lin, F.J.; Lee, C.T.; DeMayo, F.J.; Tsai, M.J.; Tsai, S.Y. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 2005, 435, 98–104. [Google Scholar] [CrossRef]
- Hinshelwood, M.M.; Repa, J.J.; Shelton, J.M.; Richardson, J.A.; Mangelsdorf, D.J.; Mendelson, C.R. Expression of LRH-1 and SF-1 in the mouse ovary: Localization in different cell types correlates with differing function. Mol. Cell. Endocrinol. 2003, 207, 39–45. [Google Scholar] [CrossRef]
- Labelle-Dumais, C.; Pare, J.F.; Belanger, L.; Farookhi, R.; Dufort, D. Impaired progesterone production in Nr5a2+/- mice leads to a reduction in female reproductive function. Biol. Reprod. 2007, 77, 217–225. [Google Scholar] [CrossRef]
- Duggavathi, R.; Volle, D.H.; Mataki, C.; Antal, M.C.; Messaddeq, N.; Auwerx, J.; Murphy, B.D.; Schoonjans, K. Liver receptor homolog 1 is essential for ovulation. Genes Dev. 2008, 22, 1871–1876. [Google Scholar] [CrossRef]
- Takamoto, N.; Kurihara, I.; Lee, K.; Demayo, F.J.; Tsai, M.J.; Tsai, S.Y. Haploinsufficiency of chicken ovalbumin upstream promoter transcription factor II in female reproduction. Mol. Endocrinol. 2005, 19, 2299–2308. [Google Scholar] [CrossRef]
- Mendoza-Villarroel, R.E.; Robert, N.M.; Martin, L.J.; Brousseau, C.; Tremblay, J.J. The nuclear receptor NR2F2 activates star expression and steroidogenesis in mouse MA-10 and MLTC-1 Leydig cells. Biol. Reprod. 2014, 91, 26. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Chen, F.; Shi, Z. Suppression of Notch Signaling Stimulates Progesterone Synthesis by Enhancing the Expression of NR5A2 and NR2F2 in Porcine Granulosa Cells. Genes 2020, 11, 120. https://doi.org/10.3390/genes11020120
Guo R, Chen F, Shi Z. Suppression of Notch Signaling Stimulates Progesterone Synthesis by Enhancing the Expression of NR5A2 and NR2F2 in Porcine Granulosa Cells. Genes. 2020; 11(2):120. https://doi.org/10.3390/genes11020120
Chicago/Turabian StyleGuo, Rihong, Fang Chen, and Zhendan Shi. 2020. "Suppression of Notch Signaling Stimulates Progesterone Synthesis by Enhancing the Expression of NR5A2 and NR2F2 in Porcine Granulosa Cells" Genes 11, no. 2: 120. https://doi.org/10.3390/genes11020120
APA StyleGuo, R., Chen, F., & Shi, Z. (2020). Suppression of Notch Signaling Stimulates Progesterone Synthesis by Enhancing the Expression of NR5A2 and NR2F2 in Porcine Granulosa Cells. Genes, 11(2), 120. https://doi.org/10.3390/genes11020120



