Mutations in Collagen Genes in the Context of an Isolated Population
Abstract
:1. Introduction
2. Materials and Methods
3. Results
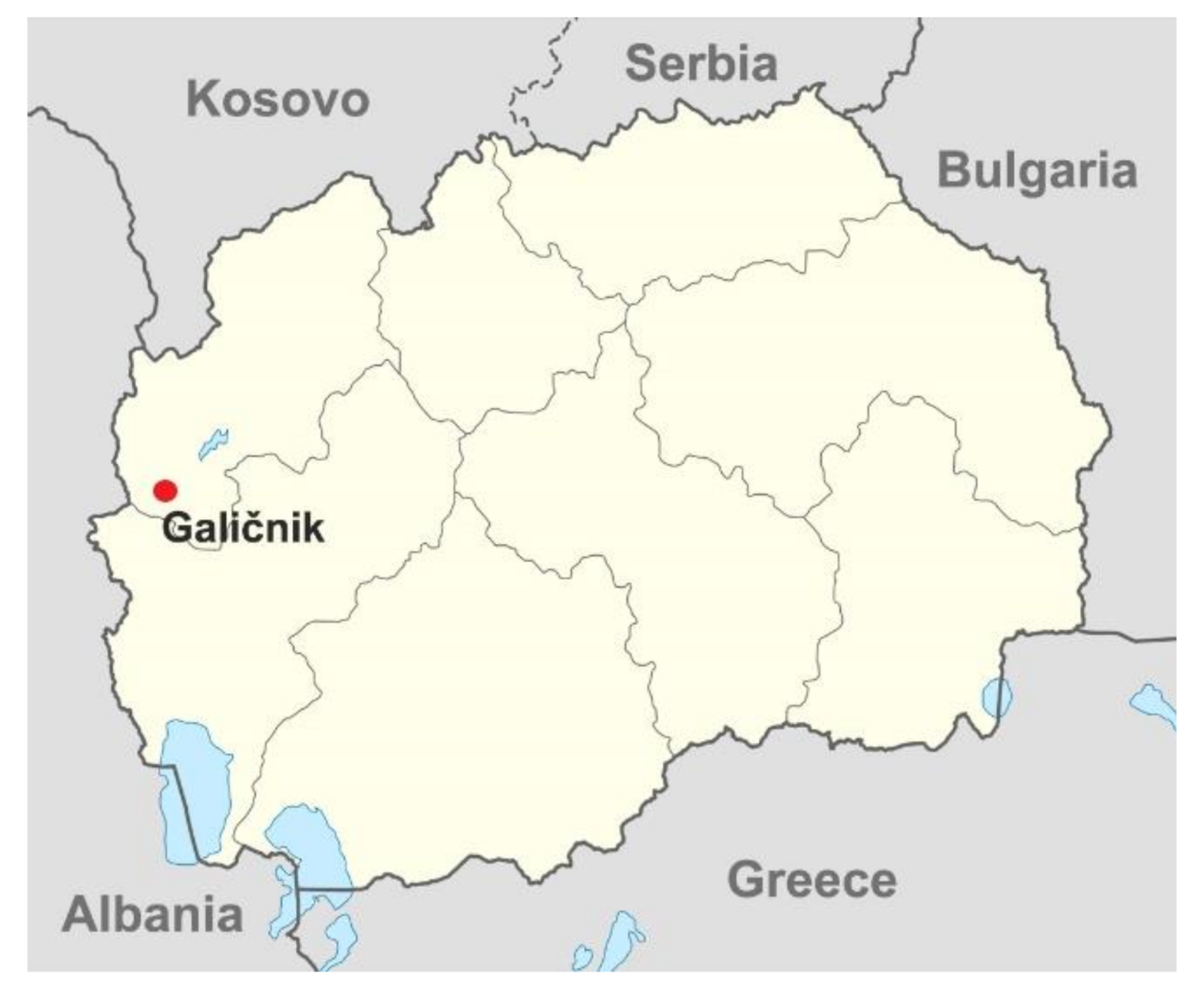
3.1. Genetic Context of Nephropathy in the Galičnik Population
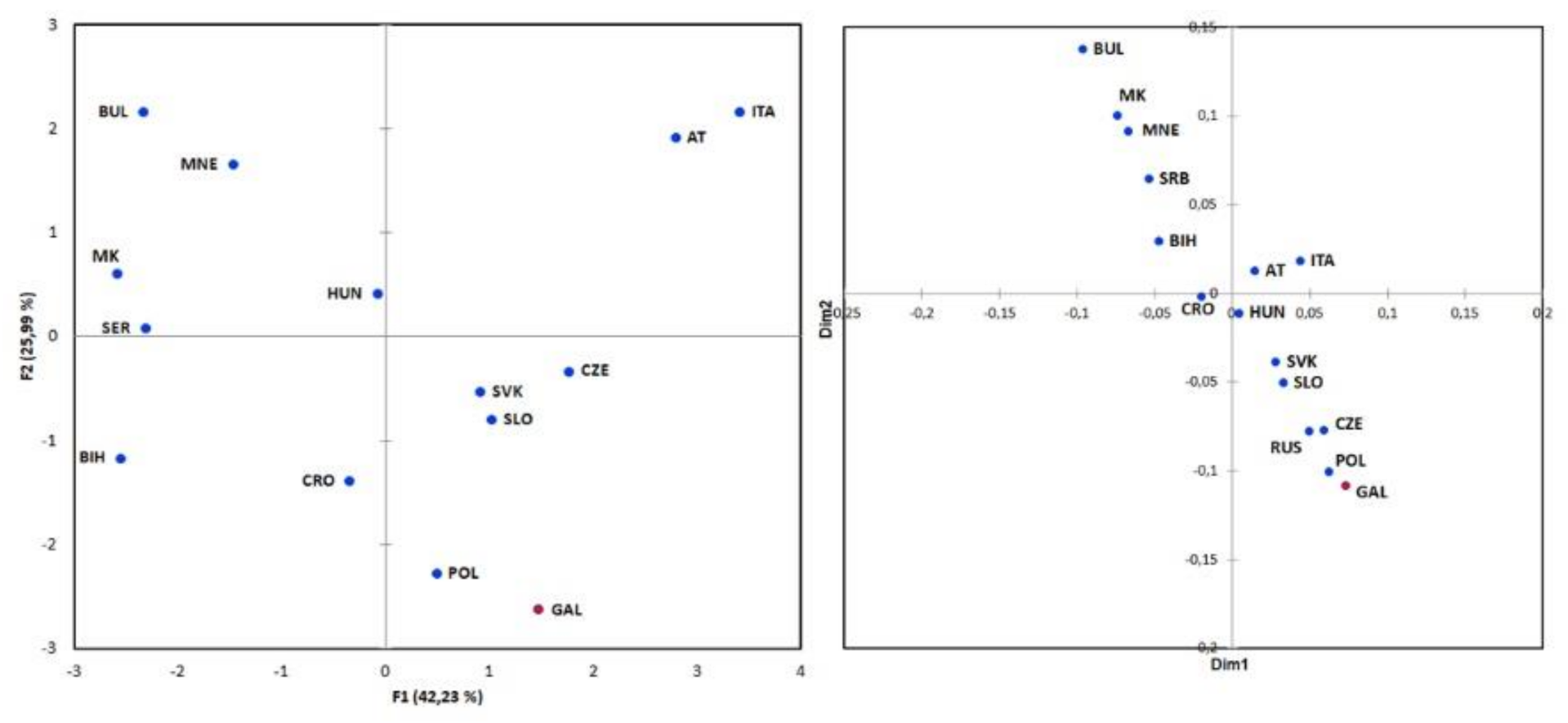
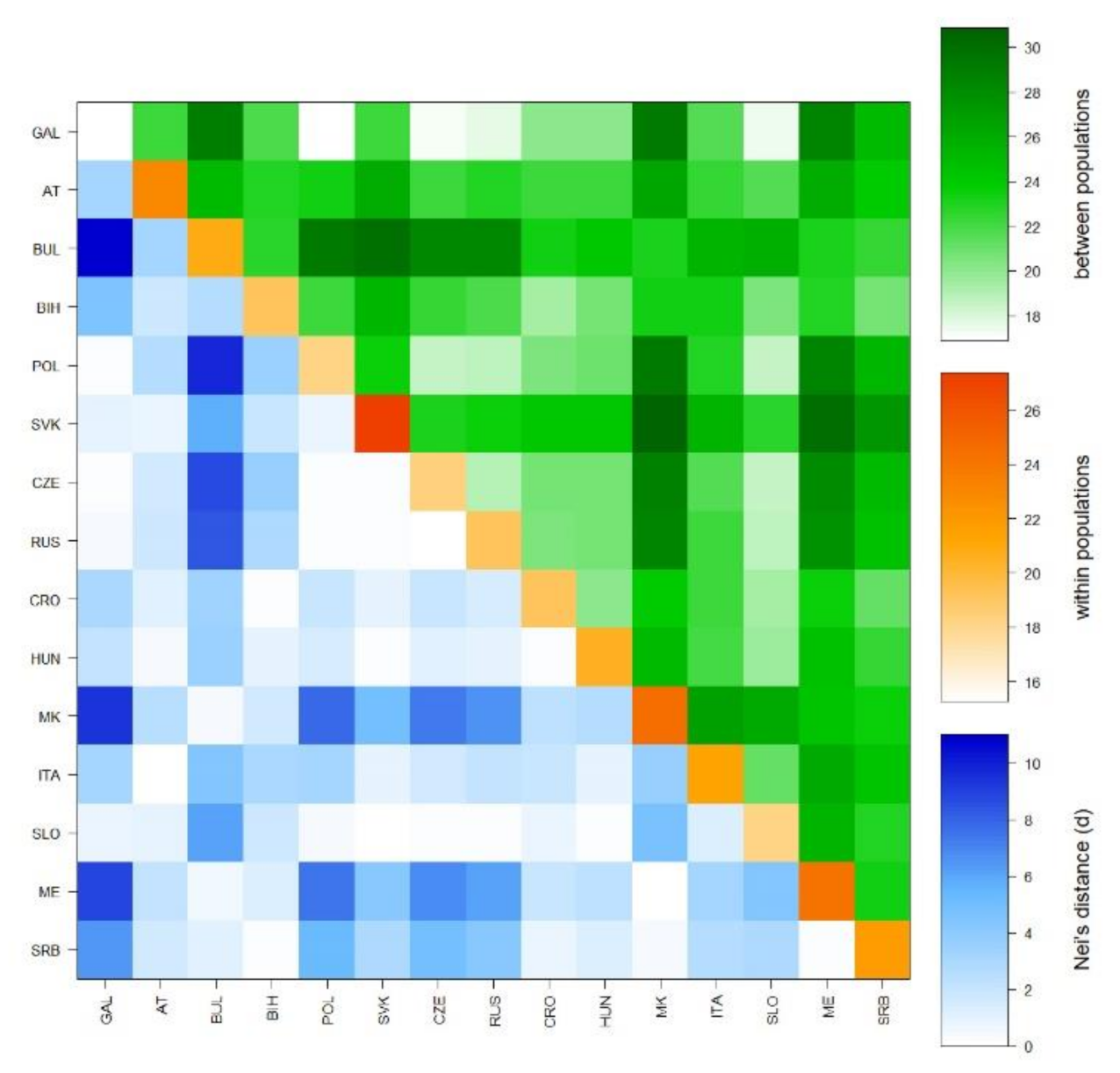
3.2. Genetic Context of the Galičnik Population
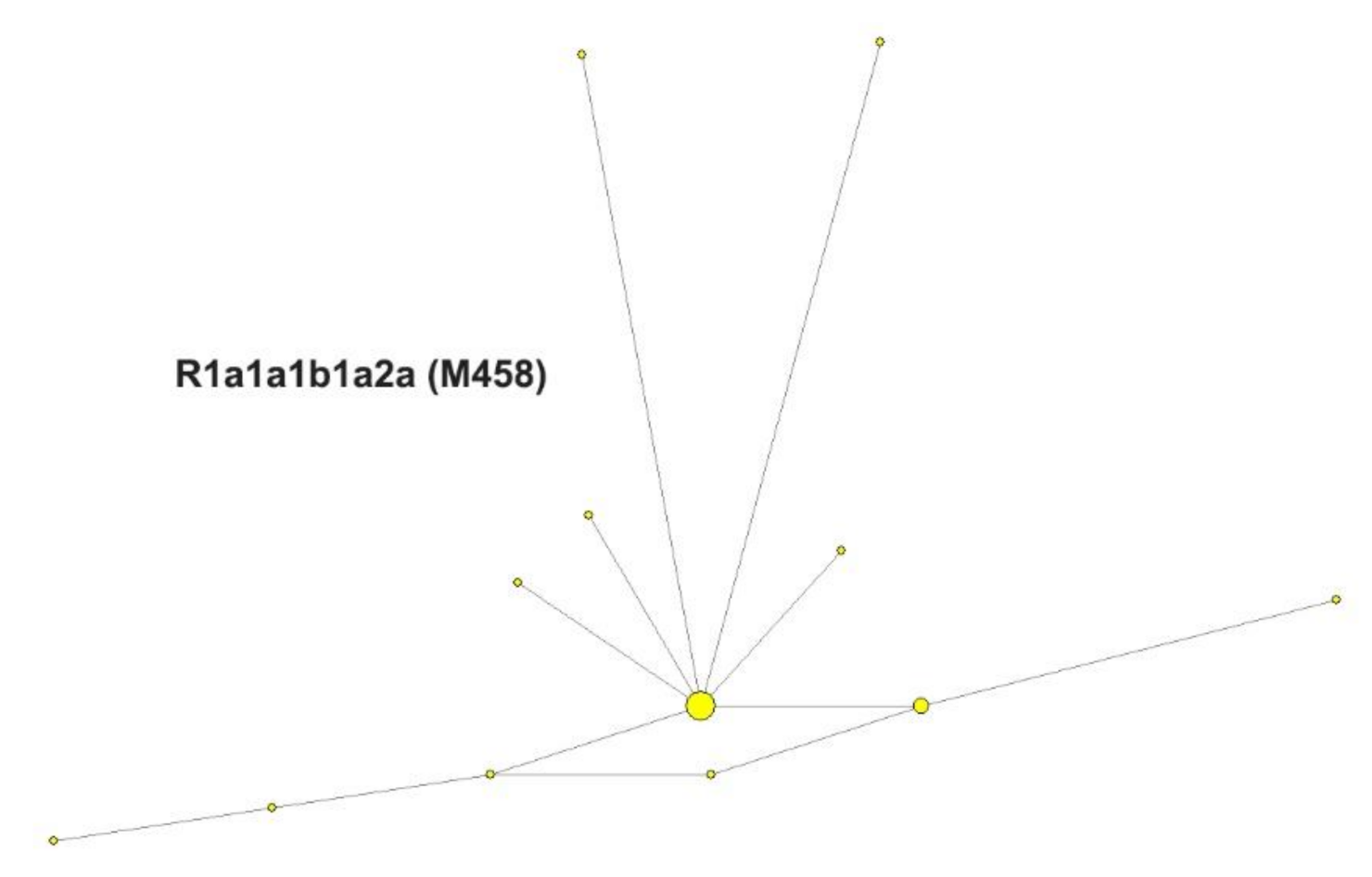
3.2.1. Y Chromosome
3.2.2. mtDNA
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Weiss, K.M. Genetic Variation and Human Disease: Principles and Evolutionary Approaches; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Rosenberg, N.A. Genetic Structure of Human Populations. Science 2002, 298, 2381–2385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli-Sfortza, L.L. The History and Geography of Human Genes; Princeton University Press: Princeton, NJ, USA, 1994. [Google Scholar]
- Cavalli-Sfortza, L.L. Genes, Peoples and Languages; University of California Press: Berkeley, CA, USA, 2001. [Google Scholar]
- Kashtan, C.E.; Ding, J.; Garosi, G.; Heidet, L.; Massella, L.; Nakanishi, K.; Nozu, K.; Renieri, A.; Rheault, M.; Wang, F.; et al. Alport syndrome: A unified classification of genetic disorders of collagen IV α345: A position paper of the Alport Syndrome Classification Working Group. Kidney Int. 2018, 93, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Šlajpah, M.; Gorinšek, B.; Berginc, G.; Vizjak, A.; Ferluga, D.; Hvala, A.; Meglič, A.; Jakša, I.; Furlan, P.; Gregorič, A.; et al. Sixteen novel mutations identified in COL4A3, COL4A4, and COL4A5 genes in Slovenian families with Alport syndrome and benign familial hematuria. Kidney Int. 2007, 71, 1287–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashtan, C.E. Alport Syndrome: An Inherited Disorder of Renal, Ocular, and Cochlear Basement Membranes. Medicine 1999, 78, 338–360. [Google Scholar] [CrossRef]
- Jais, J.P.; Knebelmann, B.; Giatras, I.; De Marchi, M.; Rizzoni, G.; Renieri, A.; Weber, M.; Gross, O.; Netzer, K.O.; Flinter, F.; et al. X-linked Alport syndrome: Natural history in 195 families and genotype-phenotype correlations in males. J. Am. Soc. Nephrol. 2000, 11, 649–657. [Google Scholar]
- Nozu, K.; Nakanishi, K.; Abe, Y.; Udagawa, T.; Okada, S.; Okamoto, T.; Kaito, H.; Kanemoto, K.; Kobayashi, A.; Tanaka, E.; et al. A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin. Exp. Nephrol. 2019, 23, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Imafuku, A.; Nozu, K.; Sawa, N.; Hasegawa, E.; Hiramatsu, R.; Kawada, M.; Hoshino, J.; Tanaka, K.; Ishii, Y.; Takaichi, K.; et al. Autosomal dominant form of type IV collagen nephropathy exists among patients with hereditary nephritis difficult to diagnose clinicopathologically. Nephrology 2018, 23, 940–947. [Google Scholar] [CrossRef] [Green Version]
- Bekheirnia, M.R.; Reed, B.; Gregory, M.C.; McFann, K.; Shamshirsaz, A.A.; Masoumi, A.; Schrier, R.W. Genotype–Phenotype Correlation in X-Linked Alport Syndrome. J. Am. Soc. Nephrol. 2010, 21, 876–883. [Google Scholar] [CrossRef] [Green Version]
- Mencarelli, M.A.; Heidet, L.; Storey, H.; Van Geel, M.; Knebelmann, B.; Fallerini, C.; Miglietti, N.; Antonucci, M.F.; Cetta, F.; Sayer, J.A.; et al. Evidence of digenic inheritance in Alport syndrome. J. Med. Genet. 2015, 52, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Pericić, M.; Lauc, L.B.; Klarić, I.M.; Rootsi, S.; Janićijević, B.; Rudan, I.; Terzić, R.; Čolak, I.; Kvesić, A.; Popović, D.; et al. High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations. Mol. Biol. Evol. 2005, 22, 1964–1975. [Google Scholar] [CrossRef]
- Battaglia, V.; Fornarino, S.; Al-Zahery, N.; Olivieri, A.; Pala, M.; Myres, N.M.; King, R.J.; Rootsi, S.; Marjanovic, D.; Primorac, A.; et al. Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe. Eur. J. Hum. Genet. 2009, 17, 853. [Google Scholar] [CrossRef]
- Völgyi, A.; Zalán, A.; Szvetnik, E.; Pamjav, H. Hungarian population data for 11 Y-STR and 49 Y-SNP markers. Forensic Sci. Int. Genet. 2009, 3, e27–e28. [Google Scholar] [CrossRef] [PubMed]
- Niederstätter, H.; Rampl, G.; Erhart, D.; Pitterl, F.; Oberacher, H.; Neuhuber, F.; Hausner, I.; Gassner, C.; Schennach, H.; Berger, B.; et al. Pasture Names with Romance and Slavic Roots Facilitate Dissection of Y Chromosome Variation in an Exclusively German-Speaking Alpine Region. PLoS ONE 2012, 7, e41885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Cruz, B.; Ioana, M.; Calafell, F.; Arauna, L.R.; Sanz, P.; Ionescu, R.; Boengiu, S.; Kalaydjieva, L.; Pamjav, H.; Makukh, H.; et al. Y-Chromosome Analysis in Individuals Bearing the Basarab Name of the First Dynasty of Wallachian Kings. PLoS ONE 2012, 7, e41803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myres, N.M.; Rootsi, S.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Pshenichnov, A.; Yunusbayev, B.; et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 2010, 19, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Underhill, P.A.; Myres, N.M.; Rootsi, S.; Metspalu, M.; Zhivotovsky, L.A.; King, R.J.; Lin, A.A.; Chow, C.-E.T.; Semino, O.; Battaglia, V.; et al. Separating the post-glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. Genet. 2010, 18, 1074. [Google Scholar] [CrossRef] [Green Version]
- Berger, B.; Lindinger, A.; Niederstätter, H.; Grubwieser, P.; Parson, W. Y-STR typing of an Austrian population sample using a 17-loci multiplex PCR assay. Int. J. Leg. Med. 2005, 119, 241–246. [Google Scholar] [CrossRef]
- Zaharova, B.; Andonova, S.; Gilissen, A.; Cassiman, J.-J.; Decorte, R.; Kremensky, I. Y-chromosomal STR haplotypes in three major population groups in Bulgaria. Forensic Sci. Int. 2001, 124, 182–186. [Google Scholar] [CrossRef]
- Klaric, I.M.; Lauc, L.B.; Pericic, M.; Janicijevic, B.; Terzić, R.; Čolak, I.; Kvesic, A.; Rudan, P. Evaluation of Y-STR variation in Bosnian and Herzegovinian population. Forensic Sci. Int. 2005, 154, 252–256. [Google Scholar] [CrossRef]
- Woźniak, M.; Grzybowski, T.; Starzyński, J.; Marciniak, T. Continuity of Y chromosome haplotypes in the population of Southern Poland before and after the Second World War. Forensic Sci. Int. Genet. 2007, 1, 134–140. [Google Scholar] [CrossRef]
- Woźniak, M.; Malyarchuk, B.; Derenko, M.; Vanecek, T.; Lazur, J.; Gomolcak, P.; Grzybowski, T. Similarities and distinctions in Y chromosome gene pool of Western Slavs. Am. J. Phys. Anthr. 2010, 142, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Derenko, M.; Malyarchuk, B.; Denisova, G.; Woźniak, M.; Grzybowski, T.; Dambueva, I.; Zakharov, I. Y-chromosome haplogroup N dispersals from south Siberia to Europe. J. Hum. Genet. 2007, 52, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Derenko, M.; Malyarchuk, B.; Denisova, G.A.; Wozniak, M.; Dambueva, I.; Dorzhu, C.; Luzina, F.; Miścicka-Śliwka, D.; Zakharov, I. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Qual. Life Res. 2006, 118, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Barać, L.; Pericić, M.; Klarić, I.M.; Janićijević, B.; Parik, J.; Rootsi, S.; Rudan, P. Y chromosome STRs in Croatians. Forensic Sci. Int. 2003, 138, 127–133. [Google Scholar] [CrossRef]
- Jakovski, Z.; Nikolova, K.; Jankova-Ajanovska, R.; Marjanovic, D.; Pojskic, N.; Janeska, B. Genetic data for 17 Y-chromosomal STR loci in Macedonians in the Republic of Macedonia. Forensic Sci. Int. Genet. 2011, 5, e108–e111. [Google Scholar] [CrossRef]
- Turrina, S.; Atzei, R.; De Leo, D. Y-chromosomal STR haplotypes in a Northeast Italian population sample using 17plex loci PCR assay. Int. J. Leg. Med. 2005, 120, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Mirabal, S.; Varljen, T.; Gayden, T.; Regueiro, M.; Vujovic, S.; Popovic, D.; Djurić, M.; Stojković, O.; Herrera, R.J. Human Y-chromosome short tandem repeats: A tale of acculturation and migrations as mechanisms for the diffusion of agriculture in the Balkan Peninsula. Am. J. Phys. Anthr. 2010, 142, 380–390. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Dunnen, J.T.D.; Antonarakis, S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum. Mutat. 2000, 15, 7–12. [Google Scholar] [CrossRef]
- Karafet, T.M.; Mendez, F.L.; Meilerman, M.B.; Underhill, P.A.; Zegura, S.L.; Hammer, M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008, 18, 830–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zupan, A.; Vrabec, K.; Glavač, D. The paternal perspective of the Slovenian population and its relationship with other populations. Ann. Hum. Biol. 2013, 40, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Charrow, J. Ashkenazi Jewish genetic disorders. Fam. Cancer 2004, 3, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Bullich, G.; Domingo-Gallego, A.; Vargas, I.; Ruiz, P.; Lorente-Grandoso, L.; Furlano, M.; Fraga, G.; Madrid, Á.; Ariceta, G.; Borregán, M.; et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018, 94, 363–371. [Google Scholar] [CrossRef]
- Longo, I.; Porcedda, P.; Mari, F.; Giachino, D.; Meloni, I.; Deplano, C.; Brusco, A.; Bosio, M.; Massella, L.; Lavoratti, G.; et al. COL4A3/COL4A4 mutations: From familial hematuria to autosomal-dominant or recessive Alport syndrome. Kidney Int. 2002, 61, 1947–1956. [Google Scholar] [CrossRef] [Green Version]
- Demosthenous, P.; Voskarides, K.; Stylianou, K.; Hadjigavriel, M.; Arsali, M.; Patsias, C.; Georgaki, E.; Zirogiannis, P.; Stavrou, C.; Daphnis, E.; et al. X-linked Alport syndrome in Hellenic families: Phenotypic heterogeneity and mutations near interruptions of the collagen domain in COL4A5. Clin. Genet. 2011, 81, 240–248. [Google Scholar] [CrossRef]
- Kovács, G.; Kalmár, T.; Endreffy, E.; Ondrik, Z.; Iványi, B.; Rikker, C.; Haszon, I.; Túri, S.; Sinkó, M.; Bereczki, C.; et al. Efficient Targeted Next Generation Sequencing-Based Workflow for Differential Diagnosis of Alport-Related Disorders. PLoS ONE 2016, 11, e0149241. [Google Scholar] [CrossRef] [Green Version]
- Voskarides, K.; Pierides, A.; Deltas, C. COL4A3/COL4A4 Mutations Link Familial Hematuria and Focal Segmental Glomerulosclerosis. Glomerular Epithelium Destruction via Basement Membrane Thinning? Connect. Tissue Res. 2008, 49, 283–288. [Google Scholar] [CrossRef]
- Voskarides, K.; Stefanou, C.; Pieri, M.; Demosthenous, P.; Felekkis, K.; Arsali, M.; Athanasiou, Y.; Xydakis, D.; Stylianou, K.; Daphnis, E.; et al. A functional variant in NEPH3 gene confers high risk of renal failure in primary hematuric glomerulopathies. Evidence for predisposition to microalbuminuria in the general population. PLoS ONE 2017, 12, e0174274. [Google Scholar] [CrossRef]
- Tonna, S.; Wang, Y.Y.; Wilson, D.; Rigby, L.; Tabone, T.; Cotton, R.; Savige, J. The R229Q mutation in NPHS2 may predispose to proteinuria in thin-basement-membrane nephropathy. Pediatr. Nephrol. 2008, 23, 2201–2207. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.S.; Yuldasheva, N.; Ruzibakiev, R.; Underhill, P.A.; Evseeva, I.; Blue-Smith, J.; Jin, L.; Su, B.; Pitchappan, R.; Shanmugalakshmi, S.; et al. The Eurasian Heartland: A continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. USA 2001, 98, 10244–10249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barford, P.M. The Early Slavs; Cornell University Press: New York, NY, USA, 2001. [Google Scholar]
- Haak, W.; Lazaridis, I.; Patterson, N.; Rohland, N.; Mallick, S.; Llamas, B.; Brandt, G.; Nordenfelt, S.; Harney, E.; Stewardson, K.; et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nat. Cell Biol. 2015, 522, 207–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guštin, M.; Kuzman, P.; Malenko, V. Ein keltischer Krieger in Lychnidos Ohrid, Mazedonien. Folia Archaeol. Balk. II 2012, 16, 181–196. [Google Scholar]
- Guštin, M. Celtic helmets from Hellenistic necropolises at Ohrid. In The Clash of Cultures? The Celts and the Macedonian World; Guštin, M., David, W., Eds.; KASTNER AG: Wolnzach, Germany, 2014. [Google Scholar]
- Kumar, V.; Langstieh, B.T.; Madhavi, K.V.; Naidu, V.M.; Singh, H.P.; Biswas, S.; Thangaraj, K.; Singh, L.; Reddy, B.M. Global Patterns in Human Mitochondrial DNA and Y-Chromosome Variation Caused by Spatial Instability of the Local Cultural Processes. PLoS Genet. 2006, 2, e53. [Google Scholar] [CrossRef]
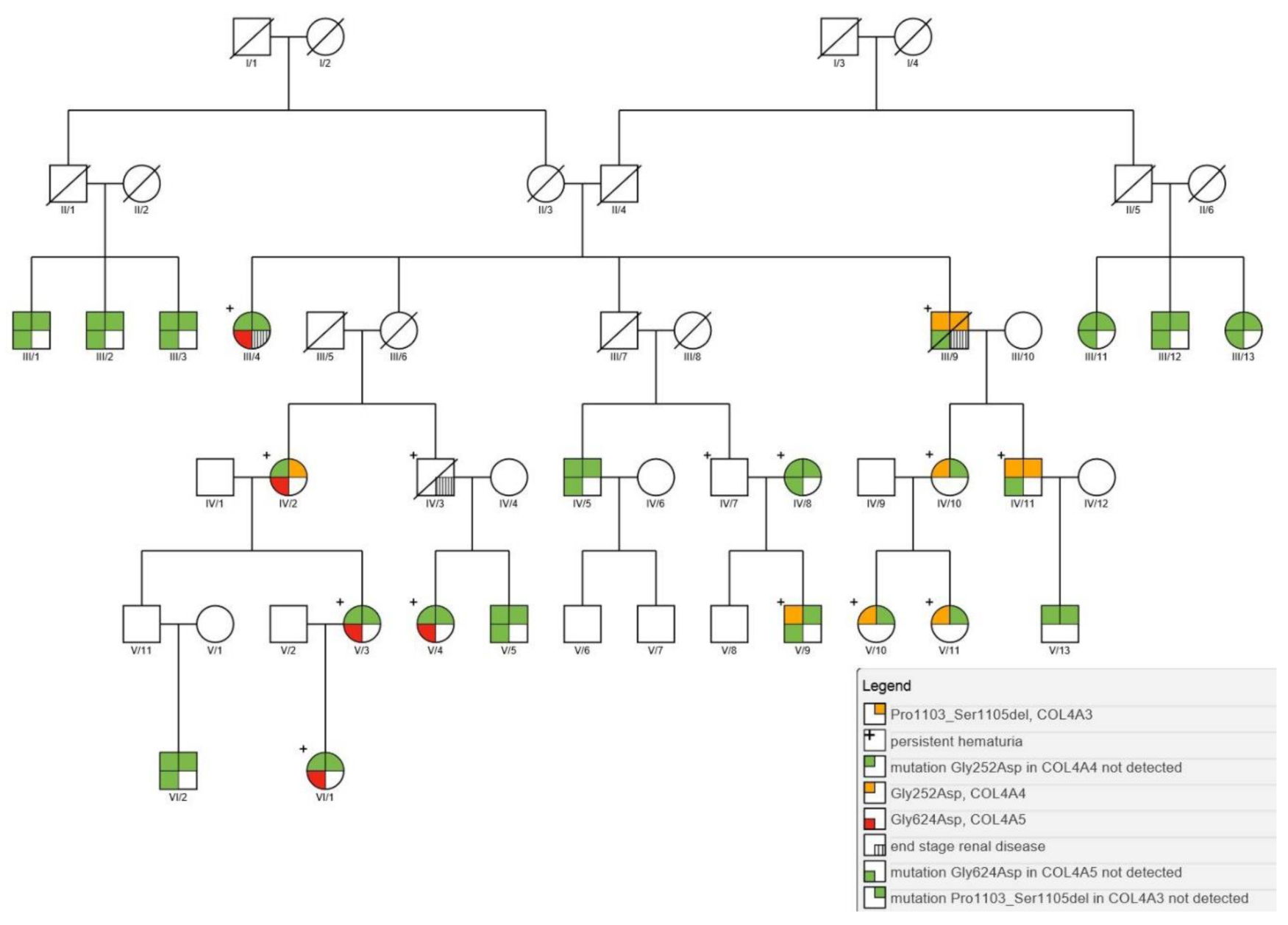
| Case Number | Sex | Age | Hematuria (Age)/Proteinuria | ESRD (Age) | Hearing Loss | Ocular Lesions | Mutation Status | ||
|---|---|---|---|---|---|---|---|---|---|
| COL4A3 | COL4A4 | COL4A5 | |||||||
| II/4 | M | deceased (61) | NA | yes (61) | NA | NA | NA | NA | NA |
| III/4 | F | 90 | yes/no | yes | yes (70) | no | wt | wt | c.1871G > A p.(Gly624Asp) |
| III/9 | M | deceased (80) | yes/yes | yes | yes (70) | no | c.3307_3315del p.(Pro1103_Ser1105del) | c.755G > A p.(Gly252Asp) | wt |
| IV/2 | F | 71 | yes (EO)/no | no | no | no | c.3307_3315del p.(Pro1103_Ser1105del) | wt | c.1871G > A p.(Gly624Asp) |
| IV/3 | M | deceased (60) | yes (EO)/yes | yes (60) | NA | NA | NA | NA | NA |
| IV/10 | F | 53 | yes/no | no | no | no | wt | c.755G > A p.(Gly252Asp) | wt |
| IV/11 | F | 56 | yes (EO)/yes | no | yes | no | c.3307_3315del p.(Pro1103_Ser1105del) | c.755G > A p.(Gly252Asp) | wt |
| V/3 | F | 48 | yes (EO)/no | no | no | no | wt | wt | c.1871G > A p.(Gly624Asp) |
| V/4 | F | 33 | yes (EO)/no | no | no | no | wt | wt | c.1871G > A p.(Gly624Asp) |
| V/9 | M | 29 | yes/no | no | no | no | wt | c.755G > A p.(Gly252Asp) | wt |
| V/10 | F | 25 | yes (8)/no | no | no | no | wt | c.755G > A p.(Gly252Asp) | wt |
| V/11 | F | 21 | yes (9)/no | no | no | no | wt | c.755G > A p.(Gly252Asp) | wt |
| VI/1 | F | 23 | yes (EO)/no | no | no | no | wt | wt | c.1871G > A p.(Gly624Asp) |
| Haplogroup | % |
| H | 28.4 |
| H * | 13.4 |
| H1 | 4.1 |
| H11 | 6.8 |
| H17 | 4.1 |
| HV | 18.9 |
| HV8 | 18.9 |
| U | 21.7 |
| U * | 1.4 |
| U2 | 4.1 |
| U3 | 1.4 |
| U5 | |
| >U5b | 2.7 |
| K | |
| >K * | 2.7 |
| >K1 | 9.4 |
| J1 | 8.1 |
| J * | 1.4 |
| J1c | 4.1 |
| J1d | 2.6 |
| T | 2.7 |
| T1 | 2.7 |
| N * | 20.2 |
| N2 * | 1.4 |
| R * | 2.7 |
| R0 | 10.7 |
| W1 | 5.4 |
| Other | 0.0 |
| Population size | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zupan, A.; Matjašič, A.; Grubelnik, G.; Tasić, V.; Momirovska, A. Mutations in Collagen Genes in the Context of an Isolated Population. Genes 2020, 11, 1377. https://doi.org/10.3390/genes11111377
Zupan A, Matjašič A, Grubelnik G, Tasić V, Momirovska A. Mutations in Collagen Genes in the Context of an Isolated Population. Genes. 2020; 11(11):1377. https://doi.org/10.3390/genes11111377
Chicago/Turabian StyleZupan, Andrej, Alenka Matjašič, Gašper Grubelnik, Velibor Tasić, and Ana Momirovska. 2020. "Mutations in Collagen Genes in the Context of an Isolated Population" Genes 11, no. 11: 1377. https://doi.org/10.3390/genes11111377





