Membrane-Anchored Hairless Protein Restrains Notch Signaling Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cloning of the Hairless Transmembrane Coding Sequence
2.2. Cloning of the GFP Transmembrane Coding Sequence
2.3. In Vivo Analysis
2.4. Western Blotting
2.5. Statistical Evaluation
3. Results
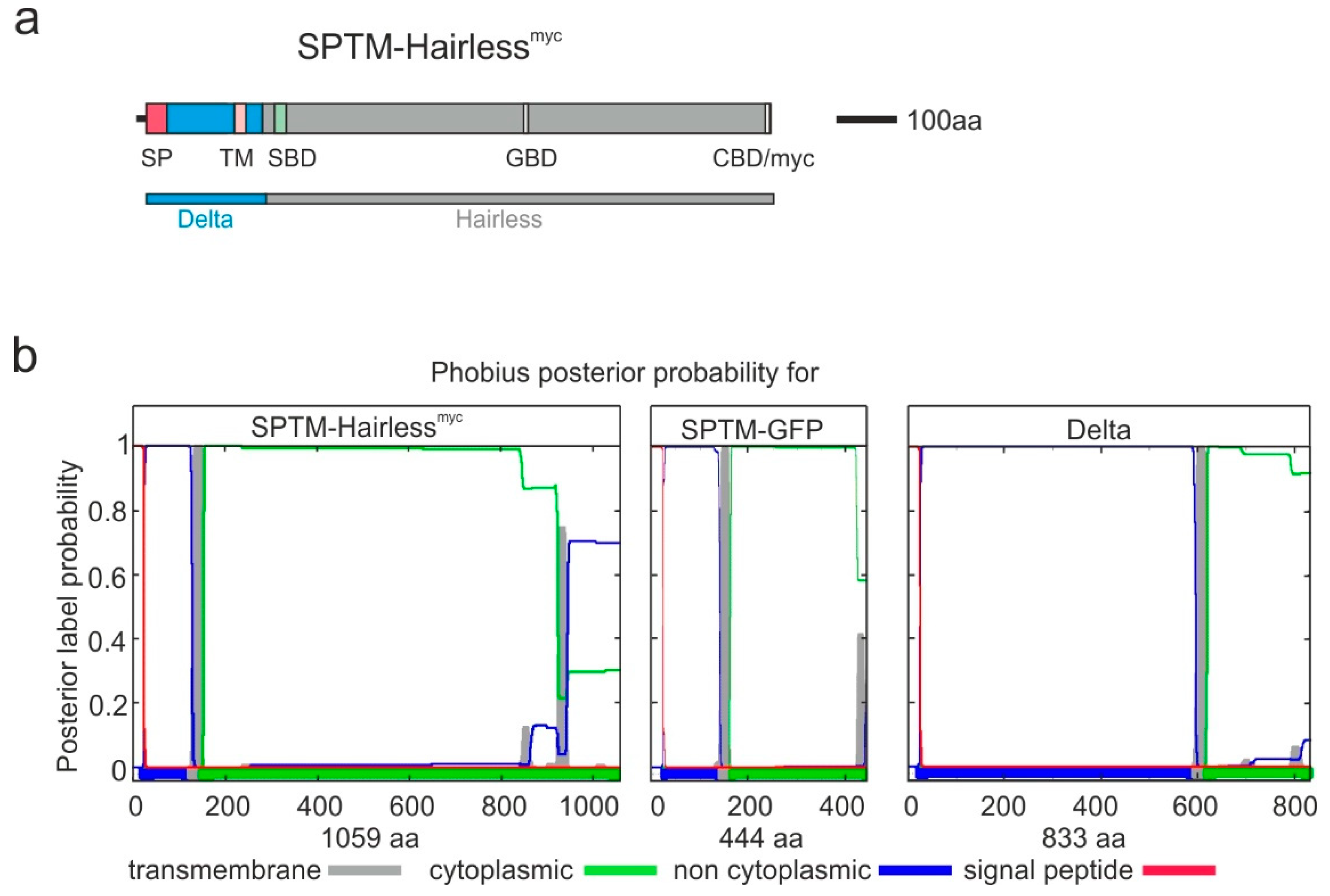
3.1. Cloning Strategy
3.2. In Vivo Consequences of the Overexpression of SPTM-Hmyc Protein
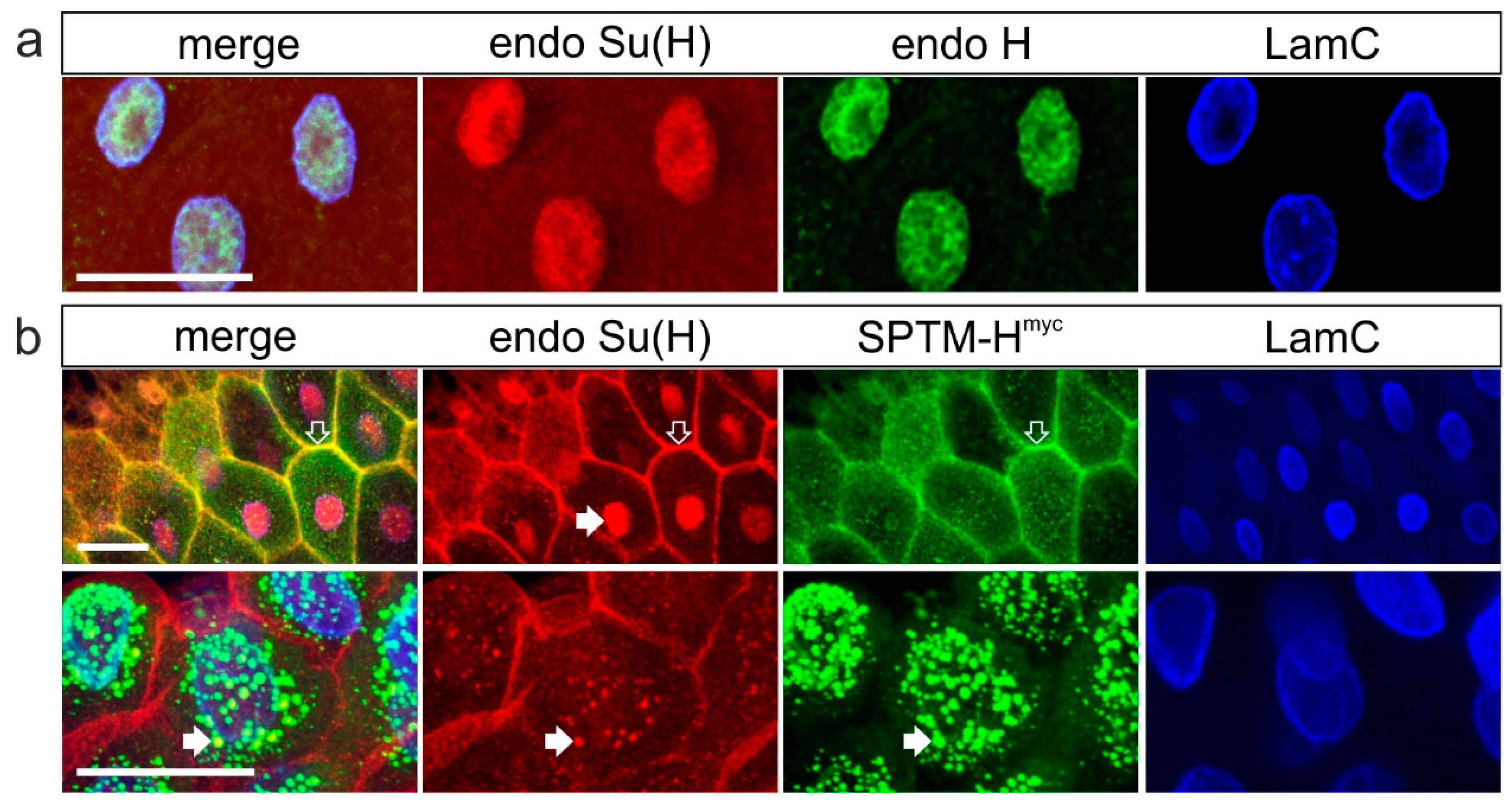
3.2.1. Subcellular Localization of Overexpressed SPTM-Hmyc Protein in Salivary Glands
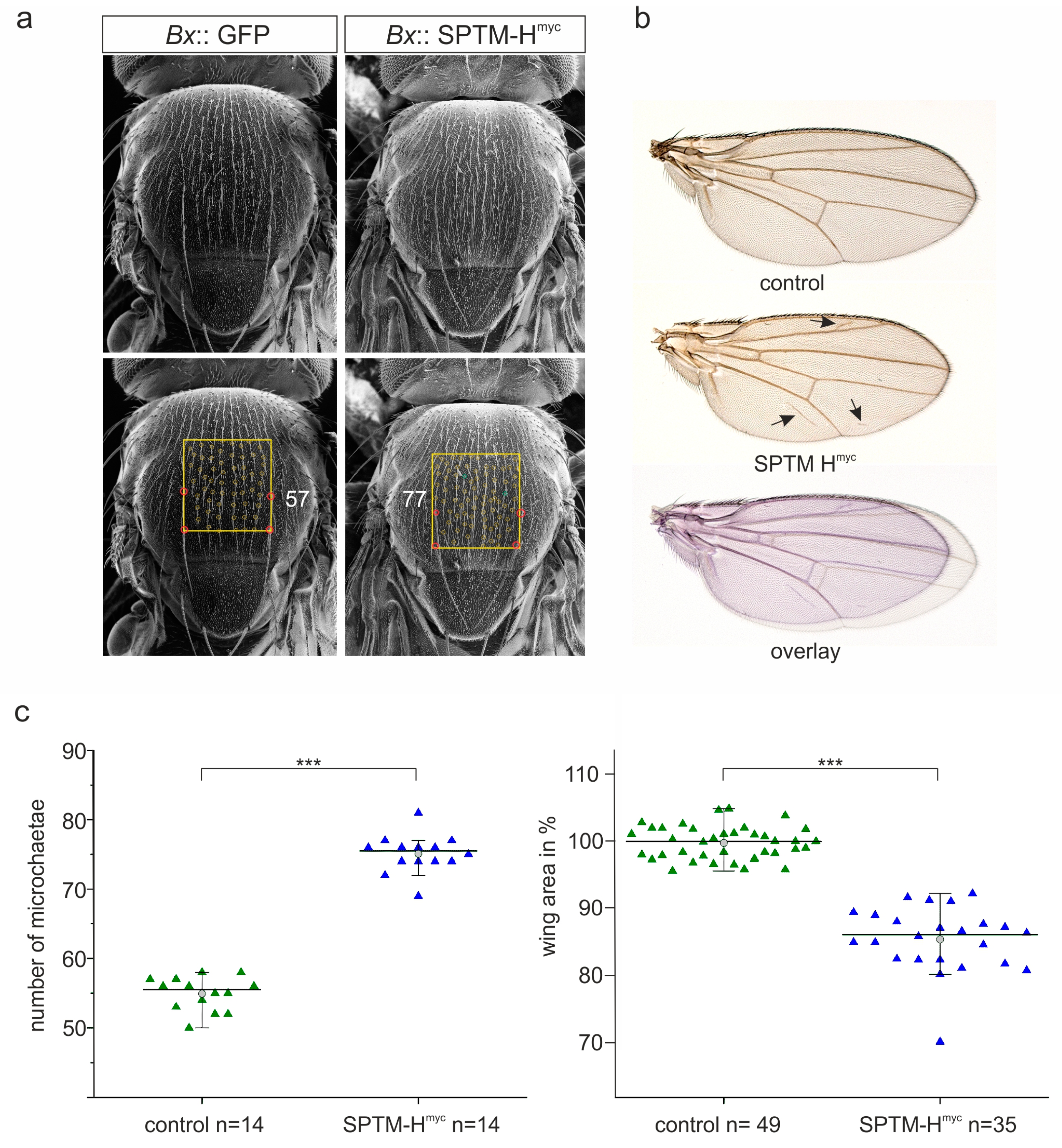
3.2.2. Phenotypic Consequences Resulting from SPTM-Hmyc Overexpression
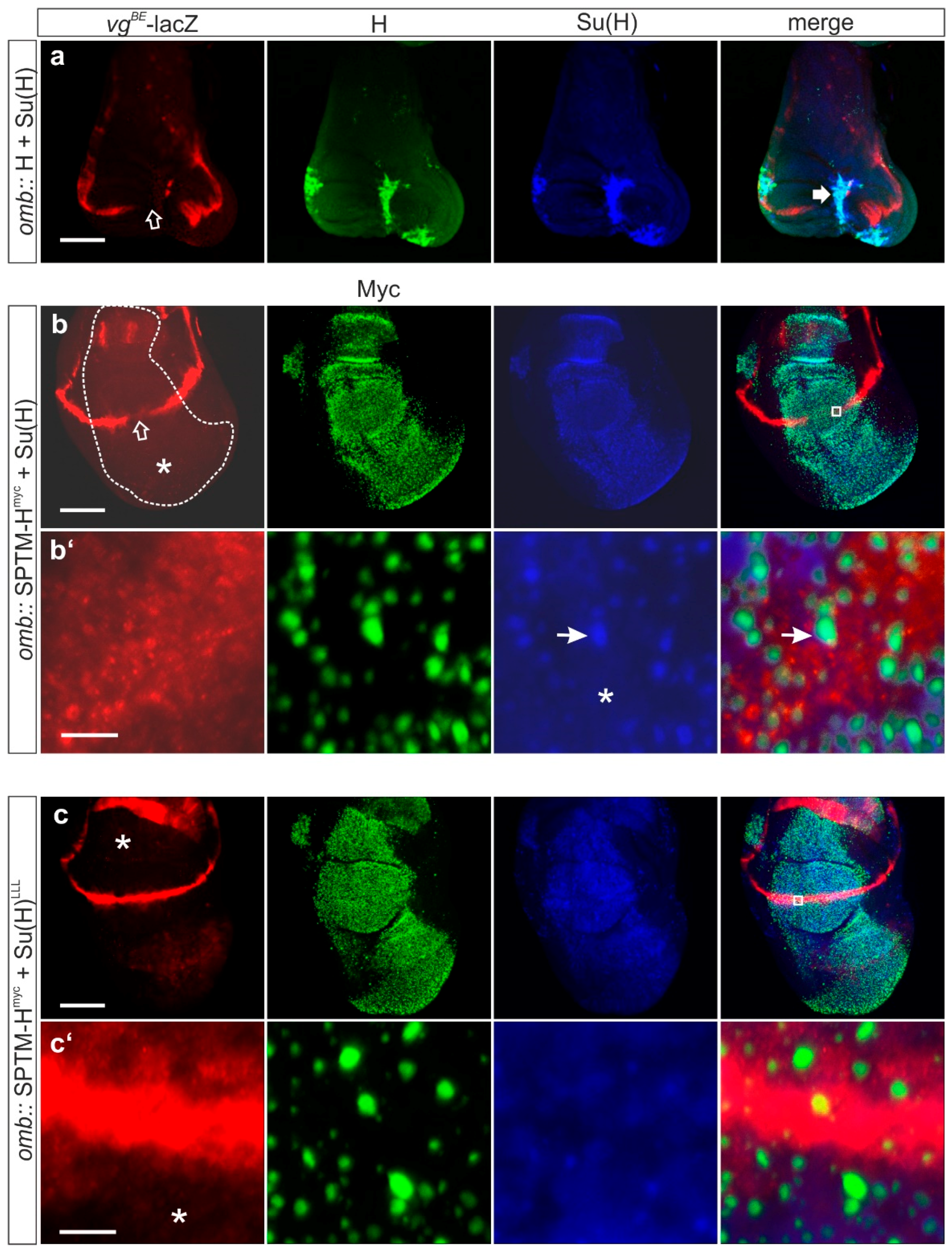
3.2.3. Influence of SPTM-Hmyc on the Activity of Notch Target Genes in Wing Imaginal Discs
3.3. Effects of a Combined Overexpression of SPTM-Hmyc and Su(H)
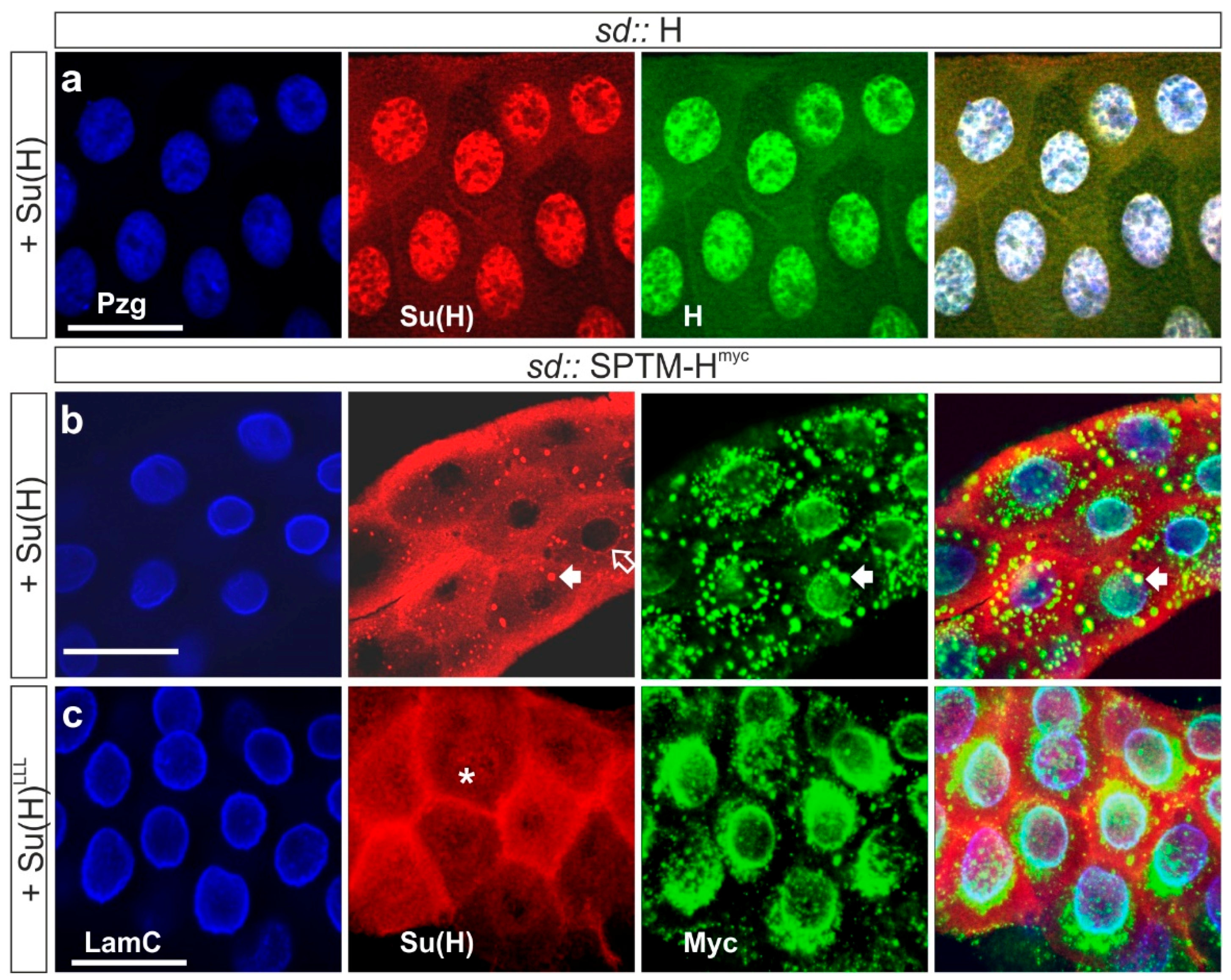
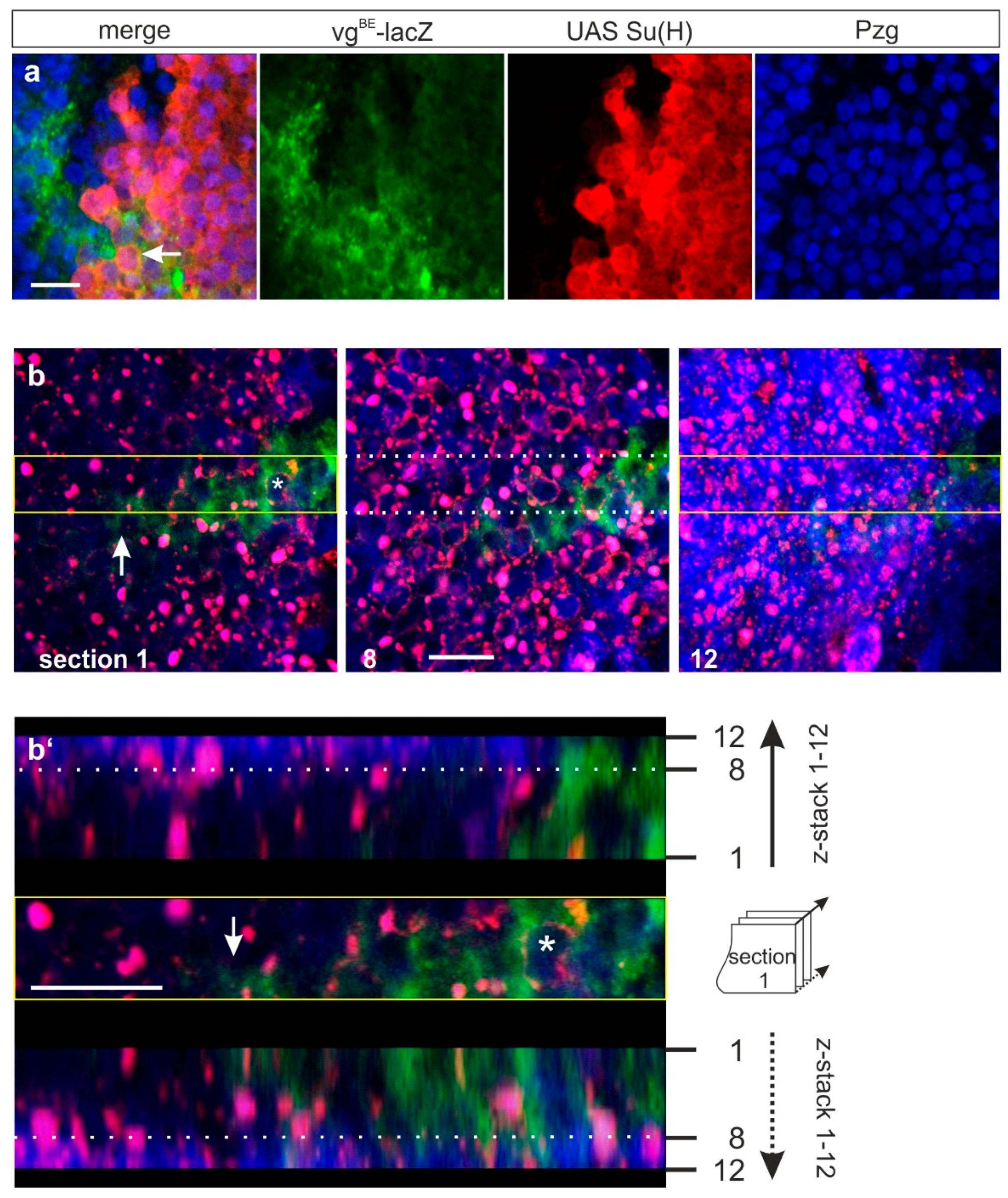
3.3.1. Co-localization of SPTM-Hmyc and Su(H) in Salivary Glands
3.3.2. Consequences of the Co-Expression of SPTM-Hmyc and Su(H) on Notch Target Gene Expression
3.3.3. Phenotypic Consequences of Co-Overexpression of SPTM-Hmyc and Su(H)
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Guruharsha, K.; Kankel, M.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012, 13, 654–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehon, R.G.; Kooh, P.J.; Rebay, I.; Regan, C.L.; Xu, T.; Muskavitch, M.A.; Artavanis-Tsakonas, S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 1990, 61, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E.; Artavanis-Tsakonas, S. The Suppressor of Hairless protein participates in Notch receptor signaling. Cell 1994, 79, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Praxenthaler, H.; Nagel, A.C.; Schulz, A.; Zimmermann, M.; Meier, M.; Schmid, H.; Preiss, A.; Maier, D. Hairless-binding deficient Suppressor of Hairless alleles reveal Su(H) protein levels are dependent on complex formation with Hairless. PLoS Genet. 2017, 13, e1006774. [Google Scholar] [CrossRef] [Green Version]
- Bray, S.; Furriols, M. Notch pathway: Making sense of Suppressor of Hairless. Curr. Biol. 2001, 11, R217–R221. [Google Scholar] [CrossRef] [Green Version]
- Schweisguth, F. Regulation of Notch signaling activity. Curr. Biol. 2004, 14, R129–R138. [Google Scholar]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef]
- Kovall, R.A.; Blacklow, S.C. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr. Top. Dev. Biol. 2010, 92, 31–71. [Google Scholar] [CrossRef]
- Hori, K.; Sen, A.; Artavanis-Tsakonas, S. Notch signaling at a glance. J. Cell Sci. 2013, 126, 2135–2140. [Google Scholar] [CrossRef] [Green Version]
- Artavanis-Tsakonas, S.; Matsuno, K.; Fortini, M.E. Notch signaling. Science 1995, 268, 225–232. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Maier, D. The evolution of transcriptional repressors in the Notch signaling pathway: A computational analysis. Hereditas 2019, 156, 5. [Google Scholar] [CrossRef]
- Furukawa, T.; Maruyama, S.; Kawaichi, M.; Honjo, T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell 1992, 69, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Schweisguth, F.; Posakony, J.W. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992, 69, 1199–1212. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.P.; Kovall, R.A. Structurally conserved binding motifs of transcriptional regulators to Notch nuclear effector CSL. Exp. Biol. Med. 2019, 244, 1520–1529. [Google Scholar] [CrossRef]
- Kaspar, M.; Klein, T. Functional analysis of murine CBF1 during Drosophila development. Dev. Dyn. 2006, 235, 918–927. [Google Scholar] [CrossRef]
- Gahr, B.M.; Brändle, F.; Zimmermann, M.; Nagel, A.C. An RBPJ-Drosophila model reveals dependence of RBPJ protein stability on the formation of transcription-regulator complexes. Cells 2019, 8, 1252. [Google Scholar] [CrossRef] [Green Version]
- Kovall, R.A.; Hendrickson, W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004, 23, 3441–3451. [Google Scholar] [CrossRef]
- Nam, Y.; Sliz, P.; Song, L.; Aster, J.C.; Blacklow, S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 2006, 124, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, J.J.; Kovall, R.A. Crystal structure of the CSL-Notch-mastermind ternary complex bound to DNA. Cell 2006, 124, 985–996. [Google Scholar] [CrossRef] [Green Version]
- Borggrefe, T.; Oswald, F. Setting the stage for Notch: The Drosophila Su(H)-Hairless repressor complex. PLoS Biol. 2016, 14, e1002524. [Google Scholar] [CrossRef] [Green Version]
- Maier, D. Hairless, the ignored antagonist of the Notch signalling pathway. Hereditas 2006, 143, 212–221. [Google Scholar] [CrossRef]
- Morel, V.; Lecourtois, M.; Massiani, O.; Maier, D.; Preiss, A.; Schweisguth, F. Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless-dCtBP complex in Drosophila. Curr. Biol. 2001, 11, 789–792. [Google Scholar] [CrossRef] [Green Version]
- Barolo, S.; Stone, T.; Bang, A.G.; Posakony, J.W. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002, 16, 1964–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, A.C.; Krejci, A.; Tenin, G.; Bravo-Patiño, A.; Bray, S.; Maier, D.; Preiss, A. Hairless-mediated repression of Notch target genes requires the combined activity of Groucho and CtBP co-repressors. Mol. Cell. Biol. 2005, 25, 10433–10441. [Google Scholar] [CrossRef] [Green Version]
- Maier, D.; Kurth, P.; Schulz, A.; Russell, A.; Yuan, Z.; Gruber, K.; Kovall, R.A.; Preiss, A. Structural and functional analysis of the repressor complex in the Notch signaling pathway of Drosophila melanogaster. Mol. Cell. Biol. 2011, 22, 3242–3252. [Google Scholar] [CrossRef]
- Yuan, Z.; Praxenthaler, H.; Tabaja, N.; Torella, R.; Preiss, A.; Maier, D.; Kovall, R.A. Structure and function of the Su(H)-Hairless repressor complex, the major antagonist of Notch signalling in Drosophila melanogaster. PLoS Biol. 2016, 14, e1002509. [Google Scholar] [CrossRef]
- Maier, D.; Nagel, A.C.; Johannes, B.; Preiss, A. Subcellular localization of Hairless protein shows a major focus of activity within the nucleus. Mech. Dev. 1999, 89, 195–199. [Google Scholar] [CrossRef]
- Wolf, D.; Smylla, T.K.; Reichmuth, J.; Hoffmeister, P.; Kober, L.; Zimmermann, M.; Turkiewicz, A.; Borggrefe, T.; Nagel, A.C.; Oswald, F.; et al. Nucleo-cytoplasmic shuttling of Drosophila Hairless/Su(H) heterodimer as a means of regulating Notch dependent transcription. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1520–1532. [Google Scholar] [CrossRef]
- Praxenthaler, H.; Smylla, T.K.; Nagel, A.C.; Preiss, A.; Maier, D. Generation of new Hairless alleles by genomic engineering at the Hairless locus in Drosophila melanogaster. PLoS ONE 2015, 10, e0140007. [Google Scholar] [CrossRef] [Green Version]
- Lindsley, D.L.; Zimm, G.G. The Genome of Drosophila Melanogaster; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- Vässin, H.; Vielmetter, J.; Campos-Ortega, J.A. Genetic interactions in early neurogenesis of Drosophila melanogaster. J. Neurogenet. 1985, 2, 291–308. [Google Scholar] [CrossRef]
- Maier, D.; Stumm, G.; Kuhn, K.; Preiss, A. Hairless, a Drosophila gene involved in neural development, encodes a novel, serine rich protein. Mech. Dev. 1992, 38, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Schweisguth, F.; Posakony, J.W. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development 1994, 120, 1433–1441. [Google Scholar]
- Bland, C.E.; Kimberly, P.; Rand, M.D. Notch-induced proteolysis and nuclear localization of the Delta ligand. J. Biol. Chem. 2003, 278, 13607–13610. [Google Scholar] [CrossRef] [Green Version]
- Bischof, J.; Maeda, R.K.; Hediger, M.; Karch, F.; Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific PhiC31 integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef] [Green Version]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar]
- Kim, J.; Sebring, A.; Esch, J.J.; Kraus, M.E.; Vorwerk, K.; Magee, J.; Carroll, S.B. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature 1996, 382, 133–138. [Google Scholar] [CrossRef]
- Maier, D.; Praxenthaler, H.; Schulz, A.; Preiss, A. Gain of function Notch phenotypes associated with ectopic expression of the Su(H) C-terminal domain illustrate separability of Notch and Hairless-mediated activities. PLoS ONE 2013, 8, e81578. [Google Scholar] [CrossRef] [Green Version]
- Kugler, S.J.; Nagel, A.C. Putzig is required for cell proliferation and regulates Notch activity in Drosophila. Mol. Biol Cell 2007, 18, 3733–3740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedel, F.; Gillingham, A.K.; Rosa-Ferreira, C.; Galindo, A.; Munro, S. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open 2016, 15, 987–992. [Google Scholar]
- Maier, D.; Nagel, A.C.; Preiss, A. Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation. Proc. Natl. Acad. Sci. USA 2002, 99, 15480–15485. [Google Scholar] [CrossRef] [Green Version]
- Smylla, T.K.; Preiss, A.; Maier, D. In vivo analysis of internal ribosome entry at the Hairless locus by genome engineering in Drosophila. Sci. Rep. 2016, 6, 34881. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biyasheva, A.; Do, T.V.; Lu, Y.; Vaskova, M.; Andres, A.J. Glue secretion in the Drosophila salivary gland: A model for steroid-regulated exocytosis. Dev. Biol. 2001, 231, 234–251. [Google Scholar] [CrossRef] [Green Version]
- Rousso, T.; Schejter, E.D.; Shilo, B.Z. Orchestrated content release from Drosophila glue-protein vesicles by a contractile actomyosin network. Nat. Cell Biol. 2016, 18, 181–190. [Google Scholar] [CrossRef]
- Gho, M.; Lecourtois, M.; Géraud, G.; Posakony, J.W.; Schweisguth, F. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 1996, 122, 1673–1682. [Google Scholar]
- Milán, M.; Diaz-Benjumea, F.J.; Cohen, S.M. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: A model for LMO oncogene function. Genes Dev. 1998, 12, 2912–2920. [Google Scholar] [CrossRef] [Green Version]
- Go, M.J.; Eastman, D.S.; Artavanis-Tsakonas, S. Cell proliferation control by Notch signaling in Drosophila development. Development 1998, 125, 2031–2040. [Google Scholar] [PubMed]
- Estella, C.; Baonza, A. Cell proliferation control by Notch signalling during imaginal discs development in Drosophila. AIMS Genet. 2015, 2, 70–96. [Google Scholar] [CrossRef]
- Slaninova, V.; Krafcikova, M.; Perez-Gomez, R.; Steffal, P.; Trantirek, L.; Bray, S.J.; Krejci, A. Notch stimulates growth by direct regulation of genes involved in the control of glycolysis and the tricarboxylic acid cycle. Open Biol. 2016, 6, 150155. [Google Scholar] [CrossRef]
- Kurth, P.; Preiss, A.; Kovall, R.A.; Maier, D. Molecular analysis of the Notch repressor-complex in Drosophila: Characterization of potential Hairless binding sites on Suppressor of Hairless. PLoS ONE 2011, 6, e27986. [Google Scholar] [CrossRef] [Green Version]
- Furriols, M.; Bray, S. Dissecting the mechanisms of Suppressor of Hairless function. Dev. Biol. 2000, 227, 520–532. [Google Scholar] [CrossRef] [Green Version]
- Fromental-Ramain, C.; Taquet, N.; Ramain, P. Transcriptional interactions between the pannier isoforms and the cofactor U-shaped during neural development in Drosophila. Mech. Dev. 2010, 127, 442–457. [Google Scholar] [CrossRef]
- Kopczynski, C.C.; Alton, A.K.; Fechtel, K.; Kooh, P.J.; Muskavitch, M.A. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988, 2, 1723–1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasquez-Del Carpio, R.; Kaplan, F.M.; Weaver, K.L.; VanWye, J.D.; Alves-Guerra, M.C.; Robbins, D.J.; Capobianco, A.J. Assembly of a Notch transcriptional activation complex requires multimerization. Mol. Cell. Biol. 2011, 31, 1396–1408. [Google Scholar] [CrossRef] [Green Version]
- Kovall, R.A. More complicated than it looks: Assembly of Notch pathway transcription complexes. Oncogene 2008, 27, 5099–5109. [Google Scholar] [CrossRef] [Green Version]
- Bray, S.J.; Gomez-Lamarca, M. Notch after cleavage. Curr. Opin. Cell Biol. 2018, 51, 103–109. [Google Scholar] [CrossRef]
- Shilo, B.Z. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp. Cell Res. 2003, 284, 140–149. [Google Scholar] [CrossRef]
- Doroquez, D.B.; Rebay, I. Signal integration during development: Mechanisms of EGFR and Notch pathway function and cross-talk. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 339–385. [Google Scholar] [CrossRef] [PubMed]
- Hasson, P.; Paroush, Z. Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE. Br. J. Cancer 2006, 94, 771–775. [Google Scholar]
- de Celis, J.F. Pattern formation in the Drosophila wing: The development of the veins. Bioessays 2003, 25, 443–451. [Google Scholar] [CrossRef]
- Blair, S.S. Wing vein pattering in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 2007, 23, 293–319. [Google Scholar] [CrossRef]
- Sotillos, S.; de Celis, J.F. Interactions between the Notch, EGFR, and decapentaplegic signaling pathways regulate vein differentiation during Drosophila pupal wing development. Dev. Dyn. 2005, 232, 738–752. [Google Scholar] [CrossRef]
- Martín-Blanco, E.; Roch, F.; Noll, E.; Baonza, A.; Duffy, J.B.; Perrimon, N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development 1999, 126, 5739–5747. [Google Scholar]
- Johannes, B.; Preiss, A. Wing vein formation in Drosophila melanogaster: Hairless is involved in the cross-talk between Notch and EGF signaling pathways. Mech. Dev. 2002, 115, 3–14. [Google Scholar] [CrossRef]
- Wang, C.; Guo, X.; Xi, R. EGFR and Notch signaling respectively regulate proliferative activity and multiple cell lineage differentiation of Drosophila gastric stem cells. Cell Res. 2014, 24, 610–627. [Google Scholar] [CrossRef] [Green Version]
- Djiane, A.; Krejci, A.; Bernard, F.; Fexova, S.; Millen, K.; Bray, S.J. Dissecting the mechanisms of Notch induced hyperplasia. EMBO J. 2013, 32, 60–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protzer, C.E.; Wech, I.; Nagel, A.C. Hairless induces cell death by downregulation of EGFR signalling activity. J. Cell Sci. 2008, 121, 3167–3176. [Google Scholar] [CrossRef] [Green Version]
- Rasheva, V.I.; Domingos, P.M. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 2009, 14, 996–1007. [Google Scholar] [CrossRef]
- Kockel, L.; Homsy, J.G.; Bohmann, D. Drosophila AP-1: Lessons from an invertebrate. Oncogene 2001, 20, 2347–2364. [Google Scholar] [CrossRef] [Green Version]
- Zecchini, V.; Brennan, K.; Martinez-Arias, A. An activity of Notch regulates JNK signalling and affects dorsal closure in Drosophila. Curr. Biol. 1999, 9, 460–469. [Google Scholar] [CrossRef]
- Klein, T.; Seugnet, L.; Haenlin, M.; Martinez Arias, A. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development 2000, 127, 3553–3566. [Google Scholar]
- Maier, D.; Chen, A.X.; Preiss, A.; Ketelhut, M. The tiny Hairless protein from Apis mellifera: A potent antagonist of Notch signaling in Drosophila melanogaster. BMC Evol. Biol. 2008, 8, 175. [Google Scholar] [CrossRef] [Green Version]
- Zehender, A.; Bayer, M.; Bauer, M.; Zeis, B.; Preiss, A.; Maier, D. Conservation of the Notch antagonist Hairless in arthropods: Functional analysis of the crustacean Daphnia pulex Hairless gene. Dev. Genes Evol. 2017, 227, 339–353. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maier, D. Membrane-Anchored Hairless Protein Restrains Notch Signaling Activity. Genes 2020, 11, 1315. https://doi.org/10.3390/genes11111315
Maier D. Membrane-Anchored Hairless Protein Restrains Notch Signaling Activity. Genes. 2020; 11(11):1315. https://doi.org/10.3390/genes11111315
Chicago/Turabian StyleMaier, Dieter. 2020. "Membrane-Anchored Hairless Protein Restrains Notch Signaling Activity" Genes 11, no. 11: 1315. https://doi.org/10.3390/genes11111315
APA StyleMaier, D. (2020). Membrane-Anchored Hairless Protein Restrains Notch Signaling Activity. Genes, 11(11), 1315. https://doi.org/10.3390/genes11111315




