Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance
Abstract
:1. Introduction
Fe2+ + H2O2 → Fe3+ + OH• + OH−
Net Reaction: O2•− + H2O2 → OH• + OH− + O2
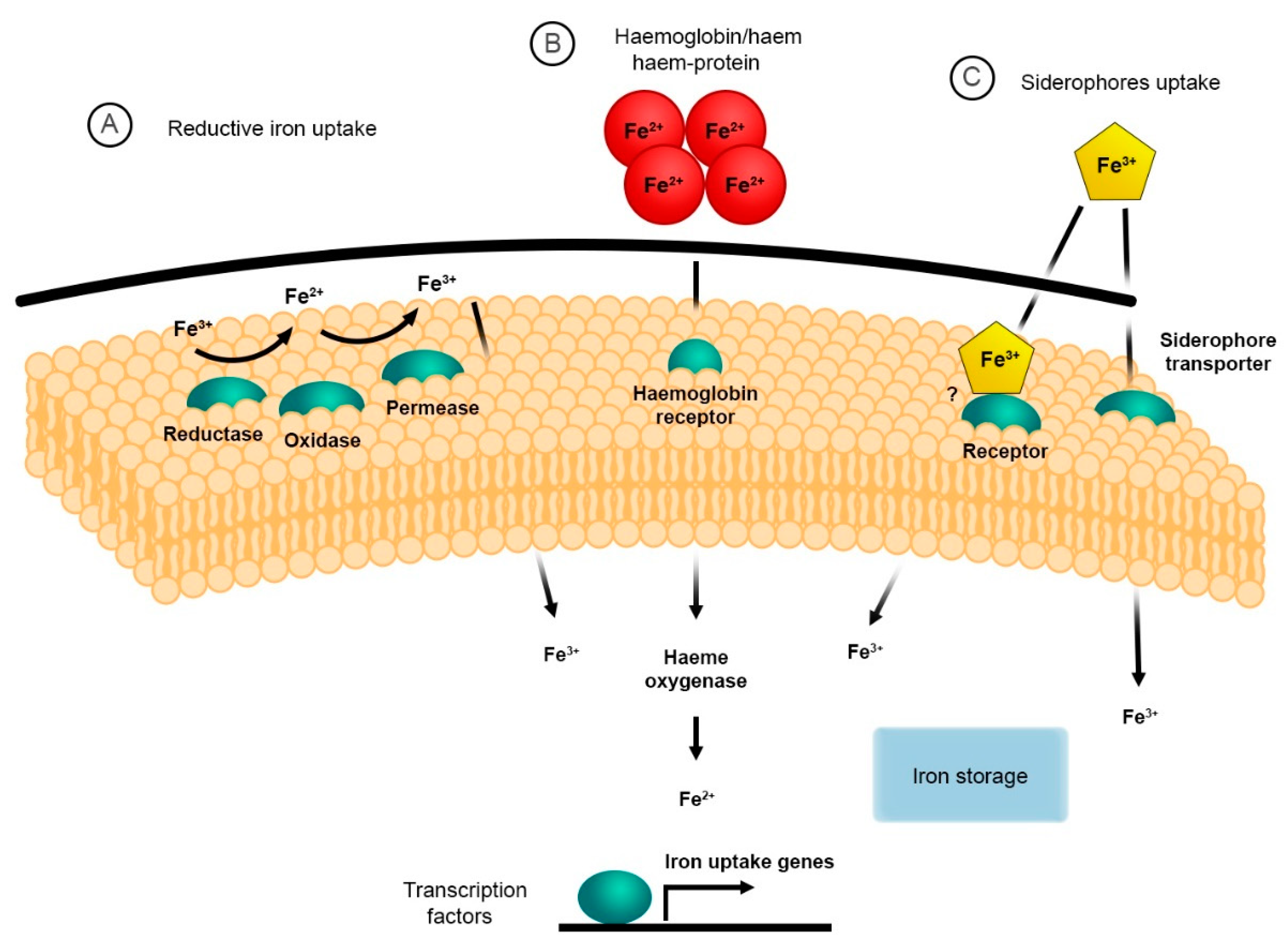
2. The Reductive System for Iron Uptake
3. Haem and Haemoglobin Utilisation
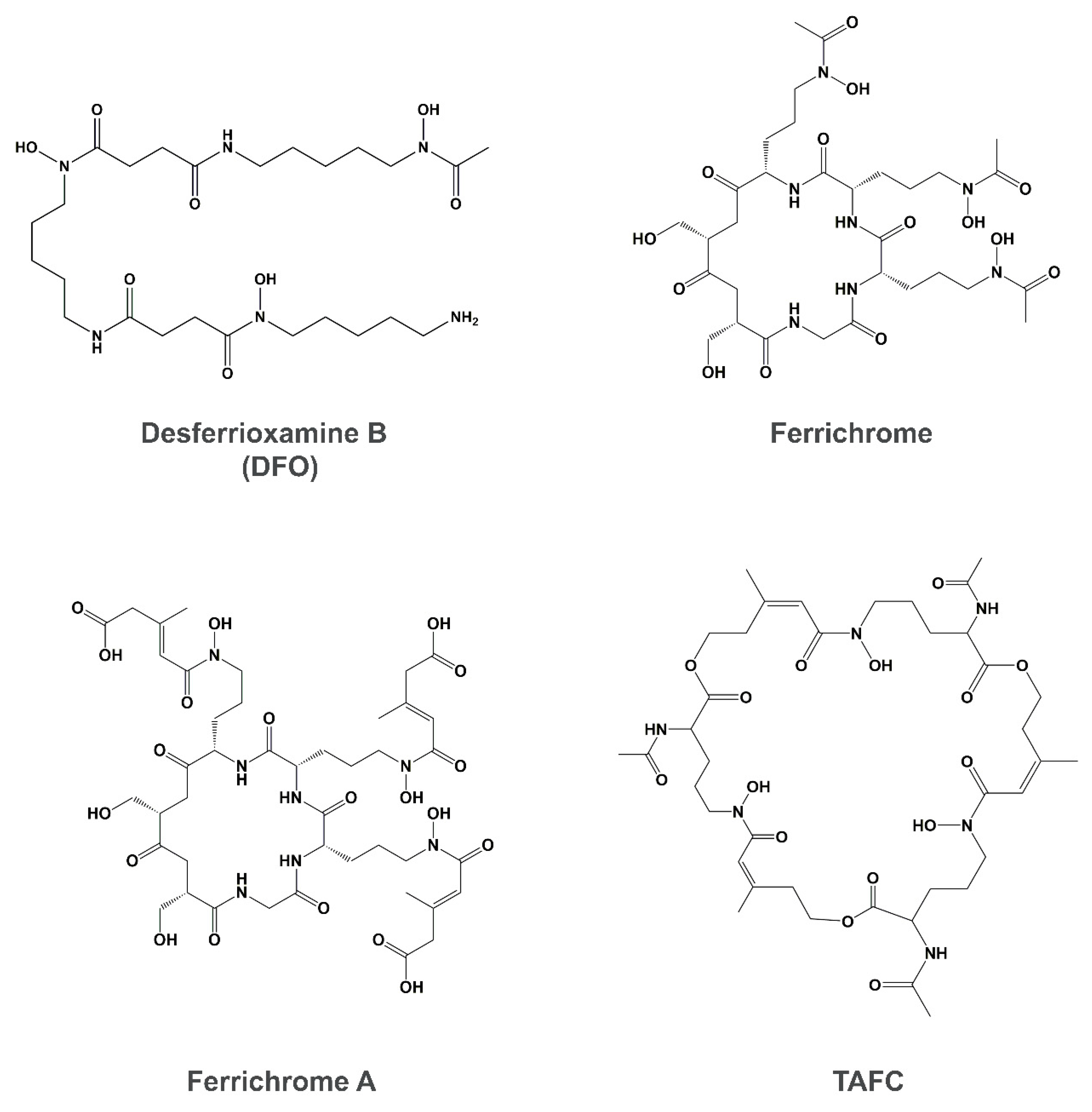
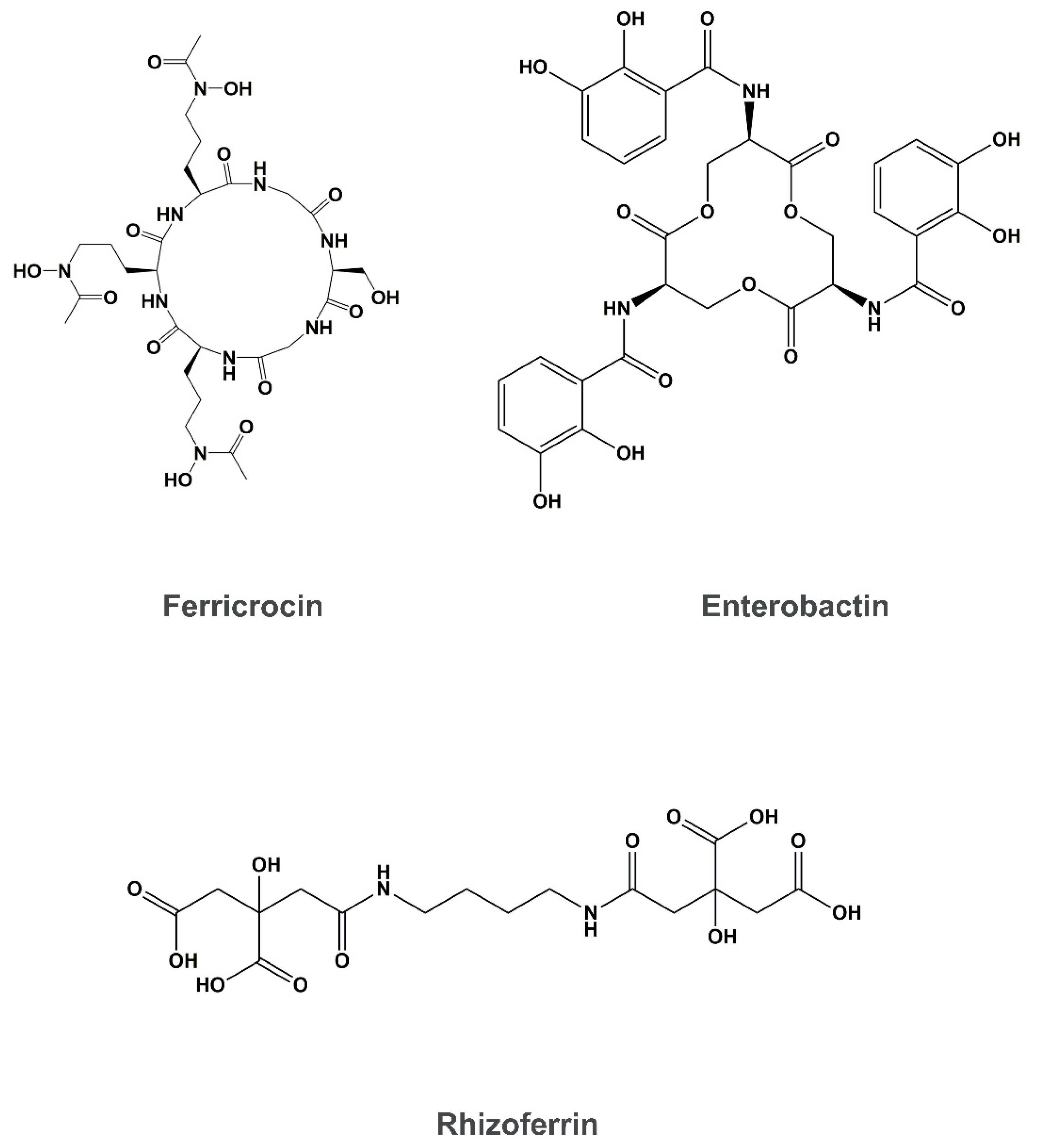
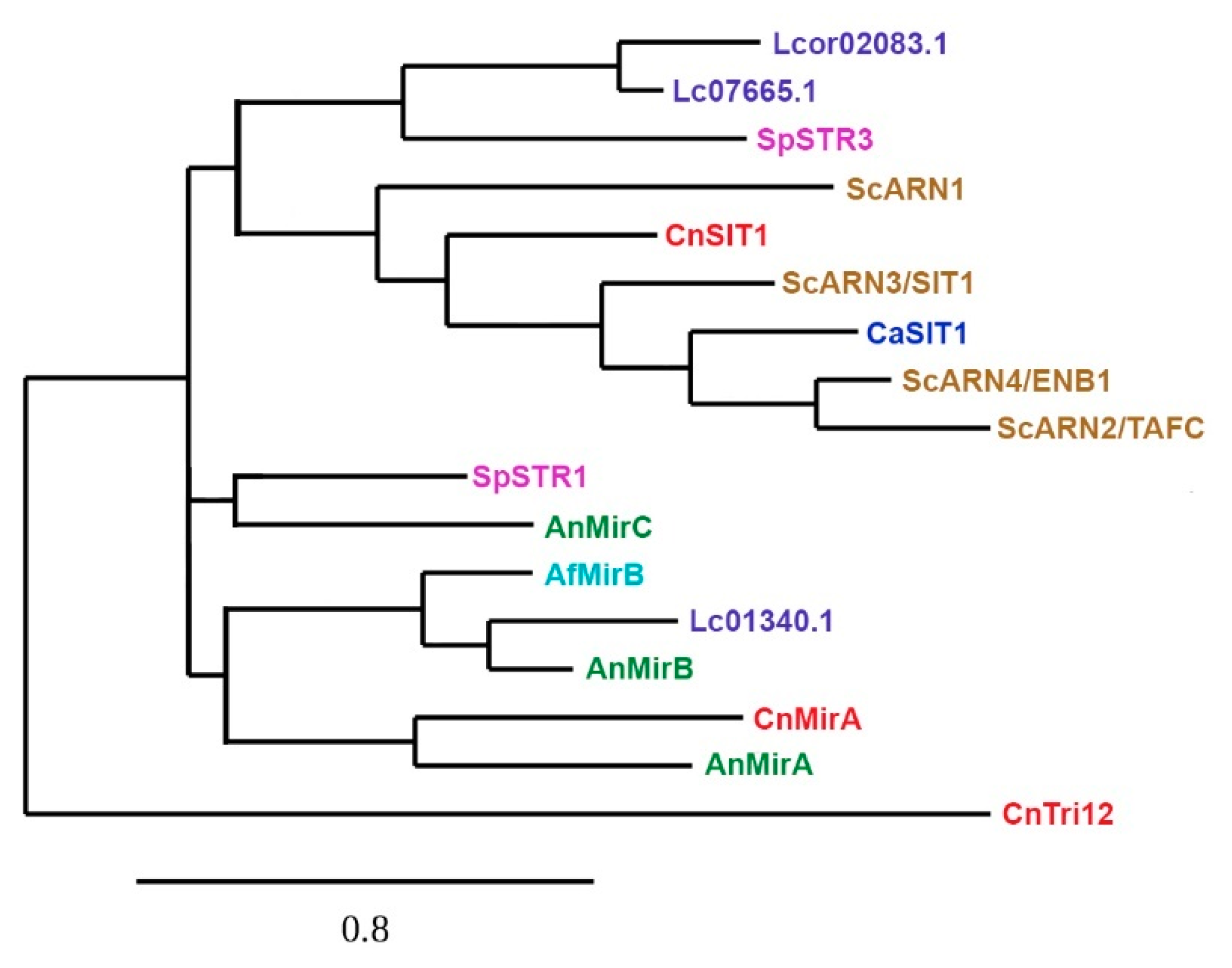
4. Siderophore Uptake
5. The Fungal Cell Wall: Composition and Role in Diagnostics
5.1. The Cell Wall Composition of Mucorales in Comparison to Other Fungi
5.2. Diagnostic Methods Based on Properties of the Fungal Cell
6. Iron Acquisition and Susceptibility to Antifungals: Implications in Therapy
6.1. Antifungal Treatment and Iron Chelation Therapy
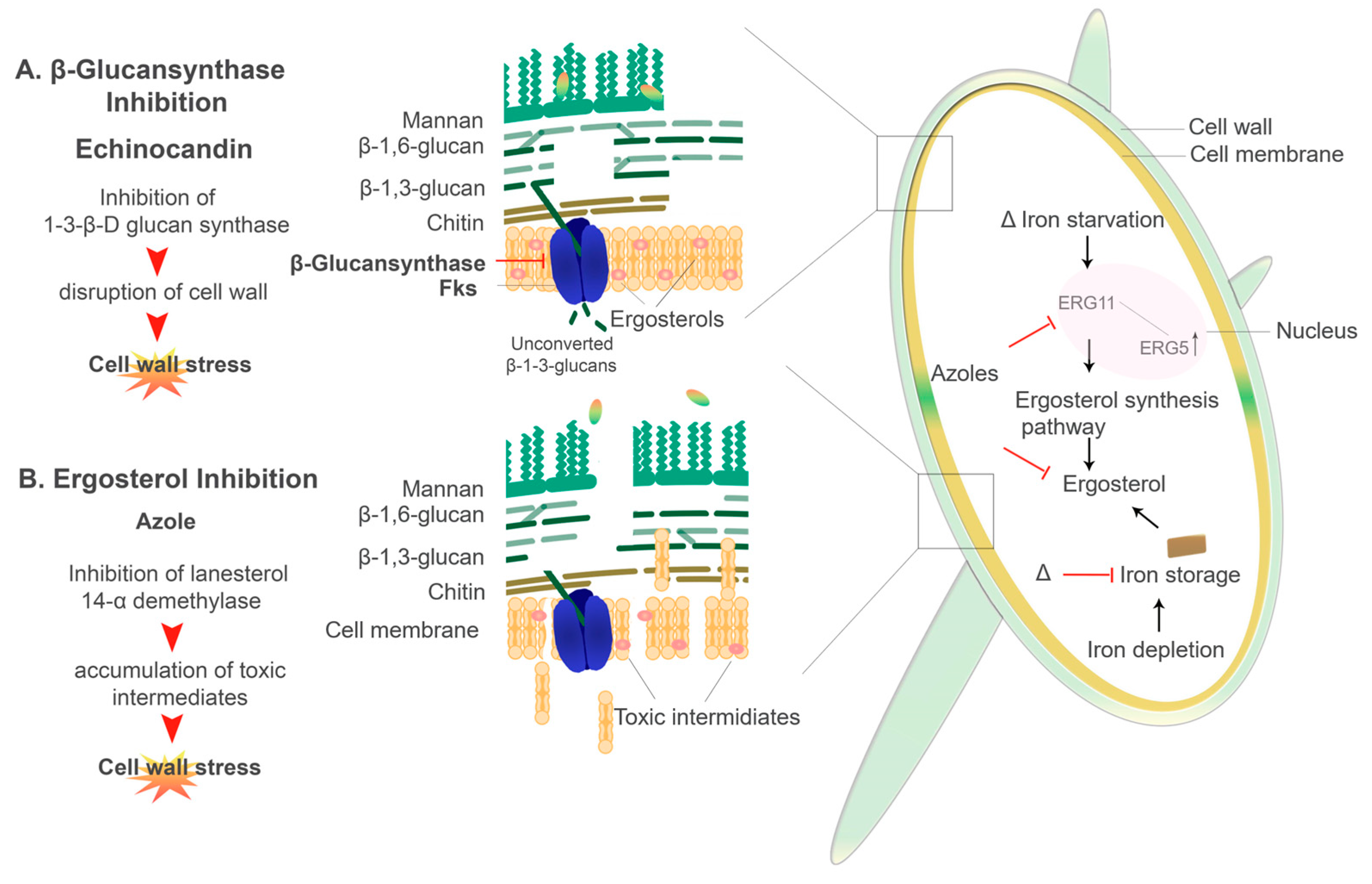
6.2. Antifungal Resistance and Iron
6.2.1. Echinocandins
6.2.2. Azoles
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Domenico, I.; Ward, D.M.; Kaplan, J. Regulation of iron acquisition and storage: Consequences for iron-linked disorders. Nat. Rev. Mol. Cell Biol. 2008, 9, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Cassat, J.E.; Skaar, E.P. Iron in Infection and Immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Philpott, C.C. Iron uptake in fungi: A system for every source. Biochim. Biophys. Acta-Mol. Cell Res. 2006, 1763, 636–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyce, K.J.; Andrianopoulos, A. Fungal dimorphism: The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol. Rev. 2015, 39, 797–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beard, J.L. Iron Biology in Immune Function, Muscle Metabolism and Neuronal Functioning. J. Nutr. 2001, 131, 568S–580S. [Google Scholar] [CrossRef] [PubMed]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 1781–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouault, T.A.; Tong, W.H. Iron–sulfur cluster biogenesis and human disease. Trends Genet. 2008, 24, 398–407. [Google Scholar] [CrossRef] [Green Version]
- Prousek, J. Fenton chemistry in biology and medicine. Pure Appl. Chem. 2007, 79, 2325–2338. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridget, J.M.C. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef]
- Brault, A.; Mourer, T.; Labbé, S. Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts. IUBMB Life 2015, 67, 801–815. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, J.; McVey Ward, D.; Crisp, R.J.; Philpott, C.C. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim. Biophys. Acta-Mol. Cell Res. 2006, 1763, 646–651. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M. Molecular strategies of microbial iron assimilation: From high-affinity complexes to cofactor assembly systems. Metallomics 2013, 5, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Carrano, C.J.; Böhnke, R.; Matzanke, B.F. Fungal ferritins: The ferritin from mycelia of Absidia spinosa is a bacterioferritin. FEBS Lett. 1996, 390, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Spellberg, B.; Walsh, T.J.; Kontoyiannis, D.P. Pathogenesis of mucormycosis. Clin. Infect. Dis. 2012, 54, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Comensoli, L.; Bindschedler, S.; Junier, P.; Joseph, E. Iron and Fungal Physiology: A Review of Biotechnological Opportunities. Adv. Appl. Microbiol. 2017, 98, 31–60. [Google Scholar] [CrossRef] [PubMed]
- Doguer, C.; Ha, J.-H.; Collins, J.F. Intersection of Iron and Copper Metabolism in the Mammalian Intestine and Liver. In Comprehensive Physiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; Volume 176, pp. 1433–1461. ISBN 2163684814. [Google Scholar]
- Kehl-Fie, T.E.; Skaar, E.P. Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 2010, 14, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.J.R.; Merlot, A.M.; Huang, M.L.H.; Bae, D.H.; Jansson, P.J.; Sahni, S.; Kalinowski, D.S.; Richardson, D.R. Cellular iron uptake, trafficking and metabolism: Key molecules and mechanisms and their roles in disease. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1130–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 2010, 36, 1–53. [Google Scholar] [CrossRef]
- Sam, Q.H.; Yew, W.S.; Seneviratne, C.J.; Chang, M.W.; Chai, L.Y.A. Immunomodulation as Therapy for Fungal Infection: Are We Closer? Front. Microbiol. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nosanchuk, J.D. Fungal diseases as neglected pathogens: A wake-up call to public health officials. PLoS Negl. Trop. Dis. 2020, 14, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Low, C.Y.; Rotstein, C. Emerging fungal infections in immunocompromised patients. F1000 Med. Rep. 2011, 3, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabra-Rizk, M.A.; Kong, E.F.; Tsui, C.; Nguyen, M.H.; Clancy, C.J.; Fidel, P.L.; Noverr, M. Candida albicans Pathogenesis: Fitting within the Host-Microbe Damage Response Framework. Infect. Immun. 2016, 84, 2724–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong-James, D.; Meintjes, G.; Brown, G.D. A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 2014, 22, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Warkentien, T.; Crum-Cianflone, N.F. An update on Cryptococcus among HIV-infected patients. Int. J. STD AIDS 2010, 21, 679–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef] [Green Version]
- Franco-Paredes, C.; Chastain, D.B.; Rodriguez-Morales, A.J.; Marcos, L.A. Cryptococcal meningoencephalitis in HIV/AIDS: When to start antiretroviral therapy? Ann. Clin. Microbiol. Antimicrob. 2017, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Haas, H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276. [Google Scholar] [CrossRef] [Green Version]
- Wiederhold, N.P. Antifungal resistance: Current trends and future strategies to combat. Infect. Drug Resist. 2017, 10, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Spellberg, B.; Edwards, J. Iron acquisition: A novel perspective on mucormycosis pathogenesis and treatment. Curr. Opin. Infect. Dis. 2008, 21, 620–625. [Google Scholar] [CrossRef] [Green Version]
- Symeonidis, A.S. The role of iron and iron chelators in zygomycosis. Clin. Microbiol. Infect. 2009, 15, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S. Host-iron assimilation: Pathogenesis and novel therapies of mucormycosis. Mycoses 2014, 57, 13–17. [Google Scholar] [CrossRef] [Green Version]
- Haas, H. Iron—A key nexus in the virulence of Aspergillus fumigatus. Front. Microbiol. 2012, 3, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, C.; Ferea, T.; Rashford, J.; Ardon, O.; Brown, P.O.; Botstein, D.; Kaplan, J.; Philpott, C.C. Desferrioxamine-mediated Iron Uptake in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 10709–10715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker Brachmann, C.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Hassett, R.F.; Romeo, A.M.; Kosman, D.J. Regulation of High Affinity Iron Uptake in the Yeast Saccharomyces cerevisiae ROLE OF DIOXYGEN AND Fe (II). J. Biol. Chem. 1998, 273, 7628–7636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, S.M.; Roy, S.; Vareechon, C.; Carrion, S.; deJesus, J.; Clark, H.; Lopez-Berges, M.S.; DiPietro, A.; Schrettl, M.; Beckmann, N.; et al. Targeting Iron Acquisition Blocks Infection with the Fungal Pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. 2013, 9, e1003436. [Google Scholar] [CrossRef]
- Lindahl, P.A. A comprehensive mechanistic model of iron metabolism in: Saccharomyces cerevisiae. Metallomics 2019, 11, 1779–1799. [Google Scholar] [CrossRef]
- Almeida, R.S.; Wilson, D.; Hube, B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009, 9, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
- Gaensly, F.; Picheth, G.; Brand, D.; Bonfim, T.M.B. The uptake of different iron salts by the yeast Saccharomyces cerevisiae. Braz. J. Microbiol. 2014, 45, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Walther, G.; Wagner, L.; Kurzai, O. Updates on the taxonomy of mucorales with an emphasis on clinically important taxa. J. Fungi 2019, 5, 106. [Google Scholar] [CrossRef] [Green Version]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.A.B.; Lesuisse, E.; Stearman, R.; Klausner, R.D.; Dancis, A. Reductive iron uptake by Candida albicans: Role of copper, iron and the TUP1 regulator. Microbiology 2002, 148, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.A.B.; Vilaire, G.; Lesuisse, E.; Dancis, A. Iron Acquisition from Transferrin by Candida albicans Depends on the Reductive Pathway. Infect. Immun. 2005, 73, 5482–5492. [Google Scholar] [CrossRef] [Green Version]
- Blatzer, M.; Binder, U.; Haas, H. The metalloreductase FreB is involved in adaptation of Aspergillus fumigatus to iron starvation. Fungal Genet. Biol. 2011, 48, 1027–1033. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Murcia, L.; Martínez-García, P.; Lax, C.; Sanchis, M.; Capilla, J.; Nicolás, F.E.; Garre, V. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Andrianaki, A.M.; Kyrmizi, I.; Thanopoulou, K.; Baldin, C.; Drakos, E.; Soliman, S.S.M.; Shetty, A.C.; McCracken, C.; Akoumianaki, T.; Stylianou, K.; et al. Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nat. Commun. 2018, 9, 3333. [Google Scholar] [CrossRef] [Green Version]
- Schwartze, V.U.; Winter, S.; Shelest, E.; Marcet-Houben, M.; Horn, F.; Wehner, S.; Linde, J.; Valiante, V.; Sammeth, M.; Riege, K.; et al. Gene Expansion Shapes Genome Architecture in the Human Pathogen Lichtheimia corymbifera: An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina). PLoS Genet. 2014, 10. [Google Scholar] [CrossRef]
- Urbanowski, J.L.; Piper, R.C. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 1999, 274, 38061–38070. [Google Scholar] [CrossRef] [Green Version]
- Haas, H.; Petrik, M.; Decristoforo, C. An Iron-Mimicking, Trojan Horse-Entering Fungi—Has the Time Come for Molecular Imaging of Fungal Infections? PLoS Pathog. 2015, 11, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Saikia, S.; Oliveira, D.; Hu, G.; Kronstad, J. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect. Immun. 2014, 82, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef] [Green Version]
- Georgatsou, E.; Alexandraki, D. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 1999, 15, 573–584. [Google Scholar] [CrossRef]
- Philpott, C.C.; Protchenko, O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 2008, 7, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.S.; Ju, X.; Gao, H.L.; Wang, T.; Thiele, D.J.; Li, J.Y.; Wang, Z.Y.; Ding, C. Reciprocal functions of Cryptococcus neoformans copper homeostasis machinery during pulmonary infection and meningoencephalitis. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterman, S.R.; Hacham, M.; Hu, G.; Zhu, X.; Park, Y.D.; Shin, S.; Panepinto, J.; Valyi-Nagy, T.; Beam, C.; Husain, S.; et al. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Investig. 2007, 117, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Iwai, Y.; Serpe, M.; Haile, D.; Yang, W.; Kosman, D.J.; Klausner, R.D.; Dancis, A. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J. Biol. Chem. 1997, 272, 17711–17718. [Google Scholar] [CrossRef] [Green Version]
- Kwok, E.Y.; Severance, S.; Kosman, D.J. Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 2006, 45, 6317–6327. [Google Scholar] [CrossRef]
- Bairwa, G.; Hee Jung, W.; Kronstad, J.W. Iron acquisition in fungal pathogens of humans. Metallomics 2017, 9, 215–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Protchenko, O.; Ferea, T.; Rashford, J.; Tiedeman, J.; Brown, P.O.; Botstein, D.; Philpott, C.C. Three Cell Wall Mannoproteins Facilitate the Uptake of Iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 49244–49250. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Xu, N.; Yu, Q.; Ding, X.; Qian, K.; Zhao, Q.; Wang, Y.; Zhang, B.; Xing, L.; Li, M. Novel insight into the expression and function of the multicopper oxidases in Candida albicans. Microbiology 2013, 159, 1044–1055. [Google Scholar] [CrossRef] [Green Version]
- Ramanan, N.; Wang, Y. A high-affinity iron permease essential for Candida albicans virulence. Science 2000, 288, 1062–1064. [Google Scholar] [CrossRef]
- Pradhan, A.; Avelar, G.M.; Bain, J.M.; Childers, D.; Pelletier, C.; Larcombe, D.E.; Shekhova, E.; Netea, M.G.; Brown, G.D.; Erwig, L.; et al. Non-canonical signalling mediates changes in fungal cell wall PAMPs that drive immune evasion. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yun, C.W.; Bauler, M.; Moore, R.E.; Klebba, P.E.; Philpott, C.C. The Role of the FRE Family of Plasma Membrane Reductases in the Uptake of Siderophore-Iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 10218–10223. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Lee, H.; Collins, M.; Tsai, H.F.; Spellberg, B.; Edwards, J.E.; Kwon-Chung, K.J.; Ibrahim, A.S. Cloning and functional characterization of the Rhizopus oryzae high affinity iron permease (rFTR1) gene. FEMS Microbiol. Lett. 2004, 235, 169–176. [Google Scholar] [CrossRef]
- Trieu, T.A.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Sanchis, M.; Capilla, J.; Navarro-Rodriguez, P.; Lopez-Fernandez, L.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M.; et al. RNAi-Based Functional Genomics Identifies New Virulence Determinants in Mucormycosis. PLoS Pathog. 2017, 13, e1006150. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Severance, S.; Kaur, N.; Wiltsie, W.; Kosman, D.J. Assembly, activation, and trafficking of the Fet3p·Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 13355–13364. [Google Scholar] [CrossRef] [Green Version]
- Severance, S.; Chakraborty, S.; Kosman, D.J. The Ftr1p iron permease in the yeast plasma membrane: Orientation, topology and structure-function relationships. Biochem. J. 2004, 380, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Stearman, R.; Yuan, D.S.; Yamaguchi-Iwai, Y.; Klausner, R.D.; Dancis, A. A Permease-Oxidase Complex Involved in High-Affinity Iron Uptake in Yeast. Science 1996, 271, 1552–1557. [Google Scholar] [CrossRef]
- Liu, M.; Lin, L.; Gebremariam, T.; Luo, G.; Skory, C.D.; French, S.W.; Chou, T.-F.; Edwards, J.E.; Ibrahim, A.S. Fob1 and Fob2 Proteins Are Virulence Determinants of Rhizopus oryzae via Facilitating Iron Uptake from Ferrioxamine. PLOS Pathog. 2015, 11, e1004842. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Gebremariam, T.; Lin, L.; Luo, G.; Husseiny, M.I.; Skory, C.D.; Fu, Y.; French, S.W.; Edwards, J.E.; Spellberg, B. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol. Microbiol. 2010, 77, 587–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Fernández, L.; Sanchis, M.; Navarro-Rodríguez, P.; Nicolás, F.E.; Silva-Franco, F.; Guarro, J.; Garre, V.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Capilla, J. Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence 2018, 9, 707–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N.; Haynes, K.; Haas, H. Siderophore Biosynthesis But Not Reductive Iron Assimilation Is Essential for Aspergillus fumigatus Virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrettl, M.; Bignell, E.; Kragl, C.; Sabiha, Y.; Loss, O.; Eisendle, M.; Wallner, A.; Arst, H.N.; Haynes, K.; Haas, H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007, 3, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Safyan, A.; Wisgott, S.; Brunke, S.; Kasper, L.; Hube, B. The Fungal Pathogen Candida glabrata Does not Depend on Surface Ferric Reductases for Iron Acquisition. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Aisen, P.; Leibman, A.; Zweier, J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J. Biol. Chem. 1978, 253, 1930–1937. [Google Scholar]
- Hare, S.A. Diverse structural approaches to haem appropriation by pathogenic bacteria. Biochim. Biophys. Acta-Proteins Proteom. 2017, 1865, 422–433. [Google Scholar] [CrossRef] [Green Version]
- Nairz, M.; Schroll, A.; Sonnweber, T.; Weiss, G. The struggle for iron - a metal at the host-pathogen interface. Cell. Microbiol. 2010, 12, 1691–1702. [Google Scholar] [CrossRef]
- Meynard, D.; Babitt, J.L.; Lin, H.Y. Review Article The liver: Conductor of systemic iron balance. Blood 2013, 123, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Weissman, Z.; Kornitzer, D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 2004, 53, 1209–1220. [Google Scholar] [CrossRef]
- Devaux, F.; Thiébaut, A. The regulation of iron homeostasis in the fungal human pathogen Candida glabrata. Microbiology 2019, 165, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Fourie, R.; Kuloyo, O.O.; Mochochoko, B.M.; Albertyn, J.; Pohl, C.H. Iron at the centre of Candida albicans interactions. Front. Cell. Infect. Microbiol. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, R.; Buisson, N.; Knight, S.; Dancis, A.; Camadro, J.; Lesuisse, E.; Inge, L. Haemin uptake and use as an iron source by Candida albicans: Role of CaHMX1 -encoded haem oxygenase. Microbiology 2003, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Weissman, Z.; Shemer, R.; Conibear, E.; Kornitzer, D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 2008, 69, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Vidanes, G.M.; Maguire, S.L.; Guida, A.; Synnott, J.M.; Andes, D.R.; Butler, G. Conserved and Divergent Roles of Bcr1 and CFEM Proteins in Candida parapsilosis and Candida albicans. PLoS ONE 2011, 6, e28151. [Google Scholar] [CrossRef] [Green Version]
- Okamoto-Shibayama, K.; Kikuchi, Y.; Kokubu, E.; Sato, Y.; Ishihara, K. Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth. FEMS Yeast Res. 2014, 14, 674–677. [Google Scholar] [CrossRef]
- Kulkarni, R.; Kelkar, H.; Dean, R. An eight-cysteine-containing CFEM domain unique toa group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 116–118. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.R.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef] [Green Version]
- Kuznets, G.; Vigonsky, E.; Weissman, Z.; Lalli, D.; Gildor, T.; Kauffman, S.J.; Turano, P.; Becker, J.; Lewinson, O.; Kornitzer, D. A Relay Network of Extracellular Heme-Binding Proteins Drives C. albicans Iron Acquisition from Hemoglobin. PLoS Pathog. 2014, 10, e1004407. [Google Scholar] [CrossRef] [Green Version]
- Pinsky, M.; Roy, U.; Moshe, S.; Weissman, Z.; Kornitzer, D. Human Serum Albumin Facilitates Heme-Iron Utilization by Fungi. MBio 2020, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 3, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Nevitt, T.; Thiele, D.J. Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.K.; Suneetha, K.J.; Kaur, R. A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem. J. 2014, 463, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Karkowska-Kuleta, J.; Satala, D.; Bochenska, O.; Rapala-Kozik, M.; Kozik, A. Moonlighting proteins are variably exposed at the cell surfaces of Candida glabrata, Candida parapsilosis and Candida tropicalis under certain growth conditions. BMC Microbiol. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Hu, G.; Caza, M.; Cadieux, B.; Bakkeren, E.; Do, E.; Jung, W.H.; Kronstad, J.W. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol. Microbiol. 2015, 96, 973–992. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Cho, Y.-J.; Do, E.; Choi, J.; Hu, G.; Cadieux, B.; Chun, J.; Lee, Y.; Kronstad, J.W.; Jung, W.H. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet. Biol. 2012, 49, 955–966. [Google Scholar] [CrossRef] [Green Version]
- Cadieux, B.; Lian, T.; Hu, G.; Wang, J.; Biondo, C.; Teti, G.; Liu, V.; Murphy, M.E.P.; Creagh, A.L.; Kronstad, J.W. The mannoprotein cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis. 2013, 207, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Caza, M.; Cadieux, B.; Chan, V.; Liu, V.; Kronstad, J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 2013, 81, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Bairwa, G.; Caza, M.; Horianopoulos, L.; Hu, G.; Kronstad, J. Role of clathrin-mediated endocytosis in the use of heme and hemoglobin by the fungal pathogen Cryptococcus neoformans. Cell. Microbiol. 2019, 21, e12961. [Google Scholar] [CrossRef]
- Spellberg, B.; Walsh, T.J.; Kontoyiannis, D.P.; Edwards, J., Jr.; Ibrahim, A.S. Recent Advances in the Management of Mucormycosis: From Bench to Bedside. Clin. Infect. Dis. 2009, 48, 1743–1751. [Google Scholar] [CrossRef]
- Schwartze, V.U.; Hoffmann, K.; Nyilasi, I.; Papp, T.; Vágvölgyi, C.; de Hoog, S.; Voigt, K.; Jacobsen, I.D. Lichtheimia species exhibit differences in virulence potential. PLoS ONE 2012, 7, e40908. [Google Scholar] [CrossRef]
- Lesuisse, E.; Blaiseau, P.L.; Dancis, A.; Camadro, J.M. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology 2001, 147, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Wilson, B.R.; Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med. 2016, 22, 1077–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldin, C.; Ibrahim, A.S. Molecular mechanisms of mucormycosis—The bitter and the sweet. PLoS Pathog. 2017, 13, e1006408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larcher, G.; Dias, M.; Razafimandimby, B.; Bomal, D.; Bouchara, J.-P. Siderophore Production by Pathogenic Mucorales and Uptake of Deferoxamine B. Mycopathologia 2013, 176, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Butler, A.; Theisen, R.M. Iron(III)–siderophore coordination chemistry: Reactivity of marine siderophores. Coord. Chem. Rev. 2010, 254, 288–296. [Google Scholar] [CrossRef] [Green Version]
- Kurth, C.; Kage, H.; Nett, M. Siderophores as molecular tools in medical and environmental applications. Org. Biomol. Chem. 2016, 14, 8212–8227. [Google Scholar] [CrossRef]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 2016, 23, 3984–3999. [Google Scholar] [CrossRef]
- Howard, D.H. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 1999, 12, 394–404. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef]
- Neilands, J.B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef] [Green Version]
- Haas, H.; Schoeser, M.; Lesuisse, E.; Ernst, J.F.; Parson, W.; Abt, B. Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J. 2003, 371, 505–513. [Google Scholar] [CrossRef] [Green Version]
- De Locht, M.; Boelaert, J.R.; Schneider, Y.-J. Iron uptake from ferrioxamine and from ferrirhizoferrin by germinating spores of Rhizopus microsporus. Biochem. Pharmacol. 1994, 47, 1843–1850. [Google Scholar] [CrossRef]
- Coale, T.H.; Moosburner, M.; Horák, A.; Oborník, M.; Barbeau, K.A.; Allen, A.E. Reduction-dependent siderophore assimilation in a model pennate diatom. Proc. Natl. Acad. Sci. USA 2019, 116, 23609–23617. [Google Scholar] [CrossRef] [Green Version]
- Haas, H. Molecular genetics of fungal siderophore biosynthesis and uptake: The role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 2003, 62, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.; Tiedeman, J.S.; Moore, R.E.; Philpott, C.C. Siderophore-iron uptake in Saccharomyces cerevisiae: Identification of ferrichrome and fusarinine transporters. J. Biol. Chem. 2000, 275, 16354–16359. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 2002, 21, 3632–3642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Froissard, M.; Belgareh-touzé, N.; Dias, M.; Buisson, N.; Camadro, J.M.; Haguenauer-tsapis, R.; Lesuisse, E. Trafficking of siderophore transporters in Saccharomyces cerevisiae and intracellular fate of ferrioxamine B conjugates. Traffic 2007, 8, 1601–1616. [Google Scholar] [CrossRef]
- Moore, R.E.; Kim, Y.; Philpott, C.C. The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 5664–5669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nies, D.H.; Janssen, P.J.; Van Houdt, R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, M.A.; Leys, N.; Vallaeys, T.; et al. The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 2016, 8, 481–507. [Google Scholar] [CrossRef] [Green Version]
- Wilson, S.; Bird, A.J. Zinc sensing and regulation in yeast model systems. Arch. Biochem. Biophys. 2016, 611, 30–36. [Google Scholar] [CrossRef]
- Howard, D.H. Iron gathering by zoopathogenic fungi. FEMS Immunol. Med. Microbiol. 2004, 40, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Heymann, P.; Gerads, M.; Schaller, M.; Dromer, F.; Winkelmann, G.; Ernst, J.F. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect. Immun. 2002, 70, 5246–5255. [Google Scholar] [CrossRef] [Green Version]
- Heymann, P.; Ernst, J.F.; Winkelmann, G. A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1p) in Saccharomyces cerevisiae. BioMetals 2000, 13, 65–72. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Chevenet, F.; Brun, C.; Bañuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Dereeper, A.; Audic, S.; Claverie, J.-M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Jeong, M.; Kang, C.; Kim, J.; Heo, D.; Chang, M.; Baek, I.; Ro, H.; Choi, I.-D.; Kim, T.-H.; Yun, C.-W. A novel function of Aft1 in regulating ferrioxamine B uptake: Aft1 modulates Arn3 ubiquitination in Saccharomyces cerevisiae. Biochem. J. 2009, 422, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.-M.; Kang, S.; Park, Y.; Yun, C. Physical interaction between Sit1 and Aft1 upregulates FOB uptake activity by inhibiting protein degradation of Sit1 in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov080. [Google Scholar] [CrossRef] [Green Version]
- Noble, S.M. Candida albicans specializations for iron homeostasis: From commensalism to virulence. Curr. Opin. Microbiol. 2013, 16, 708–715. [Google Scholar] [CrossRef] [Green Version]
- Seider, K.; Gerwien, F.; Kasper, L.; Allert, S.; Brunke, S.; Jablonowski, N.; Schwarzmüller, T.; Barz, D.; Rupp, S.; Kuchler, K.; et al. Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot. Cell 2014, 13, 170–183. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.H.; Sham, A.; Lian, T.; Singh, A.; Kosman, D.J.; Kronstad, J.W. Iron Source Preference and Regulation of Iron Uptake in Cryptococcus neoformans. PLoS Pathog. 2008, 4, e45. [Google Scholar] [CrossRef] [Green Version]
- Tangen, K.L.; Jung, W.H.; Sham, A.P.; Lian, T.; Kronstad, J.W. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 2007, 153, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, K.J.; Daveson, K.; Slavin, M.A.; van Hal, S.J.; Sorrell, T.C.; Lee, A.; Marriott, D.J.; Chapman, B.; Halliday, C.L.; Hajkowicz, K.; et al. Mucormycosis in Australia: Contemporary epidemiology and outcomes. Clin. Microbiol. Infect. 2016, 22, 775–781. [Google Scholar] [CrossRef] [Green Version]
- Chow, V.; Khan, S.; Balogun, A.; Mitchell, D.; Mühlschlegel, F.A. Invasive rhino-orbito-cerebral mucormycosis in a diabetic patient—The need for prompt treatment. Med. Mycol. Case Rep. 2015, 8, 5–9. [Google Scholar] [CrossRef]
- Boelaert, J.R.; De Locht, M.; Van Cutsem, J.; Kerrels, V.; Cantinieaux, B.; Verdonck, A.; Van Landuyt, H.W.; Schneider, Y.J. Mucormycosis during deferoxamine therapy is a siderophore-mediated infection: In vitro and in vivo animal studies. J. Clin. Investig. 1993, 91, 1979–1986. [Google Scholar] [CrossRef]
- Boelaert, J.R.; Van Cutsem, J.; De Locht, M.; Schneider, Y.J.; Crichton, R.R. Deferoxamine augments growth and pathogenicity of Rhizopus, while hydroxypyridinone chelators have no effect. Kidney Int. 1994, 45, 667–671. [Google Scholar] [CrossRef] [Green Version]
- Van Cutsem, J.; Boelaert, J.R. Effects of deferoxamine, feroxamine and iron on experimental mucormycosis (zygomycosis). Kidney Int. 1989, 36, 1061–1068. [Google Scholar] [CrossRef] [Green Version]
- Alastruey-Izquierdo, A.; Hoffmann, K.; De Hoog, G.S.; Rodriguez-Tudela, J.L.; Voigt, K.; Bibashi, E.; Walther, G. Species recognition and clinical relevance of the zygomycetous genus Lichtheimia (syn. Absidia pro parte, Mycocladus). J. Clin. Microbiol. 2010, 48, 2154–2170. [Google Scholar] [CrossRef] [Green Version]
- Schrettl, M.; Ibrahim-Granet, O.; Droin, S.; Huerre, M.; Latgé, J.P.; Haas, H. The crucial role of the Aspergillus fumigatus siderophore system in interaction with alveolar macrophages. Microbes Infect. 2010, 12, 1035–1041. [Google Scholar] [CrossRef]
- Hissen, A.H.T.; Wan, A.N.C.; Warwas, M.L.; Pinto, L.J.; Moore, M.M. The Aspergillus fumigatus Siderophore Biosynthetic Gene sidA, Encoding l-Ornithine N5-Oxygenase, Is Required for Virulence. Infect. Immun. 2005, 73, 5493–5503. [Google Scholar] [CrossRef] [Green Version]
- Schrettl, M.; Kim, H.S.; Eisendle, M.; Kragl, C.; Nierman, W.C.; Heinekamp, T.; Werner, E.R.; Jacobsen, I.; Illmer, P.; Yi, H.; et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008, 70, 27–43. [Google Scholar] [CrossRef] [Green Version]
- Burt, W.R. Identification of coprogen B and its breakdown products from Histoplasma capsulatum. Infect. Immun. 1982, 35, 990–996. [Google Scholar] [CrossRef] [Green Version]
- Silva-Bailão, M.G.; Bailão, E.F.L.C.; Lechner, B.E.; Gauthier, G.M.; Lindner, H.; Bailão, A.M.; Haas, H.; Soares, C.M.D.A. Hydroxamate production as a high affinity iron acquisition mechanism in Paracoccidioides spp. PLoS ONE 2014, 9, e105805. [Google Scholar] [CrossRef] [Green Version]
- Hwang, L.H.; Mayfield, J.A.; Rine, J.; Sil, A. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 2008, 4. [Google Scholar] [CrossRef] [Green Version]
- Beckmann, N.; Schafferer, L.; Schrettl, M.; Binder, U.; Talasz, H.; Lindner, H.; Haas, H. Characterization of the Link between Ornithine, Arginine, Polyamine and Siderophore Metabolism in Aspergillus fumigatus. PLoS ONE 2013, 8, e67426. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Kim, J.Y.; Yun, C.W. Identification of ferrichrome- and ferrioxamine B-mediated iron uptake by Aspergillus fumigatus. Biochem. J. 2016, 473, 1203–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, L.H.; Seth, E.; Gilmore, S.A.; Sil, A. SRE1 Regulates Iron-Dependent and -Independent Pathways in the Fungal Pathogen Histoplasma capsulatum. Eukaryot. Cell 2012, 11, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal Cell Wall: Emerging Antifungals and Drug Resistance. Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, R.A.; Gaffen, S.L.; Hise, A.G.; Brown, G.D. Innate defense against fungal pathogens. Cold Spring Harb. Perspect. Med. 2015, 5, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Gow, N.A.R.; Latge, J.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 3341–3354. [Google Scholar] [CrossRef] [Green Version]
- Arana, D.M.; Prieto, D.; Román, E.; Nombela, C.; Alonso-Monge, R.; Pla, J. The role of the cell wall in fungal pathogenesis. Microb. Biotechnol. 2009, 2, 308–320. [Google Scholar] [CrossRef] [Green Version]
- Bayry, J.; Beaussart, A.; Dufrêne, Y.F.; Sharma, M.; Bansal, K.; Kniemeyer, O.; Aimanianda, V.; Brakhage, A.A.; Kaveri, S.V.; Kwon-Chung, K.J.; et al. Surface structure characterization of Aspergillus fumigatus conidia mutated in the melanin synthesis pathway and their human cellular immune response. Infect. Immun. 2014, 82, 3141–3153. [Google Scholar] [CrossRef] [Green Version]
- Latgé, J.P.; Beauvais, A. Functional duality of the cell wall. Curr. Opin. Microbiol. 2014, 20, 111–117. [Google Scholar] [CrossRef]
- de Carrion, S.J.; Leal, S.M.; Ghannoum, M.A.; Aimanianda, V.; Latgé, J.-P.; Pearlman, E. The RodA Hydrophobin on Aspergillus fumigatus Spores Masks Dectin-1– and Dectin-2–Dependent Responses and Enhances Fungal Survival In Vivo. J. Immunol. 2013, 191, 2581–2588. [Google Scholar] [CrossRef] [Green Version]
- Casadevall, A.; Rosas, A.L.; Nosanchuk, J.D. Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2000, 3, 354–358. [Google Scholar] [CrossRef]
- Wheeler, R.T.; Fink, G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006, 2, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.T.; Kombe, D.; Agarwala, S.D.; Fink, G.R. Dynamic, morphotype-specific Candida albicans β-glucan exposure during infection and drug treatment. PLoS Pathog. 2008, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozza, S.; Clavaud, C.; Giovannini, G.; Fontaine, T.; Beauvais, A.; Sarfati, J.; D’Angelo, C.; Perruccio, K.; Bonifazi, P.; Zagarella, S.; et al. Immune Sensing of Aspergillus fumigatus Proteins, Glycolipids, and Polysaccharides and the Impact on Th Immunity and Vaccination. J. Immunol. 2009, 183, 2407–2414. [Google Scholar] [CrossRef] [Green Version]
- Ram, A.F.J.; Kapteyn, J.C.; Montijn, R.C.; Caro, L.H.P.; Douwes, J.E.; Baginsky, W.; Mazur, P.; van den Ende, H.; Klis, F.M. Loss of the Plasma Membrane-Bound Protein Gas1p in Saccharomyces cerevisiae Results in the Release of β1,3-Glucan into the Medium and Induces a Compensation Mechanism To Ensure Cell Wall Integrity. J. Bacteriol. 1998, 180, 1418–1424. [Google Scholar] [CrossRef] [Green Version]
- Cortés, J.C.G.; Curto, M.Á.; Carvalho, V.S.D.; Pérez, P.; Ribas, J.C. The fungal cell wall as a target for the development of new antifungal therapies. Biotechnol. Adv. 2019, 37, 107352. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Casadevall, A.; Nosanchuk, J.D.; Williamson, P.; Rodrigues, M.L. Vesicular transport across the fungal cell wall. Trends Microbiol. 2009, 17, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef]
- Lecointe, K.; Cornu, M.; Leroy, J.; Coulon, P.; Sendid, B. Polysaccharides cell wall architecture of mucorales. Front. Microbiol. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann. Transl. Med. 2014, 2, 1–14. [Google Scholar] [CrossRef]
- Masuoka, J. Surface Glycans of Candida albicans and Other Pathogenic Fungi: Physiological Roles, Clinical Uses, and Experimental Challenges. Clin. Microbiol. Rev. 2004, 17, 281–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauvais, A.; Bozza, S.; Kniemeyer, O.; Formosa, C.; Balloy, V.; Henry, C.; Roberson, R.W.; Dague, E.; Chignard, M.; Brakhage, A.A.; et al. Deletion of the α-(1,3)-Glucan Synthase Genes Induces a Restructuring of the Conidial Cell Wall Responsible for the Avirulence of Aspergillus fumigatus. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Mazur, P.; Baginsky, W. In Vitro Activity of 1,3-β-D-Glucan Synthase Requires the GTP-binding Protein Rho1. Biochemistry 1996, 271, 14604–14609. [Google Scholar]
- Aguilar-Zapata, D.; Petraitiene, R.; Petraitis, V. Echinocandins: The Expanding Antifungal Armamentarium. Clin. Infect. Dis. 2015, 61, S604–S611. [Google Scholar] [CrossRef] [Green Version]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Function and biosynthesis of cell wall α-1,3-glucan in fungi. J. Fungi 2017, 3, 63. [Google Scholar] [CrossRef]
- Perlin, D.S. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updates 2007, 10, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Lesage, G.; Bussey, H. Cell Wall Assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [Green Version]
- Latgé, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef]
- Chaudhary, P.M.; Tupe, S.G.; Deshpande, M.V. Chitin Synthase Inhibitors as Antifungal Agents. Mini-Rev. Med. Chem. 2013, 13, 222–236. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; San-Blas, G. Chitin synthesis as a target for antifungal drugs. Curr. Drug Targets-Infect. Disord. 2003, 3, 77–91. [Google Scholar] [CrossRef]
- Sorrell, T.C.; Chen, S.C.A. Fungal-derived immune modulating molecules. Adv. Exp. Med. Biol. 2009, 666, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Vecchiarelli, A. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 2000, 38, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Engel, J.; Schmalhorst, P.S.; Routier, F.H. Biosynthesis of the fungal cell wall polysaccharide galactomannan requires intraluminal GDP-mannose. J. Biol. Chem. 2012, 287, 44418–44424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Liu, L.; Tewari, R.P.; Williamson, P.R. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect. Immun. 1999, 67, 6034–6039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosanchuk, J.D.; Stark, R.E.; Casadevall, A. Fungal melanin: What do we know about structure? Front. Microbiol. 2015, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Doering, T.L. How Sweet it is! Cell Wall Biogenesis and Polysaccharide Capsule Formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247. [Google Scholar] [CrossRef] [Green Version]
- Casadevall, A. Determinants of virulence in the pathogenic fungi. Fungal Biol. Rev. 2007, 21, 130–132. [Google Scholar] [CrossRef] [Green Version]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Aisen, P.; Casadevall, A. Cryptococcus neoformans melanin and virulence: Mechanism of action. Infect. Immun. 1995, 63, 3131–3136. [Google Scholar] [CrossRef] [Green Version]
- Zaragoza, O. Basic principles of the virulence of Cryptococcus. Virulence 2019, 10, 490–501. [Google Scholar] [CrossRef] [Green Version]
- Barnes, R.A. Early diagnosis of fungal infection in immunocompromised patients. J. Antimicrob. Chemother. 2008, 61, 3–6. [Google Scholar] [CrossRef]
- De Pauw, B.; Walsh, T.J.; Donnelly, J.P.; Stevens, D.A.; Edwards, J.E.; Calandra, T.; Pappas, P.G.; Maertens, J.; Lortholary, O.; Kauffman, C.A.; et al. Revised Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer/Invasive. Clin. Infect. Dis 2008, 46, 1813–1821. [Google Scholar] [CrossRef]
- Husain, S.; Sole, A.; Alexander, B.D.; Aslam, S.; Avery, R.; Benden, C.; Billaud, E.M.; Chambers, D.; Danziger-Isakov, L.; Fedson, S.; et al. The 2015 International Society for Heart and Lung Transplantation Guidelines for the management of fungal infections in mechanical circulatory support and cardiothoracic organ transplant recipients: Executive summary. J. Hear. Lung Transplant. 2016, 35, 261–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formanek, P.E.; Dilling, D.F. Advances in the Diagnosis and Management of Invasive Fungal Disease. Chest 2019, 156, 834–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hof, H. IFI = invasive fungal infections. What is that? A misnomer, because a non-invasive fungal infection does not exist! Int. J. Infect. Dis. 2010, 14, e458–e459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlesse, F.; Daudt, L.E.; Seber, A.; Dutra, Á.P.; Melo, A.S.A.; Simões, B.P.; Macedo, C.R.D.; Bonfim, C.; Benites, E.; Gregianin, L.; et al. A consensus document for the clinical management of invasive fungal diseases in pediatric patients with hematologic cancer and/or undergoing hematopoietic stem cell transplantation in Brazilian medical centers. Braz. J. Infect. Dis. 2019, 23, 395–409. [Google Scholar] [CrossRef]
- Sipsas, N.V.; Pagoni, M.N.; Kofteridis, D.P.; Meletiadis, J.; Vrioni, G.; Papaioannou, M.; Antoniadou, A.; Petrikkos, G.; Samonis, G. Management of invasive fungal infections in adult patients with hematological malignancies in Greece during the financial crisis: Challenges and recommendations. J. Fungi 2018, 4, 94. [Google Scholar] [CrossRef] [Green Version]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef] [Green Version]
- Sipsas, N.V.; Gamaletsou, M.N.; Anastasopoulou, A.; Kontoyiannis, D.P. Therapy of mucormycosis. J. Fungi 2018, 4, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarrand, J.J.; Lichterfeld, M.; Warraich, I.; Luna, M.; Han, X.Y.; May, G.S.; Kontoyiannis, D.P. Diagnosis of invasive septate mold infections: A correlation of microbiological culture and histologic or cytologic examination. Am. J. Clin. Pathol. 2003, 119, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Cornely, O.A.; Arikan-Akdagli, S.; Dannaoui, E.; Groll, A.H.; Lagrou, K.; Chakrabarti, A.; Lanternier, F.; Pagano, L.; Skiada, A.; Akova, M.; et al. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of mucormycosis 2013. Clin. Microbiol. Infect. 2014, 20, 5–26. [Google Scholar] [CrossRef] [Green Version]
- Colombo, A.L.; de Almeida Júnior, J.N.; Slavin, M.A.; Chen, S.C.A.; Sorrell, T.C. Candida and invasive mould diseases in non-neutropenic critically ill patients and patients with haematological cancer. Lancet Infect. Dis. 2017, 17, e344–e356. [Google Scholar] [CrossRef]
- Bassetti, M.; Garnacho-Montero, J.; Calandra, T.; Kullberg, B.; Dimopoulos, G.; Azoulay, E.; Chakrabarti, A.; Kett, D.; Leon, C.; Ostrosky-Zeichner, L.; et al. Intensive care medicine research agenda on invasive fungal infection in critically ill patients. Intensiv. Care Med. 2017, 43, 1225–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanguinetti, M.; Posteraro, B.; Beigelman-Aubry, C.; Lamoth, F.; Dunet, V.; Slavin, M.; Richardson, M.D. Diagnosis and treatment of invasive fungal infections: Looking ahead. J. Antimicrob. Chemother. 2019, 74, II27–II37. [Google Scholar] [CrossRef] [Green Version]
- Lamoth, F.; Calandra, T. Early diagnosis of invasive mould infections and disease. J. Antimicrob. Chemother. 2017, 72, i19–i28. [Google Scholar] [CrossRef] [Green Version]
- Galgóczy, L. Molecular characterization of opportunistic pathogenic zygomycetes. Acta Biol. Szeged. 2005, 49, 1–7. [Google Scholar]
- Skiada, A.; Lass-Floerl, C.; Klimko, N.; Ibrahim, A.; Roilides, E.; Petrikkos, G. Challenges in the diagnosis and treatment of mucormycosis. Med. Mycol. 2018, 56, S93–S101. [Google Scholar] [CrossRef] [Green Version]
- Oz, Y.; Kiraz, N. Diagnostic methods for fungal infections in pediatric patients: Microbiological, serological and molecular methods. Expert Rev. Anti-Infect. Ther. 2011, 9, 289–298. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P.; Sumoza, D.; Tarrand, J.; Bodey, G.P.; Storey, R.; Raad, I.I. Significance of aspergillemia in patients with cancer: A 10-year study. Clin. Infect. Dis. 2000, 31, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Lowry, C.M.; Lempitski, S.J.; Kubiak, D.W.; Finkelman, M.A.; Baden, L.R. Reactivity of (1→3)-β-D-glucan assay with commonly used intravenous antimicrobials. Antimicrob. Agents Chemother. 2006, 50, 3450–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajpai, V.K.; Khan, I.; Shukla, S.; Kumar, P.; Rather, I.A.; Park, Y.H.; Huh, Y.S.; Han, Y.K. Invasive Fungal Infections and Their Epidemiology: Measures in the Clinical Scenario. Biotechnol. Bioprocess Eng. 2019, 24, 436–444. [Google Scholar] [CrossRef]
- Schwartz, S.; Kontoyiannis, D.P.; Harrison, T.; Ruhnke, M. Advances in the diagnosis and treatment of fungal infections of the CNS. Lancet Neurol. 2018, 17, 362–372. [Google Scholar] [CrossRef]
- Zheng, F.; Zha, H.; Yang, D.; Deng, J.; Zhang, Z. Diagnostic Values and Limitations of (1,3)-β-d-Glucans and Galactomannan Assays for Invasive Fungal Infection in Patients Admitted to Pediatric Intensive Care Unit. Mycopathologia 2017, 182, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Martínez, L.; Buitrago, M.J.; Castelli, M.V.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Development of a single tube multiplex real-time PCR to detect the most clinically relevant Mucormycetes species. Clin. Microbiol. Infect. 2013, 19, E1. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, P.; Cornely, O.A.; Dannaoui, E. Antifungal combinations in Mucorales: A microbiological perspective. Mycoses 2019, 62, 746–760. [Google Scholar] [CrossRef] [PubMed]
- White, P.L.; Alanio, A.; Cruciani, M.; Gorton, R.; Millon, L.; Rickerts, V.; Barnes, R.A.; Donnelly, J.P.; Loeffler, J. Nucleic Acid Tools for Invasive Fungal Disease Diagnosis. Curr. Fungal Infect. Rep. 2020, 14, 76–88. [Google Scholar] [CrossRef]
- Chong, G.L.M.; Van De Sande, W.W.J.; Dingemans, G.J.H.; Gaajetaan, G.R.; Vonk, A.G.; Hayette, M.P.; Van Tegelen, D.W.E.; Simons, G.F.M.; Rijnders, B.J.A. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J. Clin. Microbiol. 2015, 53, 868–874. [Google Scholar] [CrossRef] [Green Version]
- Ellis, M. Invasive fungal infections: Evolving challenges for diagnosis and therapeutics. Mol. Immunol. 2002, 38, 947–957. [Google Scholar] [CrossRef]
- Tissot, F.; Agrawal, S.; Pagano, L.; Petrikkos, G.; Groll, A.H.; Skiada, A.; Lass-Flörl, C.; Calandra, T.; Viscoli, C.; Herbrecht, R. ECIL-6 guidelines for the treatment of invasive candidiasis, aspergillosis and mucormycosis in leukemia and hematopoietic stem cell transplant patients. Haematologica 2017, 102, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Petrikkos, G.; Skiada, A.; Lortholary, O.; Roilides, E.; Walsh, T.J.; Kontoyiannis, D.P. Epidemiology and clinical manifestations of mucormycosis. Clin. Infect. Dis. 2012, 54, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Alastruey-Izquierdo, A.; Cuesta, I.; Walther, G.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Antifungal susceptibility profile of human-pathogenic species of Lichtheimia. Antimicrob. Agents Chemother. 2010, 54, 3058–3060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquez, L.; Quave, C.L. Prevalence and Therapeutic Challenges of Fungal Drug Resistance: Role for Plants in Drug Discovery. Antibiotics 2020, 9, 150. [Google Scholar] [CrossRef] [Green Version]
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.-H.; et al. Clinical Practice Guidelines for the Management of Cryptococcal Disease: 2010 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 291–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourad, A.; Perfect, J. Present and Future Therapy of Cryptococcus Infections. J. Fungi 2018, 4, 79. [Google Scholar] [CrossRef] [Green Version]
- Marty, F.M.; Ostrosky-Zeichner, L.; Cornely, O.A.; Mullane, K.M.; Perfect, J.R.; Thompson, G.R.; Alangaden, G.J.; Brown, J.M.; Fredricks, D.N.; Heinz, W.J.; et al. Isavuconazole treatment for mucormycosis: A single-arm open-label trial and case-control analysis. Lancet Infect. Dis. 2016, 16, 828–837. [Google Scholar] [CrossRef]
- Walsh, T.J.; Lutsar, I.; Driscoll, T.; Dupont, B.; Roden, M.; Ghahramani, P.; Hodges, M.; Groll, A.H.; Perfect, J.R. Voriconazole in the treatment of Aspergillosis, Scedosporiosis and other invasive fungal infections in children. Pediatr. Infect. Dis. J. 2002, 21, 240–248. [Google Scholar] [CrossRef]
- Shoham, S.; Magill, S.S.; Merz, W.G.; Gonzalez, C.; Seibel, N.; Buchanan, W.L.; Knudsen, T.A.; Sarkisova, T.A.; Walsh, T.J. Primary treatment of zygomycosis with liposomal amphotericin B: Analysis of 28 cases. Med. Mycol. 2010, 48, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Lax, C.; Pérez-Arques, C.; Navarro-Mendoza, M.I.; Cánovas-Márquez, J.T.; Tahiri, G.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Murcia-Flores, L.; Navarro, E.; Garre, V.; et al. Genes, Pathways, and Mechanisms Involved in the Virulence of Mucorales. Genes (Basel) 2020, 11, 317. [Google Scholar] [CrossRef] [Green Version]
- Perkhofer, S.; Lechner, V.; Lass-Flörl, C. In Vitro Activity of Isavuconazole against Aspergillus Species and Zygomycetes According to the Methodology of the European Committee on Antimicrobial Susceptibility Testing. Antimicrob. Agents Chemother. 2009, 53, 1645–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagano, L.; Caira, M.; Valentini, C.G.; Posteraro, B.; Fianchi, L. Current therapeutic approaches to fungal infections in immunocompromised hematological patients. Blood Rev. 2010, 24, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Drew, R. Potential role of aerosolized amphotericin B formulations in the prevention and adjunctive treatment of invasive fungal infections. Int. J. Antimicrob. Agents 2006, 27, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Butts, A.; Palmer, G.E.; David Rogers, P. Antifungal adjuvants: Preserving and extending the antifungal arsenal. Virulence 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, A.R.; Cardno, T.S.; Strouse, J.J.; Ivnitski-Steele, I.; Keniya, M.V.; Lackovic, K.; Monk, B.C.; Sklar, L.A.; Cannon, R.D. Targeting efflux pumps to overcome antifungal drug resistance. Futur. Med. Chem. 2016, 8, 1485–1501. [Google Scholar] [CrossRef] [Green Version]
- Zarember, K.A.; Cruz, A.R.; Huang, C.-Y.; Gallin, J.I. Antifungal Activities of Natural and Synthetic Iron Chelators Alone and in Combination with Azole and Polyene Antibiotics against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2009, 53, 2654–2656. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 2013, 57, 146–154. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, B.E.; Oltean, H.N.; Oliver, B.G.; Hoot, S.J.; Leyde, S.E.; Hedstrom, L.; White, T.C. Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 2010, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Prasad, T.; Chandra, A.; Mukhopadhyay, C.K.; Prasad, R. Unexpected Link between Iron and Drug Resistance of Candida spp.: Iron Depletion Enhances Membrane Fluidity and Drug Diffusion, Leading to Drug-Susceptible Cells. Antimicrob. Agents Chemother. 2006, 50, 3597–3606. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Gebermariam, T.; Fu, Y.; Lin, L.; Husseiny, M.I.; French, S.W.; Schwartz, J.; Skory, C.D.; Edwards, J.E.; Spellberg, B.J. The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J. Clin. Investig. 2007, 117, 2649–2657. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Gebremariam, T.; Luo, G.; Fu, Y.; French, S.W.; Edwards, J.E.; Spellberg, B. Combination Therapy of Murine Mucormycosis or Aspergillosis with Iron Chelation, Polyenes, and Echinocandins. Antimicrob. Agents Chemother. 2011, 55, 1768–1770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellberg, B.; Ibrahim, A.S.; Chin-Hong, P.V.; Kontoyiannis, D.P.; Morris, M.I.; Perfect, J.R.; Fredricks, D.; Brass, E.P. The Deferasirox–AmBisome Therapy for Mucormycosis (DEFEAT Mucor) study: A randomized, double-blinded, placebo-controlled trial. J. Antimicrob. Chemother. 2012, 67, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, J.P.; Lahav, M. Deferasirox as adjunctive therapy for mucormycosis. J. Antimicrob. Chemother. 2012, 67, 519–520. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Gebremariam, T.; French, S.W.; Edwards, J.E.; Spellberg, B. The iron chelator deferasirox enhances liposomal amphotericin B efficacy in treating murine invasive pulmonary aspergillosis. J. Antimicrob. Chemother. 2009, 65, 289–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, M.I.A.; Voigt, K. Pathogenicity patterns of mucormycosis: Epidemiology, interaction with immune cells and virulence factors. Med. Mycol. 2019, 57, S245–S256. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Spellberg, B.; Phan, Q.T.; Fu, Y.Y.; Fu, Y.Y.; Lee, A.S.; Edwards, J.E.; Filler, S.G.; Ibrahim, A.S. The endothelial cell receptor GRP78 is required for mucormycosis pathogenesis in diabetic mice. J. Clin. Investig. 2010, 120, 1914–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denning, D.W. Echinocandin antifungal drugs. Lancet 2003, 362, 1142–1151. [Google Scholar] [CrossRef]
- Sucher, A.J.; Chahine, E.B.; Balcer, H.E. Echinocandins: The newest class of antifungals. Ann. Pharmacother. 2009, 43, 1647–1657. [Google Scholar] [CrossRef]
- Perlin, D.S. Echinocandin Resistance in Candida. Clin. Infect. Dis. 2015, 61, S612–S617. [Google Scholar] [CrossRef] [Green Version]
- Onishi, J.; Meinz, M.; Thompson, J.; Curotto, J.; Dreikorn, S.; Rosenbach, M.; Douglas, C.; Abruzzo, G.; Flattery, A.; Kong, L.; et al. Discovery of Novel Antifungal (1,3)-β-d-Glucan Synthase Inhibitors. Antimicrob. Agents Chemother. 2000, 44, 368–377. [Google Scholar] [CrossRef] [Green Version]
- Hori, Y.; Shibuya, K. Role of FKS Gene in the Susceptibility of Pathogenic Fungi to Echinocandins. Med. Mycol. J. 2018, 59, E31–E40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.C.; Hicks, P.S.; Kurtz, M.B.; Rosen, H.; Schmatz, D.M.; Liberator, P.A.; Douglas, C.M. The antifungal echinocandin caspofungin acetate kills growing cells of Aspergillus fumigatus in vitro. Antimicrob. Agents Chemother. 2002, 46, 3001–3012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, S.P.; VandeWalle, K.; Ramage, G.; Patterson, T.F.; Wickes, B.L.; Graybill, J.R.; López-Ribot, J.L. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob. Agents Chemother. 2002, 46, 3591–3596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, D.M.; George, T.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Antifungal Susceptibility of Candida Biofilms: Unique Efficacy of Amphotericin B Lipid Formulations and Echinocandins. Antimicrob. Agents Chemother. 2002, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, C.; Graninger, W.; Presterl, E.; Joukhadar, C. The echinocandins: Comparison of their pharmacokinetics, pharmacodynamics and clinical applications. Pharmacology 2006, 78, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Samanta, P.; Cheng, S.; Squires, K.; Nguyen, M.-H. 1726. Candida albicans Virulence Genes Induced During Intra-abdominal Candidiasis (IAC) in the Absence of Antifungal Exposure Mediate Echinocandin Resistance. Open Forum Infect. Dis. 2019, 6, S633. [Google Scholar] [CrossRef]
- Lee, K.K.; MacCallum, D.M.; Jacobsen, M.D.; Walker, L.A.; Odds, F.C.; Gow, N.A.R.; Munro, C.A. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 2012, 56, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Scorneaux, B.; Angulo, D.; Borroto-Esoda, K.; Ghannoum, M.; Peel, M.; Wring, S. SCY-078 Is Fungicidal against Candida Species in Time-Kill Studies. Antimicrob. Agents Chemother. 2017, 61, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Larkin, E.L.; Long, L.; Isham, N.; Borroto-Esoda, K.; Barat, S.; Angulo, D.; Wring, S.; Ghannoum, M. A novel 1,3-beta-D-glucan inhibitor, IbrexafungeRP (formerly SCY-078), shows potent activity in the lower pH environment of vulvovaginitis. Antimicrob. Agents Chemother. 2019, 63, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Seo, K.; Akiyoshi, H.; Ohnishi, Y. Alteration of cell wall composition leads to amphotericin B resistance in Aspergillus flavus. Microbiol. Immunol. 1999, 43, 1017–1025. [Google Scholar] [CrossRef]
- Mesa-Arango, A.C.; Rueda, C.; Román, E.; Quintin, J.; Terrón, M.C.; Luque, D.; Netea, M.G.; Pla, J.; Zaragoza, O. Cell Wall Changes in Amphotericin B-Resistant Strains from Candida tropicalis and Relationship with the Immune Responses Elicited by the Host. Antimicrob. Agents Chemother. 2016, 60, 2326–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kontoyiannis, D.P.; Lewis, R.E. Antifungal drug resistance of pathogenic fungi. Lancet 2002, 359, 1135–1144. [Google Scholar] [CrossRef]
- Sheehan, D.J.; Hitchcock, C.A.; Sibley, C.M. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 1999, 12, 40–79. [Google Scholar] [CrossRef] [Green Version]
- Mast, N.; Zheng, W.; Stout, C.D.; Pikuleva, I.A. Antifungal azoles: Structural insights into undesired tight binding to cholesterol-metabolizing cyp46a1s. Mol. Pharmacol. 2013, 84, 86–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perea, S.; Patterson, T.F. Antifungal Resistance in Pathogenic Fungi. Clin. Infect. Dis. 2002, 35, 1073–1080. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.; Chazli, Y.E.; Babu, A.F.; Coste, A.T. Azole resistance in Aspergillus fumigatus: A consequence of antifungal use in agriculture? Front. Microbiol. 2017, 8, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warrilow, A.G.; Nishimoto, A.T.; Parker, J.E.; Price, C.L.; Flowers, S.A.; Kelly, D.E.; David Rogers, P.; Kelly, S.L. The evolution of Azole resistance in Candida albicans Sterol 14-demethylase (CYP51) through incremental amino acid substitutions. Antimicrob. Agents Chemother. 2019, 63, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Caramalho, R.; Tyndall, J.D.A.; Monk, B.C.; Larentis, T.; Lass-Flörl, C.; Lackner, M. Intrinsic short-Tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci. Rep. 2017, 7, 3–12. [Google Scholar] [CrossRef]
- Vale-Silva, L.A.; Coste, A.T.; Ischer, F.; Parker, J.E.; Kelly, S.L.; Pinto, E.; Sanglard, D. Azole Resistance by Loss of Function of the Sterol Δ 5,6 -Desaturase Gene (ERG3) in Candida albicans Does Not Necessarily Decrease Virulence. Antimicrob. Agents Chemother. 2012, 56, 1960–1968. [Google Scholar] [CrossRef] [Green Version]
- Florio, A.; Ferrari, S.; De Carolis, E.; Torelli, R.; Fadda, G.; Sanguinetti, M.; Sanglard, D.; Posteraro, B. Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol. 2011, 11, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martel, C.M.; Parker, J.E.; Warrilow, A.G.S.; Rolley, N.J.; Kelly, S.L.; Kelly, D.E. Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14α-demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 2010, 54, 4920–4923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Q.Z.; Yan, L.; Jiang, Y.Y. The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 2016, 7, 649–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces cerevisiae. MBio 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hameed, S.; Dhamgaye, S.; Singh, A.; Goswami, S.K.; Prasad, R. Calcineurin Signaling and Membrane Lipid Homeostasis Regulates Iron Mediated MultiDrug Resistance Mechanisms in Candida albicans. PLoS ONE 2011, 6, e18684. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, C.J.; Schneider, S.; Barker, K.S.; Rogers, P.D.; Morschhäuser, J. An A643T Mutation in the Transcription Factor Upc2p Causes Constitutive ERG11 Upregulation and Increased Fluconazole Resistance in Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Riemsma, R.; Hagen, S.; Kirschner-Hermanns, R.; Norton, C.; Wijk, H.; Andersson, K.E.; Chapple, C.; Spinks, J.; Wagg, A.; Hutt, E.; et al. Can incontinence be cured? A systematic review of cure rates. BMC Med. 2017, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Misslinger, M.; Gsaller, F.; Hortschansky, P.; Müller, C.; Bracher, F.; Bromley, M.J.; Haas, H. The cytochrome: B 5 CybE is regulated by iron availability and is crucial for azole resistance in A. fumigatus. Metallomics 2017, 9, 1655–1665. [Google Scholar] [CrossRef] [Green Version]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Dolan, M.A.; Ghannoum, M.A.; Kwon-Chung, K.J. Identification of a Cryptococcus neoformans Cytochrome P450 Lanosterol 14α-Demethylase (Erg11) Residue Critical for Differential Susceptibility between Fluconazole/Voriconazole and Itraconazole/Posaconazole. Antimicrob. Agents Chemother. 2012, 56, 1162–1169. [Google Scholar] [CrossRef] [Green Version]
- Vitale, R.G.; De Hoog, G.S.; Schwarz, P.; Dannaoui, E.; Deng, S.; Machouart, M.; Voigt, K.; Van De Sande, W.W.J.J.; Dolatabadi, S.; Meis, J.F.; et al. Antifungal susceptibility and phylogeny of opportunistic members of the order Mucorales. J. Clin. Microbiol. 2012, 50, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Alastruey-Izquierdo, A.; Castelli, M.V.; Cuesta, I.; Zaragoza, O.; Monzón, A.; Mellado, E.; Rodríguez-Tudela, J.L. In vitro activity of antifungals against zygomycetes. Clin. Microbiol. Infect. 2009, 15, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritz, N.; Ammann, R.A.; Aebischer, C.C.; Gugger, M.; Jaton, K.; Schmid, R.A.; Aebi, C. Failure of voriconazole to cure disseminated zygomycosis in an immunocompromised child. Eur. J. Pediatr. 2005, 164, 231–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almyroudis, N.G.; Sutton, D.A.; Fothergill, A.W.; Rinaldi, M.G.; Kusne, S. In vitro susceptibilities of 217 clinical isolates of zygomycetes to conventional and new antifungal agents. Antimicrob. Agents Chemother. 2007, 51, 2587–2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
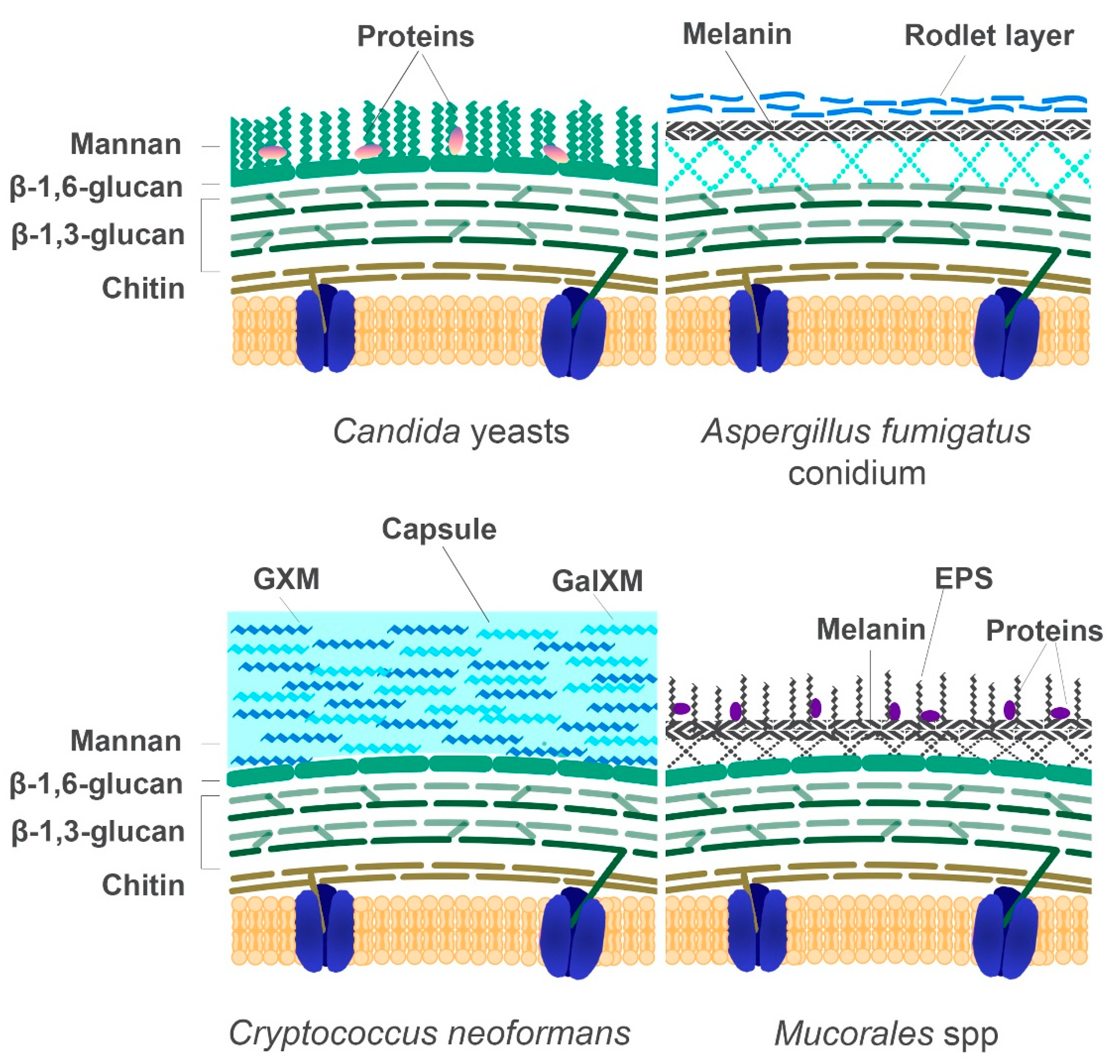
| Component | Species | Gene | Functions | Ref |
|---|---|---|---|---|
| Ferric reductases | Saccharomyces cerevisiae | FRE1, FRE2 | Ferric iron reduction at the cell surface | [3,36,65] |
| Rhizopus spp. | FRE (homolog) | Putative protein—ferric iron reduction at the cell surface | [48,66] | |
| Mucor circinelloides | FRE (homolog) | Putative protein—ferric iron reduction at the cell surface | [47,67] | |
| Lichtheimia corymbifera | FRE5 (homolog)–three copies | Putative protein—ferric iron reduction at the cell surface | [49] | |
| Multicopper ferroxidase | S. cerevisiae | FET3 | Multicopper-oxidase Ferrous iron oxidation and high-affinity uptake coupled with Ftr1 (permease) | [3,55,65,68] |
| Rhizopus spp. | FET3 homolog | Putative multicopper oxidase | [48] | |
| M. circinelloides | FETA, FETB, FETC | Ferrous iron oxidation and high-affinity iron uptake | [47] | |
| L. corymbifera | FET3/5 homolog–three copies | Putative multicopper oxidase | [49] | |
| Iron permease | S. cerevisiae | FTR1 | High-affinity iron uptake, coupled with FET3 (multicopper oxidase) | [3,59,68,69,70] |
| Rhizopus spp. | FTR1 | High affinity iron permease | [65,71,72] | |
| M. circinelloides | FTR1 (homolog) | Putative iron permease | [47,73] | |
| L. corymbifera | FTR1 (homolog)—four copies | Putative iron permease | [49] |
| Organism | Transporter | Function | Siderophore Substrate | Publication |
|---|---|---|---|---|
| S. cerevisiae | Arn1 | Ferrichrome and Ferrichrome A transporter | Ferrichrome and Ferrichrome A | [3,103,118,119,120] |
| Arn2/Taf1p | Triacetylfusarinine C (TAFC) transporter | TAFC | [3,118,119] | |
| Arn3/Sit1p | Ferrichrome and Ferrichrome A transporter | Ferrioxamine B, Ferrichrome A, Ferrichromes, Ferricrocin, Ferrichrycin, Ferrirhodin and Ferrirubin | [3,118,119] | |
| Arn4p/Enb1p | Enterobactin transporter | Enterobactin | [3,118,119,121] | |
| R.arrhizus (syn. R. oryzae, R. delemar) | Fob1, Fob2 | Ferrioxamine binding | Ferrioxamine B | [48,71] |
| L. corymbifera | Fob1 (putative protein) | Ferrioxamine binding | Ferrioxamine B | [49] |
| Method | Organism | Comment | Publications | |
|---|---|---|---|---|
| Microscopy | Direct histology and cytology | Candida spp.; Cryptococcus spp.; Aspergillus spp.; Mucorales | Gold standard, demonstration of tissue invasion | [199,205] |
| Cultures | Mycological culture | Cryptococcus spp. Candida spp.; Aspergillus spp.; Mucorales | Slow turn-around time | [206,207,208,209,210] |
| Blood cultures | Candida spp.; A. fumigatus, A. terreus; | Gold standard for candidemia; | [211,212] | |
| Serological methods | 1,3-β-D-glucan (BDG) * | Candida spp.; Aspergillus spp. | Exceptions: Mucorales and Cryptococcus spp. | [195,197,199,205,213,214,215] |
| Galactomannan (GM) enzyme immunoassay * | Aspergillus spp. | [216] | ||
| Molecular approaches | PCR (18s rDNA, 28s rDNA, ITS, mtDNA | Candida spp.; Cryptococcus spp.; Aspergillus spp.; Mucorales | - | [217,218,219,220] |
| Imaging technologies | X-rays, CT and CTPA | Aspergillus spp.; Mucorales | - | [218,221] |
| MRI and PET scan | Cryptococcus spp.; Aspergillus spp.; Mucorales | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanford, F.A.; Voigt, K. Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance. Genes 2020, 11, 1296. https://doi.org/10.3390/genes11111296
Stanford FA, Voigt K. Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance. Genes. 2020; 11(11):1296. https://doi.org/10.3390/genes11111296
Chicago/Turabian StyleStanford, Felicia Adelina, and Kerstin Voigt. 2020. "Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance" Genes 11, no. 11: 1296. https://doi.org/10.3390/genes11111296
APA StyleStanford, F. A., & Voigt, K. (2020). Iron Assimilation during Emerging Infections Caused by Opportunistic Fungi with emphasis on Mucorales and the Development of Antifungal Resistance. Genes, 11(11), 1296. https://doi.org/10.3390/genes11111296






