Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Description of the Greenhouse Trials
2.3. Phenotyping
2.4. Genotyping and Phylogenetic Analysis
2.5. Sequencing
3. Results
3.1. Description of the Core Collection
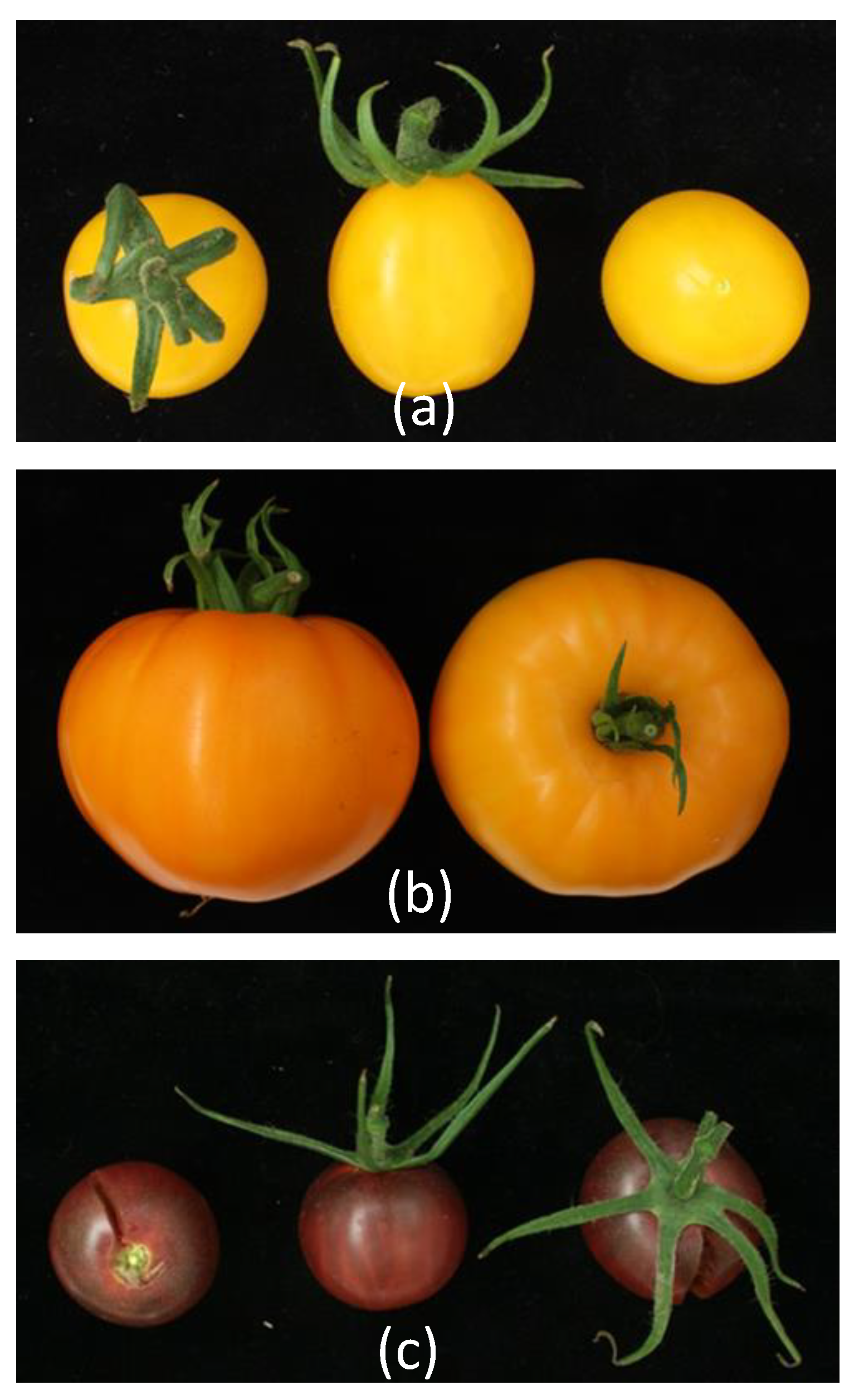
3.2. Phenotyping of the Core Collection
3.3. Plant Architecture Traits
3.4. Plant Growth Traits
3.5. Yield-related Traits
3.6. Fruit Quality Traits
3.7. Genotyping for Known Mutations or Variants Affecting Plant Architecture, Fruit Size or Shape, or Fruit Colour
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bai, Y.; Lindhout, P. Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef]
- Foolad, M.R.; Merk, H.L.; Ashrafi, H. Genetics, genomics and breeding of late blight and early blight resistance in tomato. CRC Crit. Rev. Plant Sci. 2008, 27, 75–107. [Google Scholar] [CrossRef]
- Rick, C.M.; Chetelat, R.T. Utilization of wild species for tomato improvement. Acta Hortic. 1995, 412, 21–38. [Google Scholar] [CrossRef]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J.; Francis, D.; Causse, M.; van der Knaap, E.; Cañizares, J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [Green Version]
- Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N.; Mao, L.; et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014, 80, 136–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippman, Z.; Tanksley, S.D. Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [PubMed]
- Van der Knaap, E.; Tanksley, S.D. The making of a bell pepper-shaped tomato fruit: Identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor. Appl. Genet. 2003, 107, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Grandillo, S.; Ku, H.M.; Tanksley, S.D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 1999, 99, 978–987. [Google Scholar] [CrossRef]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16 (Suppl. S1), S181–S189. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Muños, S.; Ranc, N.; Botton, E.; Bérard, A.; Rolland, S.; Duffé, P.; Carretero, Y.; Le Paslier, M.-C.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, H.M.; Doganlar, S.; Chen, K.Y.; Tanksley, S.D. The genetic basis of pear-shaped tomato fruit. Theor. Appl. Genet. 1999, 99, 844–850. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef]
- Ku, H.M.; Grandillo, S.; Tanksley, S.D. fs8.1, a major QTL, sets the pattern of tomato carpel shape well before anthesis. Theor. Appl. Genet. 2000, 101, 873–878. [Google Scholar] [CrossRef]
- Sun, L.; Rodriguez, G.R.; Clevenger, J.P.; Illa-Berenguer, E.; Lin, J.; Blakeslee, J.J.; Liu, W.; Fei, Z.; Wijeratne, A.; Meulia, T.; et al. Candidate gene selection and detailed morphological evaluations of fs8.1, a quantitative trait locus controlling tomato fruit shape. J. Exp. Bot. 2015, 66, 6471–6482. [Google Scholar] [CrossRef] [Green Version]
- Fray, R.G.; Grierson, D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 1993, 22, 589–602. [Google Scholar] [CrossRef]
- Tomes, M.L. Flower color modification associated with the gene t. Rep. Tomato Genet. Coop. 1952, 2, 12. [Google Scholar]
- Kerr, E. Green flesh, gf. Rep. Tomato Genet. Coop. 1956, 6, 17. [Google Scholar]
- Thompson, A.E.; Tomes, M.L.; Wann, E.V.; McCollum, J.; Stoner, A.K. Characterization of crimson tomato fruit color. J. Am. Soc. Hortic. Sci. 1965, 86, 610–616. [Google Scholar]
- Lindstrom, W. Inheritance in tomatoes. Genetics 1925, 10, 305–317. [Google Scholar] [PubMed]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Causse, M.; Desplat, N.; Pascual, L.; Le Paslier, M.-C.; Sauvage, C.; Bauchet, G.; Bérard, A.; Bounon, R.; Tchoumakov, M.; Brunel, D.; et al. Whole genome resequencing in tomato reveals variation associated with introgression and breeding events. BMC Genom. 2013, 14, 791. [Google Scholar] [CrossRef] [Green Version]
- Finkers, R.; Causse, M.; Giuliano, G.; Klein Lankhorst, R.; Zamir, D. SolCap Genotyping of Approx. 400 Tomato and Tomato Crop Wild Relative Genotypes. Available online: https://zenodo.org/record/2385441 (accessed on 28 October 2020).
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 4. [Google Scholar]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, S.-C.; Van Deynze, A.; Stoffel, K.; Douches, D.S.; Zarka, D.; Ganal, M.W.; Chetelat, R.T.; Hutton, S.F.; Scott, J.W.; Gardner, R.G.; et al. High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS ONE 2012, 7, e45520. [Google Scholar] [CrossRef] [Green Version]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL. Development 1998, 125, 1979–1989. [Google Scholar]
- Powell, A.L.T.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernández-Muñoz, R.; Vicente, A.; et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef] [Green Version]
- Lippman, Z.B.; Cohen, O.; Alvarez, J.P.; Abu-Abied, M.; Pekker, I.; Paran, I.; Eshed, Y.; Zamir, D. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008, 6, e288. [Google Scholar] [CrossRef] [PubMed]
- Busch, B.L.; Schmitz, G.; Rossmann, S.; Piron, F.; Ding, J.; Bendahmane, A.; Theres, K. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 2011, 23, 3595–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domínguez, M.; Dugas, E.; Benchouaia, M.; Leduque, B.; Jiménez-Gómez, J.M.; Colot, V.; Quadrana, L. The impact of transposable elements on tomato diversity. Nat. Commun. 2020, 11, 4058. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 2017, 169, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roldan, M.V.G.; Périlleux, C.; Morin, H.; Huerga-Fernandez, S.; Latrasse, D.; Benhamed, M.; Bendahmane, A. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 2017, 7, 4402. [Google Scholar] [CrossRef] [PubMed]
- Frary, A.A.; Nesbitt, T.C.; Grandillo, S.; Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; Tanksley, S.D. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Q.; Huang, Z.; Chakrabarti, M.; Illa-Berenguer, E.; Liu, X.; Wang, Y.; Ramos, A.; van der Knaap, E. Fruit weight is controlled by Cell Size Regulator encoding a novel protein that is expressed in maturing tomato fruits. PLoS Genet. 2017, 13, e1006930. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M.; et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Isaacson, T.; Ronen, G.; Zamir, D.; Hirschberg, J. Cloning of tangerine from tomato reveals a Carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 2002, 14, 333–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronen, G.; Carmel-Goren, L.; Zamir, D.; Hirschberg, J. An alternative pathway to ß-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. USA 2000, 97, 11102–11107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, C.S.; Pandey, P. A survey of cultivated heirloom tomato varieties identifies four new mutant alleles at the green-flesh locus. Mol. Breed. 2009, 24, 269–276. [Google Scholar] [CrossRef]
- Ballester, A.R.; Molthoff, J.; de Vos, R.; Hekkert, B.L.; Orzaez, D.; Fernández-Moreno, J.P.; Tripodi, P.; Grandillo, S.; Martin, C.; Heldens, J.; et al. Biochemical and molecular analysis of pink tomatoes: Deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 2010, 152, 71–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tigchelaar, E.; Tomes, M.; Kerr, E.; Barman, R. A new fruit ripening mutant, non-ripening (nor). Rep. Tomato Genet. Coop. 1973, 23, 33–34. [Google Scholar]
- Wang, R.; da Rocha Tavano, E.C.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef]
- Fridman, E.; Liu, Y.S.; Carmel-Goren, L.; Gur, A.; Shoresh, M.; Pleban, T.; Eshed, Y.; Zamir, D. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genom. 2001, 266, 821–826. [Google Scholar] [CrossRef]
- Sahu, K.K.; Chattopadhyay, D. Genome-wide sequence variations between wild and cultivated tomato species revisited by whole genome sequence mapping. BMC Genom. 2017, 18, 430. [Google Scholar] [CrossRef] [Green Version]
- Rowland, S.D.; Zumstein, K.; Nakayama, H.; Cheng, Z.; Flores, A.M.; Chitwood, D.H.; Maloof, J.N.; Sinha, N.R. Leaf shape is a predictor of fruit quality and cultivar performance in tomato. New Phytol. 2020, 226, 851–865. [Google Scholar] [CrossRef] [Green Version]
- Gonzalo, M.J.; Van Der Knaap, E. A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape. Theor. Appl. Genet. 2008, 116, 647–656. [Google Scholar] [CrossRef]
- Brewer, M.T.; Moyseenko, J.B.; Monforte, A.J.; van der Knaap, E. Morphological variation in tomato: A comprehensive study of quantitative trait loci controlling fruit shape and development. J. Exp. Bot. 2007, 58, 1339–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonge, M.; Wang, X.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 2020, 182, 145–161.e23. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.H.; Jang, J.C.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 2019, 3, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G.; Bartley, G.E.; Scolnik, P.A. Regulation of carotenoid biosynthesis during tomato development. Plant Cell 1993, 5, 379–387. [Google Scholar] [PubMed] [Green Version]
- Bramley, P.M. Carotenoid Biosynthesis and Chlorophyll Degradation. In The Molecular Biology and Biochemistry of Fruit Ripening; Seymour, G., Mervin Poole, M., Giovannoni, J., Tucker, G.A., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2013; pp. 75–116. [Google Scholar]
- Fernandez-Moreno, J.-P.; Tzfadia, O.; Forment, J.; Presa, S.; Rogachev, I.; Meir, S.; Orzaez, D.; Aharoni, A.; Granell, A. Characterization of a new pink-fruited tomato mutant results in the identification of a null allele of the SlMYB12 transcription factor. Plant Physiol. 2016, 171, 1821–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Origin | Type | Number of Accessions | Phenotyped | Sequenced Genomes for Genotyping |
|---|---|---|---|---|
| 150 genome project | Cultivated | 52 | 50 | 52 |
| Wild | 32 | 19 | ||
| This Study | Cultivated | 38 | 38 | 14 |
| Reference (Heinz 1706) | Cultivated | 1 | ||
| Total | 122 | 107 | 67 |
| Accession | ID | Name | fas | fw2.2 | lc | SUN | fw3.2 | ovate | fw11.3 | c | gf | nor | ogc | r | s | sp | t3183 | u | ug | y | j-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RF_001 | LA2706 | Moneymaker | - | + | - | - | + | - | + | - | - | - | - | - | - | - | - | + | - | - | - |
| RF_002 | LA2838A | Ailsa Craig | - | + | - | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_003 | PI406760 | Gardeners Delight | - | + | p | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_004 | LA1090 | Rutgers | - | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_005 | - | Galina (Galina’s yellow) | - | + | - | - | - | - | + | + | - | - | - | + | - | - | - | - | - | - | - |
| RF_006 | - | Ponderosa | + | + | + | - | + | - | + | - | - | - | - | - | - | - | +/- | - | - | - | - |
| RF_007 | - | Katinka Cherry | - | + | p | - | + | - | - | - | - | - | - | - | - | - | + | + | - | - | - |
| RF_008 | - | John’s big orange | - | + | + | - | + | - | + | - | - | - | - | - | - | + | + | + | - | - | - |
| RF_011 | LA2463 | All Round | - | + | - | - | + | - | + | - | - | - | - | - | - | - | - | + | - | - | - |
| RF_012 | LYC 1969 | Sonato | - | + | - | - | + | - | + | - | - | - | - | - | - | - | - | + | - | - | - |
| RF_013 | LYC 3897 | Cross Country | - | + | - | - | - | + | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_014 | LYC 3476 | Lidi | - | + | - | - | + | + | -* | - | - | - | - | + | + | - | - | - | - | - | - |
| RF_015 | - | Momatero | - | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_016 | CGN15464 | Rote Beere | - | p/+ | p | - | - | - | -* | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_017 | LYC 3340 | “Lycopersicon esculentum Mill.” | - | + | p | - | - | - | -* | - | - | - | - | +/- | - | - | - | - | - | - | - |
| RF_018 | - | DANA | + | + | +/- | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_019 | - | Large Pink | + | + | + | + | + | - | + | + | - | - | - | - | - | - | - | - | - | + | - |
| RF_020 | TLYC 3153 | “L. esculentum Mill.” | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_021 | T 828 | Bolivar | + | + | + | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_022 | PI 129097 | “L. esculentum” | - | + | x/- | + | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_023 | PI 272654 | “L. esculentum” | - | x | - | - | - | - | -* | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_024 | - | Jersey Devil | - | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_026 | - | Polish Joe | + | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_027 | CGN20815 | Cal J TM VF | - | + | - | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | + |
| RF_028 | PI 303721 | The Dutchman | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | + | - | + | - |
| RF_029 | LA4451 | Black Cherry | - | p | p | - | + | - | - | - | + | - | + | - | - | - | - | - | - | + | - |
| RF_030 | V710092 | ANTO | + | + | - | - | + | + | + | - | - | - | - | - | - | - | - | - | + | - | - |
| RF_031 | PC711092 | Winter Tipe (nor) | - | + | - | - | - | - | + | - | - | + | - | - | - | - | - | - | - | - | - |
| RF_032 | PI 93302 | Chang Li | - | + | - | - | - | + | -* | - | - | - | - | + | - | - | - | - | - | - | - |
| RF_033 | SG 16 | Belmonte | + | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_034 | - | Tiffen mennonite | + | + | + | + | + | - | + | + | - | - | - | - | - | - | - | - | - | + | - |
| RF_035 | PI 203232 | Wheatley’s Frost Resistant | - | + | - | - | - | + | -* | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_036 | PI 311117 | “L. esculentum” | - | x | - | + | - | + | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_037 | LA1324 | “L. esculentum” | - | p | p | - | - | - | -/+ | - | - | - | - | - | - | - | - | + | - | - | - |
| RF_038 | PI 158760 | Chih-Mu-Tao-Se | - | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_039 | LA0113 | “L. esculentum” | - | p- | + | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_040 | LYC 1410 | ES 58 Heinz | - | + | + | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_041 | PI 169588 | Dolmalik | + | + | + | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_042 | LYC 2962 | Ventura | - | p | p | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_077 | - | Large Red Cherry | - | + | - | - | - | - | -* | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_078 | - | Porter | - | + | - | + | + | + | -* | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_088 | - | Bloody Butcher | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_089 | - | Brandywine | + | + | + | - | + | - | + | + | - | - | - | - | - | - | - | - | - | + | - |
| RF_090 | - | Dixy Golden Giant | + | + | + | - | + | - | + | + | - | - | - | - | - | - | + | - | - | - | - |
| RF_091 | - | Giant Belgium | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_093 | - | Kentucky Beefsteak | + | + | + | - | + | - | + | - | - | - | - | - | - | - | + | - | - | + | - |
| RF_094 | LA1504 | Marmande VFA | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_096 | - | Thessaloniki | - | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_097 | - | Watermelon Beefsteak | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_102 | LA4133 | “var. cerasiforme” | - | + | - | - | - | - | -* | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_103 | LA1421 | “var. cerasiforme” | - | p/- | x/- | ? | - | - | -/-* | - | - | - | - | - | - | - | - | - | - | +/- | - |
| RF_105 | LA1479 | “var. cerasiforme” | -* | p | p | - | - | - | -* | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_201 | - | Blondokee | + | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_203 | - | Snowstorm | - | + | +/- | - | +/- | - | + | - | - | - | - | r/- | - | - | - | - | + | +/- | - |
| RF_206 | - | ABC Potato Leaf | + | + | +/- | - | - | - | + | + | - | - | - | - | - | - | - | - | - | - | - |
| RF_214 | LA4345 | Heinz 1706 (reference) | - | + | - | - | + | + | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_216 | CGN15882 | Sonora | - | + | + | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_226 | PI 320468 | DL/67/248 | - | p | - | - | + | - | -* | - | - | - | - | - | + | - | - | - | - | - | - |
| RF_227 | PI 324065 | Nagcarlan | + | x | - | - | + | + | -* | - | - | - | - | - | - | + | - | - | - | + | - |
| RF_229 | PI 372385 | Morne a L’Eau | - | + | + | - | + | - | -* | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_232 | 2K6-6003 | OH88119 | - | + | - | - | + | - | + | - | - | - | - | - | - | + | - | + | + | - | + |
| RF_233 | 2K6-6036 | NCEBR2 | - | + | + | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | + |
| RF_234 | 2K6-6040 | 981136 | - | + | - | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | + |
| RF_235 | T 519 | Kecskemeti Koria Bibor | - | + | + | - | + | - | + | - | - | - | - | - | - | + | - | + | - | - | - |
| RF_236 | - | Grosse Cotelee | + | + | + | - | + | - | + | - | - | - | - | - | - | - | - | - | - | + | - |
| RF_237 | PI 379007 | “var. cerasiforme” | n | p | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| RF_238 | - | RZ26 | n | p | - | - | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roohanitaziani, R.; de Maagd, R.A.; Lammers, M.; Molthoff, J.; Meijer-Dekens, F.; van Kaauwen, M.P.W.; Finkers, R.; Tikunov, Y.; Visser, R.G.F.; Bovy, A.G. Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits. Genes 2020, 11, 1278. https://doi.org/10.3390/genes11111278
Roohanitaziani R, de Maagd RA, Lammers M, Molthoff J, Meijer-Dekens F, van Kaauwen MPW, Finkers R, Tikunov Y, Visser RGF, Bovy AG. Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits. Genes. 2020; 11(11):1278. https://doi.org/10.3390/genes11111278
Chicago/Turabian StyleRoohanitaziani, Raana, Ruud A. de Maagd, Michiel Lammers, Jos Molthoff, Fien Meijer-Dekens, Martijn P. W. van Kaauwen, Richard Finkers, Yury Tikunov, Richard G. F. Visser, and Arnaud G. Bovy. 2020. "Exploration of a Resequenced Tomato Core Collection for Phenotypic and Genotypic Variation in Plant Growth and Fruit Quality Traits" Genes 11, no. 11: 1278. https://doi.org/10.3390/genes11111278






