Genomic Diversity in Sporadic Breast Cancer in a Latin American Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and Genotyping.
2.3. Statistical Analysis
2.4. Randomized Analysis
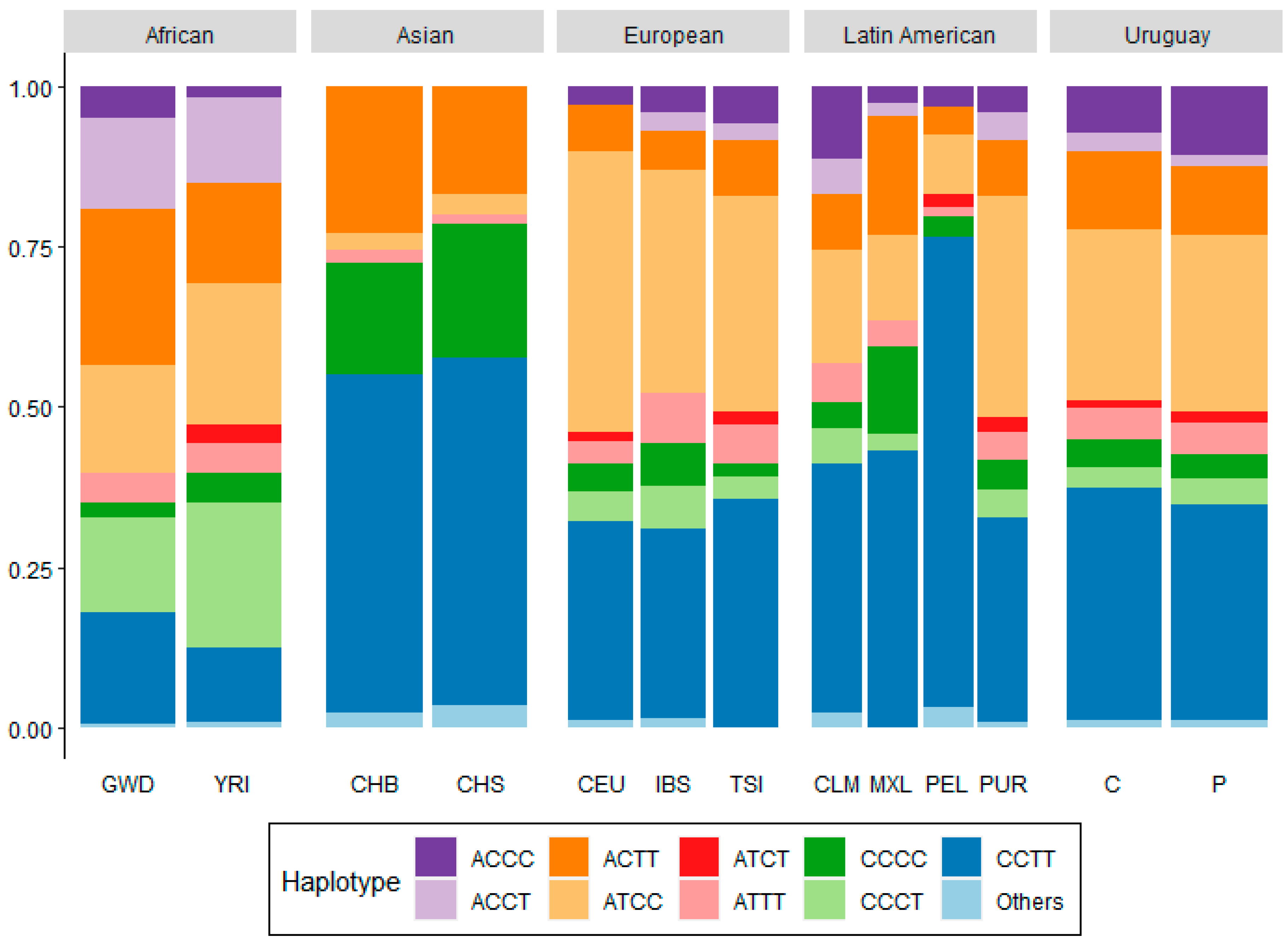
2.5. Haplotype Analysis
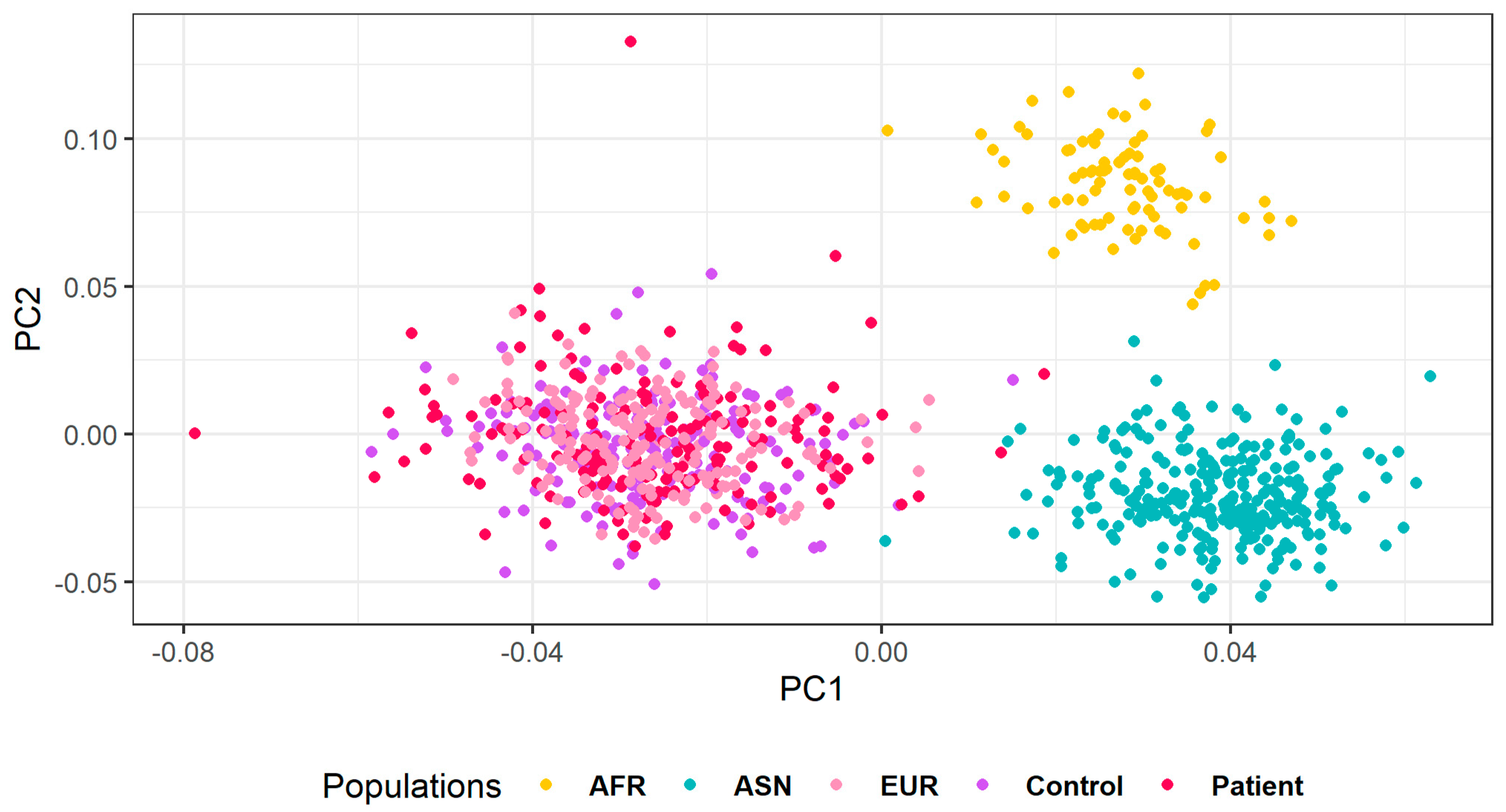
2.6. Population Structure
3. Results
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Wendt, C.; Margolin, S. Identifying breast cancer susceptibility genes–a review of the genetic background in familial breast cancer. Acta Oncol. 2019, 58, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017. [Google Scholar] [CrossRef] [Green Version]
- Sierra, M.S.; Soerjomataram, I.; Antoni, S.; Laversanne, M.; Piñeros, M.; de Vries, E.; Forman, D. Cancer patterns and trends in Central and South America. Cancer Epidemiol. 2016, 44, S23–S42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoniou, A.; Pharoah, P.D.P.; Narod, S.; Risch, H.A.; Eyfjord, J.E.; Hopper, J.L.; Loman, N.; Olsson, H.; Johannsson, O.; Borg, Å.; et al. Average Risks of Breast and Ovarian Cancer Associated with BRCA1 or BRCA2 Mutations Detected in Case Series Unselected for Family History: A Combined Analysis of 22 Studies. Am. J. Hum. Genet. 2003, 72, 1117–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedenfalk, I.; Duggan, D.; Chen, Y.; Radmacher, M.; Bittner, M.; Simon, R.; Meltzer, P.; Gusterson, B.; Esteller, M.; Raffeld, M.; et al. Gene-Expression Profiles in Hereditary Breast Cancer. N. Engl. J. Med. 2001, 344, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Di Sibio, A.; Abriata, G.; Forman, D.; Sierra, M.S. Female breast cancer in Central and South America. Cancer Epidemiol. 2016, 44, S110–S120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefani, D.; Deneo-pellegrini, H. Meat Intake, Heterocyclic and Risk of Breast A Case-Control Study in Uruguay. Cancer Epidemol. Biomark. Prev. 1997, 6, 573–581. [Google Scholar]
- Ruiz-Linares, A.; Adhikari, K.; Acuña-Alonzo, V.; Quinto-Sanchez, M.; Jaramillo, C.; Arias, W.; Fuentes, M.; Pizarro, M.; Everardo, P.; de Avila, F.; et al. Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7342 Individuals. PLoS Genet. 2014, 10, e1004572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sans, M.; Sanz, M. Admixture Studies in Latin America: From the 20th to the 21st Century. Hum. Biol. 2000, 72, 155–177. [Google Scholar]
- Belbin, G.M.; Nieves-Colón, M.A.; Kenny, E.E.; Moreno-Estrada, A.; Gignoux, C.R. Genetic diversity in populations across Latin America: Implications for population and medical genetic studies. Curr. Opin. Genet. Dev. 2018, 53, 98–104. [Google Scholar] [CrossRef]
- Dutil, J.; Golubeva, V.A.; Pacheco-Torres, A.L.; Diaz-Zabala, H.J.; Matta, J.L.; Monteiro, A.N. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: A clinical perspective. Breast Cancer Res. Treat. 2015, 154, 441–453. [Google Scholar] [CrossRef] [Green Version]
- Zavala, V.A.; Serrano-Gomez, S.J.; Dutil, J.; Fejerman, L. Genetic Epidemiology of Breast Cancer in Latin America. Genes 2019, 10, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, C.; Cardoso-Filho, C.; Bossi, L.; Lourenço, G.; Costa-Gurgel, M.; Lima, C. Association of CYP1A1 A4889G and T6235C polymorphisms with the risk of sporadic breast cancer in Brazilian women. Clinics 2015, 70, 680–685. [Google Scholar] [CrossRef]
- Martínez-Ramírez, O.C.; Pérez-Morales, R.; Castro, C.; Flores-Díaz, A.; Soto-Cruz, K.E.; Astorga-Ramos, A.; Gonsebatt, M.E.; Casas, L.; Valdés-Flores, M.; Rubio, J. Polymorphisms of catechol estrogens metabolism pathway genes and breast cancer risk in Mexican women. Breast 2013, 22, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Soto-Quintana, O.; Zúñiga-González, G.M.; Ramírez-Patiño, R.; Ramos-Silva, A.; Figuera, L.E.; Carrillo-Moreno, D.I.; Gutiérrez-Hurtado, I.A.; Puebla-Pérez, A.M.; Sánchez-Llamas, B.; Gallegos-Arreola, M.P. Association of the GSTM1 null polymorphism with breast cancer in a mexican population. Genet. Mol. Res. 2015, 14, 13066–13075. [Google Scholar] [CrossRef] [PubMed]
- Back, L.K.D.C.; Farias, T.D.J.; da Cunha, P.A.; Muniz, Y.C.N.; Ribeiro, M.C.M.; Fernandes, B.L.; Fernandes, C.K.; de Souza, I.R. Functional polymorphisms of interleukin-18 gene and risk of breast cancer in a Brazilian population. Tissue Antigens 2014, 84, 229–233. [Google Scholar] [CrossRef]
- Bonilla, C.; Bertoni, B.; Hidalgo, P.C.; Artagaveytia, N.; Ackermann, E.; Barreto, I.; Cancela, P.; Cappetta, M.; Egaña, A.; Figueiro, G.; et al. Breast cancer risk and genetic ancestry: A case–control study in Uruguay. BMC Womens Health 2015, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fejerman, L.; Romieu, I.; John, E.M.; Lazcano-Ponce, E.; Beckman, K.B.; Pérez-Stable, E.J.; Burchard, E.G.; Ziv, E.; Torres-Mejía, G. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1074–1082. [Google Scholar] [CrossRef] [Green Version]
- Renehan, A.G.; Tyson, M.; Egger, M.; Heller, R.F.; Zwahlen, M. Body-mass index and incidence of cancer: A systematic review and meta-analysis of prospective observational studies. Lancet 2008, 371, 569–578. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Douglas, I.; Forbes, H.; Dos-Santos-Silva, I.; Leon, D.A.; Smeeth, L. Body-mass index and risk of 22 specific cancers: A population-based cohort study of 5·24 million UK adults. Lancet 2014, 384, 755–765. [Google Scholar] [CrossRef] [Green Version]
- Delgado, L.; Fernández, G.; Grotiuz, G.; Cataldi, S.; González, A.; Lluveras, N.; Heguaburu, M.; Fresco, R.; Lens, D.; Sabini, G.; et al. BRCA1 and BRCA2 germline mutations in Uruguayan breast and breast-ovarian cancer families. Identification of novel mutations and unclassified variants. Breast Cancer Res. Treat. 2011, 128, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Solano, A.; Aceto, G.; Delettieres, D.; Veschi, S.; Neuman, M.; Alonso, E.; Chialina, S.; Chacón, R.; Renato, M.-C.; Podestá, E. BRCA1 And BRCA2 analysis of Argentinean breast/ovarian cancer patients selected for age and family history highlights a role for novel mutations of putative south-American origin. Springerplus 2012, 1, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alemar, B.; Herzog, J.; Brinckmann Oliveira Netto, C.; Artigalás, O.; Schwartz, I.V.D.; Matzenbacher Bittar, C.; Ashton-Prolla, P.; Weitzel, J.N. Prevalence of Hispanic BRCA1 and BRCA2 mutations among hereditary breast and ovarian cancer patients from Brazil reveals differences among Latin American populations. Cancer Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.; Gut, I.G. Genotyping single-nucleotide polymorphisms by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 782, 73–87. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Tsao, M.; Ling, X. Subsampling Method for Robust Estimation of Regression Models. Open J. Stat. 2012, 2, 281–296. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; Reshef, Y.A.; Finucane, H.K.; Schoenherr, S.; Forer, L.; McCarthy, S.; Abecasis, G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- Porchia, L.M. Common BRCA1 and BRCA2 Mutations among Latin American Breast Cancer Subjects: A Meta-Analysis. J. Carcinog. Mutagen. 2015, 6. [Google Scholar] [CrossRef]
- Bahcall, O. Fine mapping at the 11q13 locus. Nat. Genet. 2013, 45, 352. [Google Scholar] [CrossRef]
- Winkler, C.A.; Nelson, G.W.; Smith, M.W. Admixture mapping comes of age. Annu. Rev. Genomics Hum. Genet. 2010, 11, 65–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, J.; Fejerman, L.; Hu, D.; Huntsman, S.; Li, M.; John, E.M.; Torres-Mejia, G.; Kushi, L.; Ding, Y.C.; Weitzel, J.; et al. Identification of novel common breast cancer risk variants at the 6q25 locus among Latinas. Breast Cancer Res. 2019, 21, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connor, A.E.; Visvanathan, K.; Baumgartner, K.B.; Baumgartner, R.N.; Boone, S.D.; Hines, L.M.; Wolff, R.K.; John, E.M.; Slattery, M.L. Pre-diagnostic breastfeeding, adiposity, and mortality among parous Hispanic and non-Hispanic white women with invasive breast cancer: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2016. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Gómez, S.J.; Sanabria-Salas, M.C.; Garay, J.; Baddoo, M.C.; Hernández-Suarez, G.; Mejía, J.C.; García, O.; Miele, L.; Fejerman, L.; Zabaleta, J. Ancestry as a potential modifier of gene expression in breast tumors from Colombian women. PLoS ONE 2017, 12, e0183179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochmann, J.; Cappetta, M.; Pérez, J.; Colistro, V.; Nicolleti, S.; Larre Borges, A.; Ribas, G.; Martínez Asuaga, M.; Bertoni, B. Melanoma, ancestry and MC1R variants in the Uruguayan admixed population. BAG J. Basic Appl. Genet. 2016, 27, 7–18. [Google Scholar]
- Ossio, R.; Roldán-Marín, R.; Martínez-Said, H.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma: A global perspective. Nat. Rev. Cancer 2017, 17, 393–394. [Google Scholar] [CrossRef]
- Sans, M.; Da Luz, J.; Kimura, E.M.; Costa, F.F.; De Fatima Sonati, M. β-globin gene cluster haplotypes in afro-uruguayans from two geographical regions (south and north). Am. J. Hum. Biol. 2010, 22, 124–128. [Google Scholar] [CrossRef]
- Mimbacas, A.; Trujillo, J.; Gascue, C.; Javiel, G.; Cardoso, H. Prevalence of vitamin D receptor gene polymorphism in a Uruguayan population and its relation to type 1 diabetes mellitus. Genet. Mol. Res. 2007, 6, 534–542. [Google Scholar]
- Berois, N.; Touya, D.; Ubillos, L.; Bertoni, B.; Osinaga, E.; Varangot, M. Prevalence of EGFR Mutations in Lung Cancer in Uruguayan Population. J. Cancer Epidemiol. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.P.; Zhao, Q.; Wang, D.; Xie, W.Y.; Zhang, L.J.; Zhou, H.; Chen, S.Z.; Wu, L.F. The association between BRCA1 gene polymorphism and cancer risk: A meta-analysis. Oncotarget 2018, 9, 8681–8694. [Google Scholar] [CrossRef] [Green Version]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Della Valle, A.; Acevedo, C.; Esperón, P.; Neffa, F.; Artagaveytia, N.; Santander, G.; Menini, M.; Vergara, B.Q.C.; Carusso, B.Q.F.; Sapone, L.M. Cáncer de mama y ovario hereditario en Uruguay: Resultados del screening para mutaciones en genes de susceptibilidad por secuenciación de nueva generación. Rev. Med. Urug. 2017, 33, 97–102. [Google Scholar]
- Pilleron, S.; Soerjomataram, I.; Soto-Perez-de-Celis, E.; Ferlay, J.; Vega, E.; Bray, F.; Piñeros, M. Aging and the cancer burden in Latin America and the Caribbean: Time to act. J. Geriatr. Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Bernhisel, R.; Larson, K.; Caswell-Jin, J.L.; Shadyab, A.H.; Ochs-Balcom, H.; Stefanick, M.L. Prevalence of Pathogenic Variants in Cancer Susceptibility Genes among Women with Postmenopausal Breast Cancer. JAMA J. Am. Med. Assoc. 2020, 323, 995–997. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.C.; Spurdle, A.B.; Sinilnikova, O.M.; Healey, S.; Pooley, K.A.; Schmutzler, R.K.; Versmold, B.; Engel, C.; Meindl, A.; Arnold, N.; et al. Common Breast Cancer-Predisposition Alleles Are Associated with Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. Am. J. Hum. Genet. 2008, 82, 937–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Easton, D.F.; Pooley, K.A.; Dunning, A.M.; Pharoah, P.D.; Thompson, D.; Ballinger, D.G.; Struewing, J.P.; Morrison, J.; Field, H.; Luben, R.; et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007, 447, 1087–1093. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.L.; Baumgartner, K.B.; Giuliano, A.R.; Byers, T.; Herrick, J.S.; Wolff, R.K. Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res Treat 2011, 129, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Bryc, K.; Durand, E.Y.; Macpherson, J.M.; Reich, D.; Mountain, J.L. The genetic ancestry of african americans, latinos, and european Americans across the United States. Am. J. Hum. Genet. 2015, 96, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Bertoni, B.; Budowle, B.; Sans, M.; Barton, S.A.; Chakraborty, R. Admixture in Hispanics: Distribution of ancestral population contributions in the continental United States. Hum. Biol. 2003, 75, 1–11. [Google Scholar] [CrossRef]
- Dunning, A.M.; Michailidou, K.; Kuchenbaecker, K.B.; Thompson, D.; French, J.D.; Beesley, J.; Healey, C.S.; Kar, S.; Pooley, K.A.; Lopez-Knowles, E.; et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016, 48, 374–386. [Google Scholar] [CrossRef]
- Zeng, H.; Gifford, D.K. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017, 45, e99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labrie, V.; Buske, O.J.; Oh, E.; Jeremian, R.; Ptak, C.; Gasiūnas, G.; Maleckas, A.; Petereit, R.; Žvirbliene, A.; Adamonis, K.; et al. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nat. Struct. Mol. Biol. 2016, 23, 566–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappetta, M.; Berdasco, M.; Hochmann, J.; Bonilla, C.; Sans, M.; Hidalgo, P.C.; Artagaveytia, N.; Kittles, R.; Martínez, M.; Esteller, M.; et al. Effect of genetic ancestry on leukocyte global DNA methylation in cancer patients. BMC Cancer 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| GENE | Region | SNP | MAFC | MAFP | p-value | OR (95% CI) | RDM SES (%) |
|---|---|---|---|---|---|---|---|
| MAP3K1 | 5q11.2 | rs16886165T>G | 0.157 | 0.217 | 0.023 | 1.62 (1.07–2.47) | <50 |
| CNTNAP2 | 7q35 | rs2214681C>T | 0.387 | 0.478 | 0.028 | 1.45 (1.04–2.02) | 64 |
| FGFR2 | 10q26.13 | rs2981582C>T | 0.396 | 0.469 | 0.024 | 1.52 (1.06–2.18) | <50 |
| VDR | 12q13.11 | rs4237855G>A | 0.425 | 0.526 | 0.036 | 1.38 (1.02–1.86) | 50 |
| BRCA2 | 13q13.1 | rs144848A>C | 0.249 | 0.311 | 0.080 | 1.38 (0.96–1.97) | <50 |
| RANKL | 13q14.11 | rs9594759C>T | 0.423 | 0.549 | 0.014 | 1.88 (1.14–3.12) | <50 |
| BRCA1 | 17q21.31 | rs1799950A>G | 0.081 | 0.049 | 0.038 | 0.48 (0.24–0.96) | <50 |
| PTGIS | 20q13.13 | rs8183919C>T | 0.249 | 0.317 | 0.038 | 1.46 (1.02–2.10) | <50 |
| GENE | SNPS | HAPLOTYPE | FP | FC | p-value | OR | RDM SES % |
|---|---|---|---|---|---|---|---|
| ESR1 * | rs2941740|rs1999805|rs827423 | ||||||
| GCC | 0.043 | 0.029 | 0.212 | 2.06 | <50 | ||
| GCT | 0.050 | 0.048 | 0.619 | 1.28 | <50 | ||
| GTC | 0.212 | 0.133 | 0.005 | 2.05 | 91 | ||
| GTT | 0.206 | 0.180 | 0.228 | 1.35 | <50 | ||
| ACC | 0.164 | 0.183 | 0.463 | 0.83 | <50 | ||
| ACT | 0.173 | 0.241 | 0.004 | 0.49 | 52 | ||
| ATC | 0.050 | 0.072 | 0.162 | 0.53 | <50 | ||
| ATT | 0.102 | 0.113 | 0.598 | 0.84 | <50 | ||
| FGFR2 * | rs1219648|rs2981582 | ||||||
| AC | 0.500 | 0.531 | 0.264 | 0.80 | <50 | ||
| GC | 0.012 | 0.058 | 0.023 | 0.22 | 99 | ||
| GT | 0.488 | 0.412 | 0.098 | 1.40 | <50 | ||
| VDR | rs2238136|rs4237855 | ||||||
| AA | 0.031 | 0.017 | 0.278 | 1.67 | <50 | ||
| AG | 0.200 | 0.217 | 0.628 | 0.89 | <50 | ||
| GA | 0.495 | 0.406 | 0.031 | 1.36 | 55 | ||
| GG | 0.274 | 0.360 | 0.026 | 0.69 | 60 | ||
| BRCA2 | rs144848|rs4987117 | ||||||
| GC | 0.360 | 0.239 | 0.007 | 1.75 | 89 | ||
| TC | 0.640 | 0.761 | 0.008 | 0.58 | 86 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brignoni, L.; Cappetta, M.; Colistro, V.; Sans, M.; Artagaveytia, N.; Bonilla, C.; Bertoni, B. Genomic Diversity in Sporadic Breast Cancer in a Latin American Population. Genes 2020, 11, 1272. https://doi.org/10.3390/genes11111272
Brignoni L, Cappetta M, Colistro V, Sans M, Artagaveytia N, Bonilla C, Bertoni B. Genomic Diversity in Sporadic Breast Cancer in a Latin American Population. Genes. 2020; 11(11):1272. https://doi.org/10.3390/genes11111272
Chicago/Turabian StyleBrignoni, Lucía, Mónica Cappetta, Valentina Colistro, Mónica Sans, Nora Artagaveytia, Carolina Bonilla, and Bernardo Bertoni. 2020. "Genomic Diversity in Sporadic Breast Cancer in a Latin American Population" Genes 11, no. 11: 1272. https://doi.org/10.3390/genes11111272
APA StyleBrignoni, L., Cappetta, M., Colistro, V., Sans, M., Artagaveytia, N., Bonilla, C., & Bertoni, B. (2020). Genomic Diversity in Sporadic Breast Cancer in a Latin American Population. Genes, 11(11), 1272. https://doi.org/10.3390/genes11111272







