Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Genotyping
2.3. Genetic Diversity
2.4. Population Structure
3. Results
3.1. Microsatellite Markers
3.2. Genetic Diversity
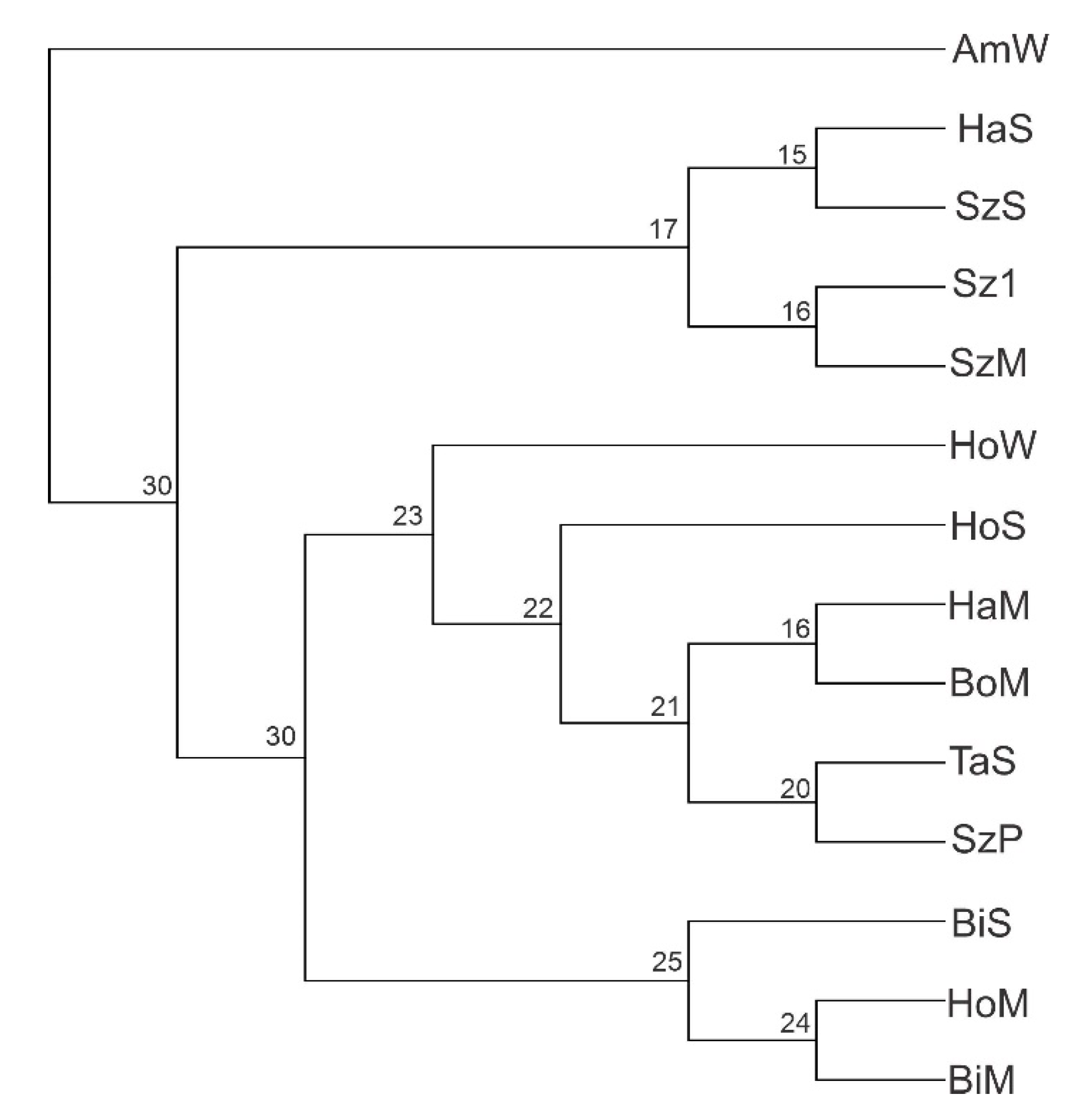
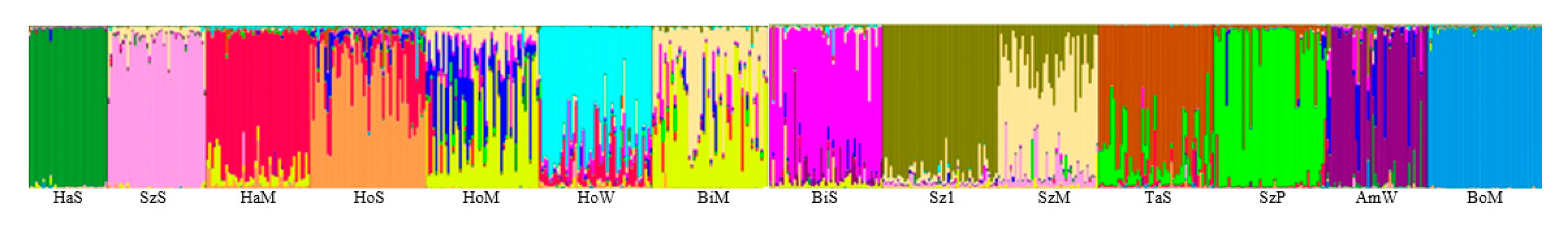
3.3. Population Structure
4. Discussion
4.1. Genetic Diversity
4.2. Population Structure
4.3. Conservation Implications
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ghelichpour, M.; Shabani, A.; Shabanpour, B. Microsatellite variations and genetic structure of common carp (Cyprinus carpio) populations in Gomishan bay and Gorganroud River (Southeast of the Caspian Sea). Int. J. Aquat. Biol. 2013, 1, 22–27. [Google Scholar]
- Power, M.; Dempson, J.B.; Reist, J.D.; Schwarz, C.J.; Power, G. Latitudinal variation in fecundity among Arctic charr populations in eastern North America. J. Fish. Biol. 2005, 67, 255–273. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation. Genetics 2002. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Ryder, O.A. Conservation action for gazelles: An urgent need. Trends Ecol. Evol. 1987, 2, 143–144. [Google Scholar] [CrossRef]
- Balon, K. Origin and domestication of the wild carp, Cyprinus carpio: From Roman gourmets to the swimming flowers. Aquaculture 1995, 129, 3–48. [Google Scholar] [CrossRef]
- Flajs’hans, M.; Hulata, G. Common Carp-Cyprinus Carpio. Genimpact Final Scientific Report; University of South Bohemia: Vodnany, Czech Republic, 2006. [Google Scholar]
- Teletchea, F.; Fontaine, P. Levels of domestication in fish: Implications for the sustainable future of aquaculture. Fish. Fish. 2014, 15, 181–195. [Google Scholar] [CrossRef]
- Kohlmann, K.; Gross, R.; Murakaeva, A.; Kersten, P. Genetic variation and structure of common carp populations throughout the distribution range inferred from allozyme, microsatellite and mtDNA marker. Aquat. Living Resour. 2003, 16, 421–431. [Google Scholar] [CrossRef]
- FAO. The state of world fisheries and aquaculture. Int. J. Fish. Aquac. 2018, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef] [Green Version]
- Bakos, J.; Gorda, S. Genetic improvement of common carp strains using intraspecific hybridization. Aquaculture 1995, 129, 183–186. [Google Scholar] [CrossRef]
- David, L.; Rosenberg, N.A.; Lavi, U.; Feldman, M.W.; Hillel, J. Genetic diversity and population structure inferred from the partially duplicated genome of domesticated carp, Cyprinus carpio L. Genet. Sel. Evol. 2007, 39, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napora-Rutkowski, Ł.; Rakus, K.; Nowak, Z.; Szczygieł, J.; Pilarczyk, A.; Ostaszewska, T.; Irnazarow, I. Genetic diversity of common carp (Cyprinus carpio L.) strains breed in Poland based on microsatellite, AFLP, and mtDNA genotype data. Aquaculture 2017, 473, 433–442. [Google Scholar] [CrossRef]
- Nielsen, H.M.; Ødegård, J.; Olesen, I.; Gjerde, B.; Ardo, L.; Jeney, G.; Jeney, Z. Genetic analysis of common carp (Cyprinus carpio) strains: I: Genetic parameters and heterosis for growth traits and survival. Aquaculture 2010, 304, 14–21. [Google Scholar] [CrossRef]
- Ødegård, J.; Olesen, I.; Dixon, P.; Jeney, Z.; Nielsen, H.M.; Way, K.; Joiner, C.; Jeney, G.; Ardó, L.; Rónyai, A.; et al. Genetic analysis of common carp (Cyprinus carpio) strains. II: Resistance to koi herpesvirus and Aeromonas hydrophila and their relationship with pond survival. Aquaculture 2010, 304, 7–13. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for Methods Utilizing Environmental DNA for Detection of Fish Species. Genes 2020, 11, 296. [Google Scholar] [CrossRef] [Green Version]
- Lehoczky, I.; Kovács, B.; Kovács, G.; Gorda, S.; Péteri, A.; Bakos, J. A ponty genetikája és erőforrásai. Vármédia-Print Kft. Gödöllő. 9-34. In: A ponty (Cyprinus carpio L.) biológiája és tenyésztése. Csorbai, B., Urbányi, B. Szent István Egyetem, Mezőgazdaság- és Környezettudományi Kar, Akvakultúra és Környezetbiztonsági Intézet, Halgazdálkodási Tanszék megbízásából Vármédia-Print Kft. Gödöllő 2018, 203. [Google Scholar]
- Bakos, J.; Gorda, S.; Váradi, L.; Balogh, J. Tenyésztő szervezetek szerepe a magyar pontyfajták fenntartásában és nemesítésében. Xxi Halászati Tudományos Tanácskozás Szarvas 1997, 32, 25–26. [Google Scholar]
- Gorda, S.; Borbély, A. Ponty Teljesítményvizsgálat Eredménye. Ph.D. Thesis, École Polytechnique, Paris, France, 2014; pp. 1–27. [Google Scholar]
- Lehoczky, I. A HAKI Ponty élő Génbankjának Populációgenetikai Vizsgálata Mikroszatellit DNS Markerekkel és PCR-RFLP Módszerrel. Ph.D. Thesis, University of Kaposvár, Kaposvár, Hungary, 2006. [Google Scholar]
- Lehoczky, I.; Magyary, I.; Hancz, C.; Weiss, S. Preliminary studies on the genetic variability of six Hungarian common carp strains using microsatellite DNA markers. Hydrobiologia 2005, 533, 223–228. [Google Scholar] [CrossRef]
- Kongchum, P.; Palti, Y.; Hallerman, E.M.; Hulata, G.; David, L. SNP discovery and development of genetic markers for mapping innate immune response genes in common carp (Cyprinus carpio). Fish. Shellfish Immunol. 2010, 29, 356–361. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, L.; Jiang, P.; Li, D.; Lu, C.; Sun, X. Genome evolution trend of common carp (Cyprinus carpio L.) as revealed by the analysis of microsatellite loci in a gynogentic family. J. Genet. Genom. 2008, 35, 97–103. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, Q.; Wang, Z.; Ye, Y. Genetic variation analysis within and among six varieties of common carp (Cyprinus carpio L.) in China using microsatellite markers. Genomics 2004, 40, 1389–1393. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Voronova, N.V. Genetic evolution and diversity of common carp Cyprinus carpio L. Cent. Eur. J. Biol. 2009, 4, 304–312. [Google Scholar] [CrossRef]
- Mabuchi, K.; Miya, M.; Senou, H.; Suzuki, T.; Nishida, M. Complete mitochondrial DNA sequence of the Lake Biwa wild strain of common carp (Cyprinus carpio L.) further evidence for an ancient origin. Aquaculture 2006, 257, 68–77. [Google Scholar] [CrossRef]
- Mondol, R.K.; Islam, S.; Alam, S. Characterization of different strains of common carp (Cyprinus carpio L.) (Cyprinidae, Cypriniformes) in Bangladesh using microsatellite DNA markers. Genet. Mol. Biol. 2006, 29, 626–633. [Google Scholar] [CrossRef]
- Vandeputte, M. Selective breeding of quantitative traits in the common carp (Cyprinus carpio): A review. Aquat. Living Resour. 2003, 16, 399–407. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography-The History and Formation of Species. Integr. Comp. Biol. 2000, 447, 134–135. [Google Scholar]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, L.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals-a review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.J.; Cordes, J.F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]
- Schlötterer, C. The evolution of molecular markers-just a matter of fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Desvignes, J.F.; Laroche, J.; Durand, J.D.; Bouvet, Y. Genetic variability in reared stocks of common carp (Cyprinus carpio L.) based on allozymes and microsatellites. Aquaculture 2001, 194, 291–301. [Google Scholar] [CrossRef]
- Hulak, M.; Kaspar, V.; Kohlmann, K.; Coward, K.; Tešitel, J.; Rodina, M.; Gela, D.; Kocour, M.; Linhart, O. Microsatellite-based genetic diversity and differentiation of foreign common carp (Cyprinus carpio) strains farmed in the Czech Republic. Aquaculture 2010, 298, 194–201. [Google Scholar] [CrossRef]
- Ludanny, R.I.; Chrisanfova, G.G.; Vasilyev, V.A.; Prizenko, V.K.; Bogeruk, A.K.; Ryskov, A.P.; Semyenova, S.K. Genetic diversity and differentiation of Russian common carp (Cyprinus carpio L.) breeds inferred from RAPD markers. Russ. J. Genet. 2006, 42, 928–935. [Google Scholar] [CrossRef]
- Tomljanovic, T.; Treer, T.; Cubric, V.C.; Safner, T.; Sprem, N.; Piria, M.; Matulic, D.; Safner, R.; Anicic, I. Microsatellite-based genetic variability and differentation of hatchery and feral common carp Cyprinus carpio L. (Cyprinidae, Cyprinidae) populations in Croatia. Arch. Biol. Sci. Belgrade 2013, 65, 577–584. [Google Scholar] [CrossRef] [Green Version]
- Bártfai, R.; Egedi, S.; Yue, G.H.; Kovács, B.; Urbányi, B.; Tamás, G.; Horváth, L.; Orbán, L. Genetic analysis of two common carp broodstocks by RAPD and microsatellite markers. Aquaculture 2003, 219, 157–167. [Google Scholar] [CrossRef]
- Gorda, S. Vízi Genetikai Erőforrások-a Magyar Pontytenyésztési Program.; május 4; Sáregres-Rétimajor. In Proceedings of the NACEE General Assembly and Workshop on Some Specific Issues of Freshwater Aquaculture, Retimajor, Hungary, 3–4 May 2012. [Google Scholar]
- Crooijmans, R.P.M.A.; Bierbooms, V.A.F.; Komen, J.; Van der Poel, J.J.; Groenen, M.A.M. Microsatellite markers in common carp (Cyprinus carpio L.). Anim. Genet. 1997, 28, 129–134. [Google Scholar] [CrossRef]
- Yue, G.H.; Ho, M.Y.; Orban, L.; Komen, J. Microsatellites within genes and ESTs of common carp and their applicability in silver crucian carp. Aquaculture 2004, 234, 85–98. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Biol. Evol. 2006, 24, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Raymond, M.; Rousset, F. An Exact Test for Population Differentiatio. J. Artic. Evol. 1995, 49, 1280–1283. [Google Scholar]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 6–103. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Goudet, J.; Perrin, N.; Waser, P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 2002, 11, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, I.C.; Berkowitz, M.L. Ewald summation for systems with slab geometry. J. Chem. Phys. 1999, 111, 3155. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing Tables of Statistical Tests. J. Artic. 1989, 43, 223–225. [Google Scholar]
- Cavalli-Sforza, L.L.; Edwards, A.W.F. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967, 19, 233–257. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Zhao, Y.; Zhu, X.; Li, Z.; Xu, W.; Dong, J.; Wei, H.; Li, Y.; Li, X. Genetic diversity and structure of Chinese grass shrimp, Palaemonetes sinensis, inferred from transcriptome-derived microsatellite markers. BMC Genet. 2019, 20, 75. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef]
- Rannala, B.; Mountain, J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol. Ecol. 2003, 13, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resourches 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falush, D.; Stephens, M.; Pritchard, K.J. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Liu, X.; Cao, Y.; Xue, T.; Wu, R.; Zhou, Y.U.; Zhou, C.; Zanatta, D.T.; Ouyang, S.; Wu, X. Genetic structure and diversity of Nodularia douglasiae (Bivalvia: Unionida) from the middle and lower Yangtze River drainage. PLoS ONE 2017, 12, e0189737. [Google Scholar] [CrossRef] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; Holdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, A.N. CLUMPP a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlmann, K.; Kersten, P.; Flajshans, M. Microsatellite-based genetic variability and differentiation of domesticated, wild and feral common carp (Cyprinus carpio L.) populations. Aquaculture 2005, 247, 253–266. [Google Scholar] [CrossRef]
- O’Connell, M.; Wright, J.M. Microsatellite DNA in fishes. Rev. Fish. Biol. Fish. 1997, 7, 331–363. [Google Scholar] [CrossRef]
- Ruzzante, D.E. A comparison of several measures of genetic distance and population structure with microsatellite data: Bias and sampling variance. Can. J. Fish. Aquat. Sci. 1998, 55, 1–14. [Google Scholar] [CrossRef]
- Hale, M.L.; Burg, T.M.; Steeves, T.E. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 2012, 7, 45170. [Google Scholar] [CrossRef]
- Landguth, E.L.; Fedy, B.C.; Oyler-McCance, S.J.; Garey, A.L.; Emel, S.L.; Mumma, M.; Wagner, H.H.; Fortin, M.J.; Cushman, S.A. Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol. Ecol. Resour. 2012, 12, 276–284. [Google Scholar] [CrossRef]
- Putman, A.I.; Carbone, I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014, 4, 4399–4428. [Google Scholar] [CrossRef]
- Sánchez-Montes, G.; Ariño, A.H.; Vizmanos, J.L.; Wang, J.; Martínez-Solano, Í. Effects of sample size and full sibs on genetic diversity characterization: A case study of three syntopic Iberian pond-breeding amphibians. J. Hered. 2017, 108, 535–543. [Google Scholar] [CrossRef]
- Libiger, O.; Nievergelt, C.M.; Schork, N.J. Comparison of genetic distance measures using human SNP genotype data. Hum. Biol. 2009, 81, 389–406. [Google Scholar] [CrossRef]
- Treer, T.; Safner, R.; Aničić, I.; Kolak, A.; Dražić, M. Morphological variation among four strains of common carp Cyprinus carpio in Croatia. Folia Zool. 2000, 49, 69–74. [Google Scholar]
- Pintér, K. A magyar halászat helye az Európai Unióban. Halászat 2003, 96, 47–50. [Google Scholar]
- Lengyel, P.; Udvari, Z. A haltenyésztés hatósági feladatainak átszervezése. Földművelésügyi Minisztérium, Horgászati és Halgazdálkodási Főosztály. Halászat 2017, 110, 12–16. [Google Scholar]
- Kánainé Sipos, D.; Bakos, K.; Ősz, Á.; Hegyi, Á.; Müller, T.; Urbányi, B.; Kovács, B. Development and characterization of 49 novel microsatellite markers in the African catfish, Clarias gariepinus (Burchell, 1822). Mol. Biol. Rep. 2019, 46, 6599–6608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashrafzadeh, M.R.; Djan, M.; Szendrei, L.; Paulauskas, A.; Scandura, M.; Bagi, Z.; Ilie, D.E.; Kerdikoshvili, N.; Marek, P.; Soós, N.; et al. Large-scale mitochondrial DNA analysis reveals new light on the phylogeography of Central and Eastern-European Brown hare (Lepus europaeus Pallas, 1778). PLoS ONE 2018, 13, e0204653. [Google Scholar] [CrossRef] [Green Version]
- Fraser, D.J. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol. Appl. 2008, 1, 535–586. [Google Scholar] [CrossRef]
- Ren, W.; Hu, L.; Guo, L.; Zhang, J.; Tang, L.; Zhang, E.; Chen, X. Preservation of the genetic diversity of a local common carp in the agricultural heritage rice-fish system. Proc. Natl. Acad. Sci. USA 2018, 115, E546–E554. [Google Scholar] [CrossRef] [Green Version]
- Thrupp, L.A. Linking agricultural biodiversity and food security: The valuable role of agrobiodiversity for sustainable agriculture. Int. Aff. 2000, 76, 265–281. [Google Scholar] [CrossRef]

| Hungarian Strains | Abbreviation | Sample Collection Place | Sample Collection According to Hatcheries | GPS Coordinate (N: north latitude; E: east longitude) | Number of Samples (n) |
|---|---|---|---|---|---|
| Amur wild carp | AmW | Szarvas | Department of Fish Biology, National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture | N: 46.51.33.30 E: 20.31.09.56 | 42 |
| Biharugra scaly | BiS | Biharugra | Hatchery of Biharugra Ltd. | N: 46.56.46.91 E: 21.36.46.96 | 48 |
| Biharugra mirror | BiM | Biharugra | Hatchery of Biharugra Ltd. | N: 46.56.46.91 E: 21.36.46.96 | 48 |
| Böszörmény mirror | BoM | Tiszaszentimre | CLARIAS Agriculture, Producer and Trade Lp. | N: 47.29.31.31 E: 20.34.67.56 | 48 |
| Hortobágy wild | HoW | Hortobágy | Hatchery of Hortobágy cPlc. | N: 47.37.24.13 E: 21.05.17.31 | 47 |
| Hortobágy scaly | HoS | Hortobágy | Hatchery of Hortobágy cPlc. | N: 47.37.24.13 E: 21.05.17.31 | 48 |
| Hortobágy mirror | HoM | Hortobágy | Hatchery of Hortobágy cPlc. | N: 47.37.24.13 E: 21.05.17.31 | 47 |
| Szarvas-15 | SZ1 | Szarvas | Department of Fish Biology, National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture | N: 46.51.33.30 E: 20.31.09.56 | 48 |
| Szarvas P3 | SzP | Szarvas | Department of Fish Biology, National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture | N: 46.51.33.30 E: 20.31.09.56 | 47 |
| Szeged scaly | SzS | Debrecen | Fish Biology Laboratory, Faculty of Agricultural and Food Sciences and Environmental Management, University of Debrecen | N: 47.33.02.91 E: 21.36.19.25 | 41 |
| Szeged mirror | SzM | Szarvas | Department of Fish Biology, National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture | N: 46.51.33.30 E: 20.31.09.56 | 42 |
| Hajdúszoboszló scaly | HaS | Debrecen | Fish Biology Laboratory, Faculty of Agricultural and Food Sciences and Environmental Management, University of Debrecen | N: 47.33.02.91 E: 21.36.19.25 | 33 |
| Hajdúszoboszló mirror | HaM | Hajdúszoboszló | Bocskai Fishing Ltd. | N: 47.26.39.68 E:21.20.05.88 | 43 |
| Tata scaly | TaS | Szarvas | Department of Fish Biology, National Agricultural Research and Innovation Centre, Research Institute for Fisheries and Aquaculture | N: 46.51.33.30 E: 20.31.09.56 | 48 |
| Locus | Forward and Reverse Sequence (5′-3′) | Fluorescent Dye | Annealing Temperature (°C) | Fragment Range (bp) | Multiplex for Fragment Analysis | Reference |
|---|---|---|---|---|---|---|
| Cca24 | AAATTTTCAAGACTGGGTGGTT ACAGCAAGATGACAAAATGAGTG | ATTO565 | 60 | 210-234 | 1 | [42] |
| Cca67 | GTAGCCCCAAAAGATGTAGCA TGGTCAAGTTCAGAGGCTGTAT | FAM | 60 | 209-299 | 3 | [42] |
| MFW3 | GATCAGAAGGTACAGAGAAG CCTTACAGAAAACCTGTTTGC | ATTO565 | 58 | 134-240 | 1 | [41] |
| MFW4 | TCCAAGTCAGTTTAATCACCG GGGAAGCGTTGACAACAAGC | HEX | 59 | 138-253 | 1 | [41] |
| MFW6 | ACCTGATCAATCCCTGGCTC TTGGGACTTTTAAATCACGTTG | FAM | 60 | 158-212 | 2 | [41] |
| MFW7 | GATCTGCAAGCATATCTGTCG ATCTGAACCTGCAGCTCCTC | ATTO550 | 59 | 132-152 | 2 | [41] |
| MFW11 | GCATTTGCCTTGATGGTTGTG TCGTCTGGTTTAGAGTGCTGC | ATTO565 | 60 | 110-196 | 2 | [41] |
| MFW13 | ATGATGAGAACATTGTTTACAG TGAGAGAACAATGTGGATGAC | HEX | 58 | 192-270 | 2 | [41] |
| MFW15 | CTCCTGTTTTGTTTTGTGAAA GTTCACAAGGTCATTTCCAGC | ATTO550 | 59 | 159-283 | 3 | [41] |
| MFW17 | CAGTGAGACGATTACCTTGG GTGAGCAGCCCACATTGAAC | HEX | 60 | 254-312 | 1 | [41] |
| MFW26 | CCCTGAGATAGAAACCACTG CACCATGCTTGGATGCAAAAG | ATTO550 | 60 | 151-221 | 4 | [41] |
| MFW31 | CCTTCCTCTGGCCATTCTCAC TACATCGCAGAGAATTCGTAAG | ATTO550 | 60 | 283-305 | 1 | [41] |
| Locus/Strain | HaS | SzS | HaM | HoS | HoM | HoW | BiM | BiS | Sz1 | SzM | TaS | SzP | AmW | BoM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cca24 | 0.087 | 0.000 | 0.000 | 0.001 | 0.000 | 0.021 | 0.000 | 0.000 | 0.066 | 0.000 | 0.000 | 0.000 | 0.083 | 0.000 |
| MFW3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| MFW4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.067 | 0.000 | 0.000 | 0.000 |
| MFW17 | 0.000 | 0.023 | 0.000 | 0.000 | 0.006 | 0.037 | 0.000 | 0.020 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| MFW31 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| MFW6 | 0.001 | 0.192 | 0.041 | 0.048 | 0.069 | 0.000 | 0.033 | 0.085 | 0.064 | 0.173 | 0.040 | 0.202 | 0.189 | 0.000 |
| MFW7 | 0.000 | 0.088 | 0.000 | 0.050 | 0.000 | 0.000 | 0.000 | 0.012 | 0.058 | 0.000 | 0.014 | 0.000 | 0.000 | 0.000 |
| MFW11 | 0.000 | 0.160 | 0.000 | 0.000 | 0.133 | 0.237 | 0.262 | 0.164 | 0.000 | 0.305 | 0.000 | 0.000 | 0.317 | 0.000 |
| MFW13 | 0.000 | 0.184 | 0.000 | 0.016 | 0.011 | 0.000 | 0.000 | 0.154 | 0.258 | 0.298 | 0.011 | 0.000 | 0.184 | 0.000 |
| Cca67 | 0.000 | 0.000 | 0.000 | 0.044 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.051 | 0.000 | 0.000 |
| MFW15 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| MFW26 | 0.000 | 0.000 | 0.234 | 0.212 | 0.000 | 0.086 | 0.206 | 0.192 | 0.063 | 0.000 | 0.007 | 0.000 | 0.095 | 0.000 |
| Strain | MNA | Apr | Ar | Ne | Ho | He | dHW | Fis | Assignment Test |
|---|---|---|---|---|---|---|---|---|---|
| HaS | 6.500 | 9 | 1.670 | 5.290 | 0.790 | 0.730 | 1 | −0.080 | 100 |
| SzS | 8.180 | 4 | 1.680 | 4.270 | 0.770 | 0.750 | 3 | −0.027 | 97 |
| HaM | 8.450 | 2 | 1.820 | 5.670 | 0.920 | 0.820 | 2 | −0.118 | 88 |
| HoS | 11.080 | 11 | 1.860 | 7.810 | 0.860 | 0.860 | 1 | 0.002 | 97 |
| HoM | 14.000 | 4 | 1.830 | 6.470 | 0.900 | 0.830 | 2 | −0.083 | 89 |
| HoW | 12.250 | 24 | 1.890 | 9.710 | 0.900 | 0.890 | 1 | −0.007 | 93 |
| BiM | 17.580 | 1 | 1.800 | 5.240 | 0.840 | 0.800 | 2 | −0.051 | 83 |
| BiS | 11.500 | 8 | 1.830 | 7.090 | 0.810 | 0.840 | 3 | 0.026 | 91 |
| SZ1 | 13.000 | 1 | 1.740 | 4.640 | 0.720 | 0.710 | 3 | −0.004 | 97 |
| SzM | 9.630 | 0 | 1.770 | 4.210 | 0.730 | 0.750 | 1 | 0.034 | 88 |
| TaS | 7.900 | 11 | 1.820 | 5.910 | 0.930 | 0.820 | 3 | −0.134 | 95 |
| SzP | 11.580 | 3 | 1.790 | 4.960 | 0.900 | 0.790 | 4 | −0.140 | 93 |
| AmW | 8.500 | 17 | 1.800 | 5.870 | 0.740 | 0.810 | 1 | 0.083 | 100 |
| BoM | 13.080 | 22 | 1.800 | 5.360 | 0.990 | 0.800 | 5 | −0.250 | 100 |
| Strain | HaS | SzS | HaM | HoS | HoM | HoL | BiM | BiS | SZ1 | SzM | TaS | SzP | AmW | BoM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HaS | 0.000 | 0.164 | 0.173 | 0.158 | 0.134 | 0.131 | 0.155 | 0.152 | 0.231 | 0.188 | 0.176 | 0.181 | 0.147 | 0.174 |
| SzS | 0.565 | 0.000 | 0.146 | 0.137 | 0.122 | 0.116 | 0.135 | 0.116 | 0.204 | 0.114 | 0.146 | 0.149 | 0.131 | 0.140 |
| HaM | 0.702 | 0.647 | 0.000 | 0.041 | 0.056 | 0.036 | 0.066 | 0.077 | 0.176 | 0.096 | 0.080 | 0.074 | 0.112 | 0.066 |
| HoS | 0.702 | 0.649 | 0.454 | 0.000 | 0.051 | 0.035 | 0.071 | 0.076 | 0.155 | 0.106 | 0.071 | 0.072 | 0.102 | 0.090 |
| HoM | 0.601 | 0.570 | 0.497 | 0.479 | 0.000 | 0.043 | 0.028 | 0.040 | 0.136 | 0.067 | 0.079 | 0.074 | 0.089 | 0.111 |
| HoW | 0.652 | 0.624 | 0.491 | 0.476 | 0.446 | 0.000 | 0.053 | 0.054 | 0.127 | 0.085 | 0.054 | 0.057 | 0.078 | 0.063 |
| BiM | 0.605 | 0.561 | 0.476 | 0.518 | 0.347 | 0.479 | 0.000 | 0.040 | 0.131 | 0.047 | 0.101 | 0.098 | 0.084 | 0.121 |
| BiS | 0.651 | 0.559 | 0.555 | 0.548 | 0.405 | 0.490 | 0.400 | 0.000 | 0.110 | 0.067 | 0.103 | 0.105 | 0.077 | 0.113 |
| SZ1 | 0.683 | 0.605 | 0.652 | 0.615 | 0.555 | 0.595 | 0.530 | 0.523 | 0.000 | 0.099 | 0.181 | 0.196 | 0.133 | 0.194 |
| SzM | 0.630 | 0.465 | 0.547 | 0.615 | 0.498 | 0.585 | 0.429 | 0.493 | 0.437 | 0.000 | 0.113 | 0.131 | 0.112 | 0.139 |
| TaS | 0.723 | 0.660 | 0.513 | 0.519 | 0.559 | 0.527 | 0.585 | 0.607 | 0.666 | 0.606 | 0.000 | 0.039 | 0.120 | 0.106 |
| SzP | 0.714 | 0.652 | 0.496 | 0.532 | 0.521 | 0.539 | 0.559 | 0.580 | 0.683 | 0.592 | 0.389 | 0.000 | 0.126 | 0.124 |
| AmW | 0.618 | 0.581 | 0.622 | 0.615 | 0.536 | 0.564 | 0.500 | 0.512 | 0.563 | 0.584 | 0.625 | 0.612 | 0.000 | 0.149 |
| BoM | 0.687 | 0.632 | 0.498 | 0.561 | 0.633 | 0.538 | 0.632 | 0.660 | 0.700 | 0.665 | 0.560 | 0.600 | 0.684 | 0.000 |
| Number of Groups | Source of Variation | df | Sum of Square | Variance Component | Percentage Variation | p |
|---|---|---|---|---|---|---|
| One group | Among strains | 13 | 25.270 | 0.017 | 3.790 | 0.000 |
| Within strains | 1264 | 513.670 | 0.412 | 96.030 | ||
| Total | 1259 | 538.940 | 0.429 | |||
| Six (geographical) groups | Among groups | 5 | 10.590 | 0.001 | 0.420 | 0.270 |
| Among strains within groups | 8 | 14.680 | 0.015 | 3.620 | 0.000 | |
| Within strains | 1246 | 513.67 | 0.412 | 95.960 | 0.000 | |
| Total | 1259 | 538.94 | 0.429 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tóth, B.; Khosravi, R.; Ashrafzadeh, M.R.; Bagi, Z.; Fehér, M.; Bársony, P.; Kovács, G.; Kusza, S. Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation. Genes 2020, 11, 1268. https://doi.org/10.3390/genes11111268
Tóth B, Khosravi R, Ashrafzadeh MR, Bagi Z, Fehér M, Bársony P, Kovács G, Kusza S. Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation. Genes. 2020; 11(11):1268. https://doi.org/10.3390/genes11111268
Chicago/Turabian StyleTóth, Bianka, Rasoul Khosravi, Mohammad Reza Ashrafzadeh, Zoltán Bagi, Milán Fehér, Péter Bársony, Gyula Kovács, and Szilvia Kusza. 2020. "Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation" Genes 11, no. 11: 1268. https://doi.org/10.3390/genes11111268
APA StyleTóth, B., Khosravi, R., Ashrafzadeh, M. R., Bagi, Z., Fehér, M., Bársony, P., Kovács, G., & Kusza, S. (2020). Genetic Diversity and Structure of Common Carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: Implications for Conservation. Genes, 11(11), 1268. https://doi.org/10.3390/genes11111268






