Transcription Factors of the bHLH Family Delineate Vertebrate Landmarks in the Nervous System of a Simple Chordate
Abstract
:1. Introduction
2. bHLH Transcription Factors and Organizing Centers in the Developing Ascidian Nervous System
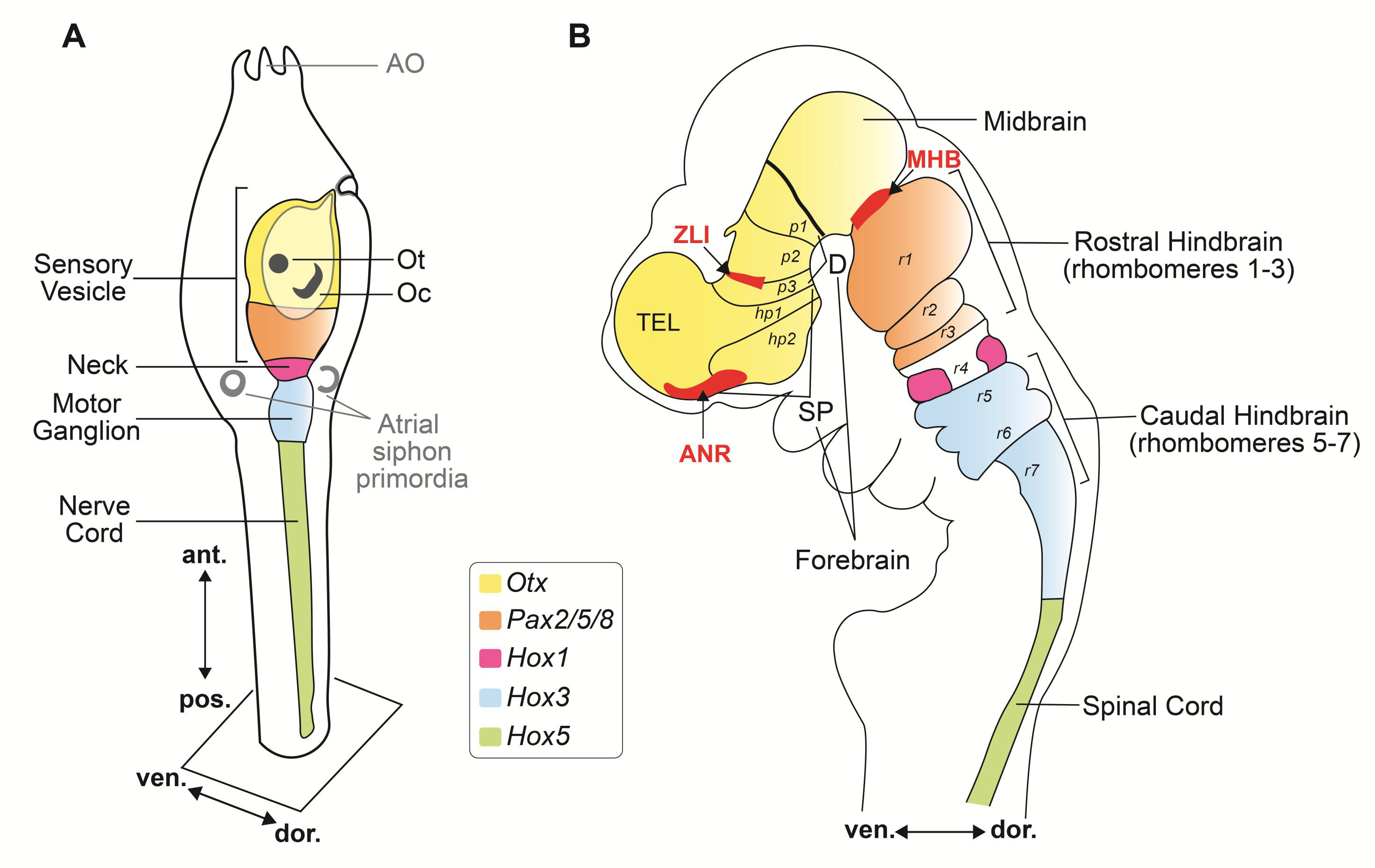
3. Anatomo-Physiological Vertebrate Landmarks in the Uncomplicated Nervous System of the Ascidian Larva
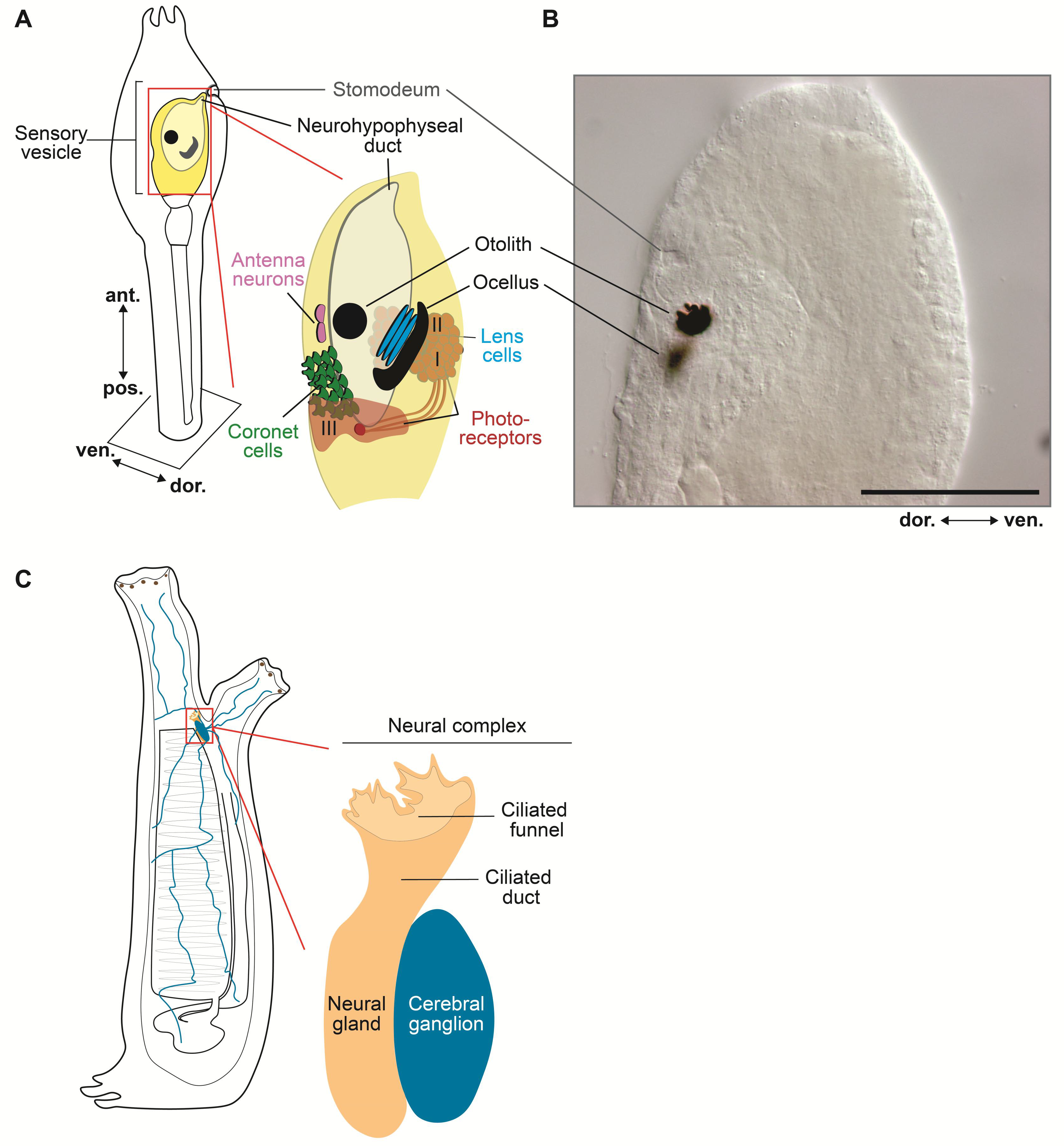
4. Cellular and Molecular Topography of the Ciona Larval Nervous System
5. bHLH Transcription Factors in the Nervous System of Adult Ascidians
6. The bHLH Family of Transcription Factors in Ciona

6.1. Group A
6.2. Group B
6.3. Group C
6.4. Group D
6.5. Group E
6.6. Group F
6.7. Outgroup
7. Cross-Regulatory Interactions among bHLH Transcription Factors in Ciona
8. Conclusions
| Gene Name | Alternative Names | Human Hits * | Gene Models KH2013 KY2019 | Expression at Mid/Late Tailbud | Reference | scRNA-Seq Data from Larvae | |
|---|---|---|---|---|---|---|---|
| [27] | [31] | ||||||
| Ascl.a (achaete-scute family bHLH transcription factor.a) | Achaete-Scute a-like2 | ASCL1; ASCL3; ASCL5 | KH.L9.13 KY.Chr2.2314 | Epid., palps | [50] | ||
| Ascl.b (achaete-scute family bHLH transcription factor.b) | Achaete-Scute b | ASCL3; ASCL4; ASCL5 | KH.C2.880 KY.Chr2.2022 | ESNs | [163] | ||
| Ascl.c (achaete-scute family bHLH transcription factor.c) | Achaete-Scute a-like1 | ASCL3; ASCL5; NEUROD1 | KH.C2.560 KY.Chr2.1484 | Anterior SV, weak mesench. and palps | [50]; Figure 3 | ||
| Atoh8 (Atonal bHLH transcription factor 8) | Net; NeuroD-like | ATOH1; ATOH8; NEUROD4 | KH.C9.872 KY.Chr9.174 | Anterior SV, MG, cESNs, trunk endod. | [50,84] Figure 3 | aSV | Ventral SV |
| Atonal | ATOH1; ATOH7; NEUROD1 | KH.C8.175 KY.Chr8.248 | ESNs, palps | [50] | cESNs rTENs | ||
| Bhlha15 (basic helix-loop-helix family member a15) | Mist | BHLHA15; NEUROG1; NEUROG2 | KH.C3.308 KY.Chr3.1309 | Mesench. | [50] | ||
| Hand (heart and neural crest derivatives expressed) | HAND1; HAND2; TCF15 | KH.C14.604 KY.Chr14.359 | Mesench. | [50] | |||
| Hand-r (heart and neural crest derivatives expressed-related) | NoTrlc | HAND1; HAND2; SCX | KH.C1.1116 KY.Chr1.2070 | Mesench., TVCs, SV | [164]; Figure 3 | ||
| Mesp (mesoderm posterior bHLH transcription factor) | MESP2; MSGN1; PTF1A | KH.C3.100 KY.Chr3.993 | Anterior ventral primary muscle, TVCs | [159] | |||
| Mrf (Myogenic regulatory factor) | MyoD; CiMDFa | MYF5; MYF6; MYOD1 | KH.C14.307 KY.Chr14.1058 | Muscle | [156] | ||
| Neurog (Neurogenin) | NEUROG1; NEUROG2; NEUROG3 | KH.C6.129 KY.Chr6.427 | SV, MG, NC Atrial ectoderm | [50,147,208] Figure 3 | aATENs, aSV, MHB, SV, MG, Epend. | Dorsolat. SV; SV wall | |
| Ptf1a (pancreas associated transcription factor 1a) | Ptfa | not determined | KH.C3.967 KY.Chr3.526 | SV | [165]; Figure 3 | ||
| Ptf1a-r (pancreas associated transcription factor 1a-related) | Ptfb | not determined | KH.L116.39 KY.Chr11.543 | SV | [50]; Figure 3 | Coronet cells | Ventral SV |
| Tcf3 (transcription factor 3) | E12/E47 E2A | not determined | KH.C3.480 KY.Chr3.781 | Diffuse signal, predominant in SV # | [50]; Figure 3 | ||
| Tcf15-r (transcription factor 15-related) | Paraxis-like | SCX; TAL2; TCF15 | KH.S781.11 KY.Chr11.73 | No expression | [160] | ||
| Twist-r.a (twist family bHLH transcription factor-related.a) | Twist-like-1a | ATOH1; NEUROD6; PTF1A | KH.C5.416 KY.Chr5.356 | TLCs | [164] | ||
| Twist-r.b (twist family bHLH transcription factor-related.b) | Twist-like-1b | ATOH1; NEUROD6; NEUROG2 | KH.C5.554 KY.Chr5.355 | Mesench. | [124] | ||
| Twist-r.c (twist family bHLH transcription factor-related.c) | Twist-like-2 | TAL2; TWIST1; TWIST2 | KH.C5.202 KY.Chr5.357 | Mesench., TLCs | [50] | Mesench. | |
| AP4 (transcription factor AP-4) | SREBF1; TFAP4; TWIST1 | KH.C14.448 KY.Chr14.930 | Mesench., SV | [50]; Figure 3 | |||
| Figla-r (Folliculogenesis specific bHLH transcription factor-related) | FIGLA; NHLH1; TAL2 | KH.C2.1152 KY.Chr2.2108 | Not analyzed | ||||
| Mad (Mothers against dpp) | Noto7 | MXD1; MXD4; MXI1 | KH.C1.661 KY.Chr1.761 | SV, MG, palps, notochord, tail epid., tail muscles | [50,179] | Neurons, notochord | Dorsolat. SV, ventral SV, Endod., Epid. |
| Max (Myc associated factor X) | Not determined | KH.C5.373 KY.Chr5.121 | Mesench. | [50] | ANB, aSV, MG | Epid., Mesench., Endod. | |
| Mitf (Microphthalmia-associated transcription factor) | MITF; TFE3; TFEB | KH.C10.106 KY.Chr10.837 | Mesench., pigmented cells (otolith, ocellus), MG | [50,120]; Figure 3 | Collocytes aATENs | Epid. | |
| Mlx (Max-like protein x) | MLXIP; MLXIPL | KH.C11.706 KY.Chr11.477 | Mesench. | [50] | |||
| Mnt-r (Max network transcriptional repressor-related) | Mnt-like | MNT; MXD1; MXD3 | KH.L20.34 KY.Chr6.608 | No expression; a faint signal in SV, palps and mesench. might be present in late tailbuds | [50] | ||
| Myc (Myelocytomatosis) | MNT; MYC; MYCN | KH.L24.23 KY.Chr1.686 | Mesench., anterior SV, trunk endod.; in juveniles: endostyle pharyngeal gills, heart, intestine, body wall muscle, neural gland epidermis, stomach, esophagus | [50,145] Figure 3 | ANB | Endod., Epid. | |
| Srebp (Sterol regulatory element-binding transcription factor 1) | SREBF1; SREBF2 | KH.L99.12 KY.Chr9.7 | SV, palps; epid. (in larvae) | [50,141] Figure 3 | NC | SV wall, ventral SV | |
| Usf (Upstream transcription factor) | USF1; USF2; USF3 | KH.C3.624 KY.Chr3.1438 | Mesench., faint signal in SV and NC | [50]; Figure 3 | Dorsolat. SV, Mesench., Epid. | ||
| AHR (Aryl hydrocarbon receptor) | AHR; AHRR; SIM2 | KH.C12.93 KY.Chr12.869 | Mesench., palps, NC | [50] | Collocytes aATENs | Epid. | |
| ARNT (Aryl hydrocarbon receptor nuclear translocator) | ARNT; ARNT2; ARNTL | KH.C5.213 KY.Chr5.617 | Weak mesench., notochord, unclear signal in epidermis; in juveniles: endostyle pharyngeal gills, neural gland, stomach, esophagus | [50,136,180] | |||
| Hif (Hypoxia inducible factor) | EPAS1; HIF1A; HIF3A | KH.C4.83 KY.Chr4.583 | Mesench., anterior SV, palps, ventral midline, tail epid.; in juveniles: endostyle pharyngeal gills, heart, intestine, body wall muscle, neural gland epidermis, stomach, esophagus | [30,50]; Figure 3 | aSV | ||
| Sim (Single-minded) | Not determined | KH.L20.56 KY.Chr6.618 | Not analyzed | ||||
| Trh (Trachealess) | NPAS1; NPAS3; SIM2 | KH.L154.23 KY.Chr11.674 | Mesench., very weak SV | [50] | |||
| Id.a (inhibitor of DNA binding.a) | Emc | ID2; ID3 | KH.C7.692 KY.Chr7.1153 | Not analyzed | SV wall, mesench., epid. | ||
| Id.b (inhibitor of DNA binding.b) | Emc2 | ID1; ID2; ID3 | KH.C7.157 KY.Chr7.1157 | SV, MG, NC; palps, tail epid., ESNs | [50] | NC, aSV | SV wall, ventral SV, Endod., Mesench. |
| Hes.a (hairy and enhancer of split.a) | E(spl)/hairy-a | HES1; HES2; HES4 | KH.C1.159 KY.Chr1.28 | Muscle, SV, epid. | [50]; Figure 3 | NC | Epid., Mesench., Endod., SV wall, dorsolat. SV |
| Hes.b (hairy and enhancer of split.b) | E(spl)/hairy-b | HES1; HES4; HES6 | KH.C3.312 KY.Chr3.580 | Patchy SV, trunk epid., rows of tail epid.; in juveniles: body wall muscle, stigmatal cells | [50,137,187] | Epend., NC, aSV, MHB, SV | Dorsolat. SV, Epid., SV wall, Endod. |
| Hes.c (hairy and enhancer of split.c) | E(spl)/hairy-c | HES1; HES2; HES4 | KH.L34.9 KY.Chr1.1234 | No distinct zygotic signal, ubiquitous staining throughout embryogenesis | [50] | Collocytes | Endod., epid. |
| Hey (hes related family bHLH transcription factor with YRPW motif) | HEY1; HEY2; HEYL | KH.L130.6 KY.Chr10.1431 | No distinct zygotic signal | [50] | Endod., epid., mesench. | ||
| Ebf (Ebf transcription factor) | COE | EBF1; EBF2; EBF3 | KH.L24.10 KY.Chr1.724 | SV, MG; neurohypophysis primordium; tail epid. | [50]; Figure 3 | Epend., Eminens cell, pSV aSV, MG | Dorsolat. SV |
| bHLH-like1 | CCDC169-SOHLH2; SOHLH1; SOHLH2 | KH.C9.380 KY.Chr9.350 | Not analyzed | ||||
| Bhlh-tun1 (Tunicate bhlh 1) | Orphan bHLH-1 | Not determined | KH.C7.269 KY.Chr7.1158 | SV, palps, notochord, epid. | [50,166]; Figure 3 | NC, aSV, PSCs related, pigment cells, MHB, SV, pSV, Epend., Notochord | SV wall, ESNs, ventral SV, epid., endod., mesench. |
| Bhlh-tun2 (Tunicate bhlh 2) | Orphan bHLH-2 | NHLH1; NHLH2; TAL2 | KH.C4.649 KY.Chr4.1008 | SV, BTNs, mesench. | [50,108]; Figure 3 | aSV, MG, Eminens cell | Dorsolat. SV |
| Bhlh-tun3 (Tunicate bhlh 3) | Orphan bHLH-3 | Not determined | KY.Chr10.1238 | Not analyzed | |||
| Bhlh-tun4 (Tunicate bhlh 4) | Orphan bHLH-4 | Not determined | KH.L41.39 KY.Chr4.1211 | No distinct zygotic signal | [50] | ||
| Tcf4 (transcription factor 4) | TFDP1; TFDP2; TFDP3 | KH.L60.12 KY.Chr1.10 | No distinct zygotic signal; weak mesench. throughout embryogenesis | [50] | |||
| Phylogenetic Group | Characteristics | C. robusta bHLH Genes |
|---|---|---|
| A | Bind to CAGCTG or CACCTG | Ascl.a, Ascl.b, Ascl.c *, Atoh8 *, Atonal, Bhlha15, Hand, Hand-r *, Mesp, Mrf, Neurog *, Ptf1a *, Ptf1a-r *, Tcf3 *, Tcf15-r, Twist-r.a, Twist-r.b, Twist-r.c |
| B | Bind to CACGTG or CATGTTG | AP4*, Figla-r #, Mad *, Max, Mitf *, Mlx, Mnt-r *, Myc *, Srebp *, Usf * |
| C | Bind to ACGTG or GCGTG. Contain a PAS domain | AHR, ARNT, Hif *, Sim #, Trh |
| D | Lack basic domain and do not bind to DNA. Act as antagonists of group A bHLH proteins | Id.a #, Id.b * |
| E | Contain an orange domain and a WRPW peptide | Hes.a *, Hes.b *, Hes.c, Hey |
| F | Contain an additional COE domain, which is involved in dimerization and DNA binding | Ebf * |
| Outgroup | Bhlh-tun1 *, Bhlh-tun2 *, Bhlh-tun3 #, Bhlh-tun4 Uncertain classification: bHLH-like1 # |
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANR | anterior neural ridge; |
| bHLH | basic helix-loop-helix |
| BTN | bipolar tail neuron |
| CNS | central nervous system; |
| FGF | Fibroblast growth factor |
| GMP | 3′,5′-cyclic guanosine monophosphate |
| GTP | guanosine-5′-triphosphate |
| hr(s) | hour(s); |
| kb | kilobase(s), or 1000 base pairs; |
| MG | motor ganglion; |
| MHB | midbrain-hindbrain boundary; |
| MO | morpholino oligonucleotide; |
| NC | nerve cord; |
| SV | sensory vesicle; |
| TF(s) | transcription factor(s); |
| WMISH | whole-mount in situ hybridization; |
| ZLI | zona limitans intrathalamica. |
References
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Chabry, L. Contribution a l’embryologie normale et teratologique des Ascidies simples. J. Anat. Physiol. (Paris) 1887, 23, 167–319. [Google Scholar]
- Conklin, E.G. The organization and cell-lineage of the ascidian egg. Acad. Nat. Sci. 1905. [CrossRef]
- Lawrence, P.A.; Levine, M. Mosaic and regulative development: Two faces of one coin. Curr. Biol. 2006, 16, R236–R239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortolani, G. Risultati definitivi sulla distribuzione dei territory presuntivi degli organi nel germe di Ascidie allo stadio VIII, determinati con le marche al carbone. Section II: History and Philosophy of the Life Sciences. Pubbl. Stn. Zool. Napoli 1954, 25, 161–187. [Google Scholar]
- Anderson, H.E.; Christiaen, L. Ciona as a Simple Chordate Model for Heart Development and Regeneration. J. Cardiovasc. Dev. Dis. 2016, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Horie, R.; Hazbun, A.; Chen, K.; Cao, C.; Levine, M.; Horie, T. Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 2018, 560, 228–232. [Google Scholar] [CrossRef]
- Lemaire, P.; Smith, W.C.; Nishida, H. Ascidians and the Plasticity of the Chordate Developmental Program. Curr. Biol. 2008, 18, R620–R631. [Google Scholar] [CrossRef] [Green Version]
- Satoh, N. Developmental Biology of Ascidians; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Satoh, N. The ascidian tadpole larva: Comparative molecular development and genomics. Nat. Rev. Genet. 2003, 4, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Smith, W.C. Ascidian notochord morphogenesis. Dev. Dyn. 2007, 236, 1748–1757. [Google Scholar] [CrossRef] [Green Version]
- Di Gregorio, A. The notochord gene regulatory network in chordate evolution: Conservation and divergence from Ciona to vertebrates. Curr. Top. Dev. Biol. 2020, 139, 325–374. [Google Scholar] [CrossRef] [PubMed]
- Maguire, J.E.; Pandey, A.; Wu, Y.; Di Gregorio, A. Investigating Evolutionarily Conserved Molecular Mechanisms Controlling Gene Expression in the Notochord. Adv. Exp. Med. Biol. 2018, 1029, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Corbo, J.C.; Erives, A.; Di Gregorio, A.; Chang, A.; Levine, M. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development 1997, 124, 2335–2344. [Google Scholar] [PubMed]
- Hashimoto, H.; Robin, F.B.; Sherrard, K.M.; Munro, E. Sequential Contraction and Exchange of Apical Junctions Drives Zippering and Neural Tube Closure in a Simple Chordate. Dev. Cell 2015, 32, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.S.; Satoh, N.; Satou, Y. Region specific gene expressions in the central nervous system of the ascidian embryo. Mech. Dev. 2002, 119 (Suppl. 1), S275–S277. [Google Scholar] [CrossRef]
- Meinertzhagen, I.A.; Okamura, Y. The larval ascidian nervous system: The chordate brain from its small beginnings. Trends Neurosci. 2001, 24, 401–410. [Google Scholar] [CrossRef]
- Stolfi, A.; Gainous, T.B.; Young, J.J.; Mori, A.; Levine, M.; Christiaen, L. Early Chordate Origins of the Vertebrate Second Heart Field. Science 2010, 329, 565–568. [Google Scholar] [CrossRef] [Green Version]
- Tolkin, T.; Christiaen, L. Development and Evolution of the Ascidian Cardiogenic Mesoderm. Curr. Top. Dev. Biol. 2012, 100, 107–142. [Google Scholar] [CrossRef]
- Razy-Krajka, F.; Gravez, B.; Kaplan, N.; Racioppi, C.; Wang, W.; Christiaen, L. An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. ELife 2018, 7. [Google Scholar] [CrossRef]
- Wang, B.T.; Yu, X.Y.; Zhu, Y.J.; Zhuang, M.; Zhang, Z.M.; Jin, L.; Jin, F.J. Research progress on the basic helix-loop-helix transcription factors of Aspergillus species. Adv. Appl. Microbiol. 2019, 109, 31–59. [Google Scholar] [CrossRef]
- Racioppi, C.; A Wiechecki, K.A.; Christiaen, L. Combinatorial chromatin dynamics foster accurate cardiopharyngeal fate choices. ELife 2019, 8, e49921. [Google Scholar] [CrossRef] [PubMed]
- Patricolo, E.; Cammarata, M.; D’Agati, P. Presence of thyroid hormones in ascidian larvae and their involvement in metamorphosis. J. Exp. Zoöl. 2001, 290, 426–430. [Google Scholar] [CrossRef]
- Nakayama, S.; Ogasawara, M. Compartmentalized expression patterns of pancreatic- and gastric-related genes in the alimentary canal of the ascidian Ciona intestinalis: Evolutionary insights into the functional regionality of the gastrointestinal tract in Olfactores. Cell Tissue Res. 2017, 370, 113–128. [Google Scholar] [CrossRef]
- Thompson, J.M.; Di Gregorio, A. Insulin-like genes in ascidians: Findings in Ciona and hypotheses on the evolutionary origins of the pancreas. Genesis 2015, 53, 82–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capellini, T.D.; Dunn, M.P.; Passamaneck, Y.J.; Selleri, L.; Di Gregorio, A. Conservation of notochord gene expression across chordates: Insights from theLeprecangene family. Genesis 2008, 46, 683–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; Lemaire, L.A.; Yoon, P.H.; Choi, Y.A.; Parsons, L.R.; Matese, J.C.; Wang, W.; Levine, M.; Chen, K. Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 2019, 571, 349–354. [Google Scholar] [CrossRef]
- José-Edwards, D.S.; Oda-Ishii, I.; Kugler, J.E.; Passamaneck, Y.J.; Katikala, L.; Nibu, Y.; Di Gregorio, A. Brachyury, Foxa2 and the cis-Regulatory Origins of the Notochord. PLoS Genet. 2015, 11, e1005730. [Google Scholar] [CrossRef] [Green Version]
- Moret, F.; Christiaen, L.; Deyts, C.; Blin, M.; Joly, J.S.; Vernier, P. The dopamine-synthesizing cells in the swimming larva of the tunicate Ciona intestinalis are located only in the hypothalamus-related domain of the sensory vesicle. Eur. J. Neurosci. 2005, 21, 3043–3055. [Google Scholar] [CrossRef]
- Moret, F.; Christiaen, L.; Deyts, C.; Blin, M.; Vernier, P.; Joly, J.S. Regulatory gene expressions in the ascidian ventral sensory vesicle: Evolutionary relationships with the vertebrate hypothalamus. Dev. Biol. 2005, 277, 567–579. [Google Scholar] [CrossRef]
- Sharma, S.; Wang, W.; Stolfi, A. Single-cell transcriptome profiling of the Ciona larval brain. Dev. Biol. 2019, 448, 226–236. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Schwinn, K.E. Gene regulation networks generate diverse pigmentation patterns in plants. Plant Signal. Behav. 2014, 9, e29526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Robe, K.; Gaymard, F.; Izquierdo, E.; Dubos, C. The Transcriptional Control of Iron Homeostasis in Plants: A Tale of bHLH Transcription Factors? Front. Plant Sci. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef]
- Richards, G.S.; Simionato, E.; Perron, M.; Adamska, M.; Vervoort, M.; Degnan, B.M. Sponge Genes Provide New Insight into the Evolutionary Origin of the Neurogenic Circuit. Curr. Biol. 2008, 18, 1156–1161. [Google Scholar] [CrossRef]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Biol. 2007, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Gyoja, F. Basic helix-loop-helix transcription factors in evolution: Roles in development of mesoderm and neural tissues. Genesis 2017, 55. [Google Scholar] [CrossRef]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef]
- Bertrand, N.; Castro, D.S.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517–530. [Google Scholar] [CrossRef]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef]
- Fu, Y.; Yuan, S.-S.; Zhang, L.-J.; Ji, Z.-L.; Quan, X.-J. Atonal bHLH transcription factor 1 is an important factor for maintaining the balance of cell proliferation and differentiation in tumorigenesis. Oncol. Lett. 2020, 20, 2595–2605. [Google Scholar] [CrossRef]
- Zhu, Q.Q.; Ma, C.; Wang, Q.; Song, Y.; Lv, T. The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumor Biol. 2016, 37, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Hattori, D.; Kitayama, A.; Ueno, N.; Taira, M. Identification of target genes for the Xenopus Hes-related protein XHR1, a prepattern factor specifying the midbrain–hindbrain boundary. Dev. Biol. 2005, 283, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vue, T.Y.; Aaker, J.; Taniguchi, A.; Kazemzadeh, C.; Skidmore, J.M.; Martin, D.M.; Martin, J.F.; Treier, M.; Nakagawa, Y. Characterization of progenitor domains in the developing mouse thalamus. J. Comp. Neurol. 2007, 505, 73–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Chan, J.A.; Schuurmans, C. Proneural bHLH Genes in Development and Disease. Curr. Top. Dev. Biol. 2014, 110, 75–127. [Google Scholar] [CrossRef]
- Corbo, J.C.; Di Gregorio, A.; Levine, M. The ascidian as a model organism in developmental and evolutionary biology. Cell 2001. [Google Scholar] [CrossRef] [Green Version]
- Passamaneck, Y.J.; Di Gregorio, A. Ciona intestinalis: Chordate development made simple. Dev. Dyn. 2005, 233, 1–19. [Google Scholar] [CrossRef]
- Ryan, K.; Lu, Z.; Meinertzhagen, I.A. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. ELife 2016, 5, e16962. [Google Scholar] [CrossRef]
- Di Gregorio, A.; Corbo, J.C.; Levine, M. The Regulation of forkhead/HNF-3β Expression in the Ciona Embryo. Dev. Biol. 2001, 229, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Imai, K.S.; Hino, K.; Yagi, K.; Satoh, N.; Satou, Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Towards a comprehensive understanding of gene networks. Development 2004, 131, 4047–4058. [Google Scholar] [CrossRef] [Green Version]
- José-Edwards, D.S.; Kerner, P.; Kugler, J.E.; Deng, W.; Jiang, D.; Di Gregorio, A. The identification of transcription factors expressed in the notochord of Ciona intestinalis adds new potential players to the brachyury gene regulatory network. Dev. Dyn. 2011, 240, 1793–1805. [Google Scholar] [CrossRef] [Green Version]
- Yasuo, H.; Satoh, N. Function of vertebrate T gene. Nature 1993, 364, 582–583. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, T.; Yoshida, N.; Satoh, N.; Saiga, H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. USA 2004, 101, 15118–15123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keys, D.N.; Lee, B.I.; Di Gregorio, A.; Harafuji, N.; Detter, J.C.; Wang, M.; Kahsai, O.; Ahn, S.; Zhang, C.; Doyle, S.A.; et al. A saturation screen for cis-acting regulatory DNA in the Hox genes of Ciona intestinalis. Proc. Natl. Acad. Sci. USA 2005, 102, 679–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albuixech-Crespo, B.; Blanch, L.L.; Burguera, D.; Maeso, I.; Sánchez-Arrones, L.; A Moreno-Bravo, J.; Somorjai, I.M.; Pascual-Anaya, J.; Puelles, E.; Bovolenta, P.; et al. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS Biol. 2017, 15, e2001573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puelles, L.; Rubenstein, J.L. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 1993, 16, 472–479. [Google Scholar] [CrossRef]
- Vieira, C.; Pombero, A.; Garcia-Lopez, R.; Gimeno, L.; Echevarria, D.; Martinez, S. Molecular mechanisms controlling brain development: An overview of neuroepithelial secondary organizers. Int. J. Dev. Biol. 2010, 54, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.; Stern, C.D. Organizers in Development. Curr. Top. Dev. Biol. 2016, 117, 435–454. [Google Scholar] [CrossRef]
- Martinez Arias, A.; Steventon, B. On the nature and function of organizers. Development 2018, 145, dev159525. [Google Scholar] [CrossRef] [Green Version]
- Pani, A.M.; Mullarkey, E.E.; Aronowicz, J.; Assimacopoulos, S.; Grove, E.A.; Lowe, C.J. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 2012, 483, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Holland, P.W. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrateorigins of midbrain development. Development 2004, 131, 3285–3294. [Google Scholar] [CrossRef] [Green Version]
- Ikuta, T.; Saiga, H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: Revisit to the tripartite model and the origin of the midbrain–hindbrain boundary region. Dev. Biol. 2007, 312, 631–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.S.; Stolfi, A.; Levine, M.; Satou, Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 2009, 136, 285–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubenstein, J.L.; Shimamura, K.; Martinez, S.; Puelles, L. Regionalization of the prosencephalic neural plate. Ann. Rev. Neurosci. 1998, 21, 445–477. [Google Scholar] [CrossRef] [PubMed]
- Dickmeis, T.; Rastegar, S.; Lam, C.S.; Aanstad, P.; Clark, M.; Fischer, N.; Rosa, F.; Korzh, V.; Strähle, U. Expression of the helix-loop-helix gene id3 in the zebrafish embryo. Mech. Dev. 2002, 113, 99–102. [Google Scholar] [CrossRef]
- Ross, S.E.; Greenberg, M.E.; Stiles, C.D. Basic Helix-Loop-Helix Factors in Cortical Development. Neuron 2003, 39, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Moret, F.; Guilland, J.C.; Coudouel, S.; Rochette, L.; Vernier, P. Distribution of tyrosine hydroxylase, dopamine, and serotonin in the central nervous system of amphioxus (Branchiostoma lanceolatum): Implications for the evolution of catecholamine systems in vertebrates. J. Comp. Neurol. 2004, 468, 135–150. [Google Scholar] [CrossRef]
- Manni, L.; Agnoletto, A.; Zaniolo, G.; Burighel, P. Stomodeal and neurohypophysial placodes in Ciona intestinalis: Insights into the origin of the pituitary gland. J. Exp. Zoöl. B Mol. Dev. Evol. 2005, 304, 324–339. [Google Scholar] [CrossRef]
- Christiaen, L.; Jaszczyszyn, Y.; Kerfant, M.; Kano, S.; Thermes, V.; Joly, J.-S. Evolutionary modification of mouth position in deuterostomes. Semin. Cell Dev. Biol. 2007, 18, 502–511. [Google Scholar] [CrossRef]
- Veeman, M.T.; Newman-Smith, E.; El-Nachef, D.; Smith, W.C. The ascidian mouth opening is derived from the anterior neuropore: Reassessing the mouth/neural tube relationship in chordate evolution. Dev. Biol. 2010, 344, 138–149. [Google Scholar] [CrossRef] [Green Version]
- Glardon, S.; Holland, L.Z.; Gehring, W.J.; Holland, N.D. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): Insights into eye and photoreceptor evolution. Development 1998, 125, 2701–2710. [Google Scholar] [PubMed]
- Candiani, S.; Holland, N.D.; Oliveri, D.; Parodi, M.; Pestarino, M. Expression of the amphioxus Pit-1 gene (AmphiPOU1F1/Pit-1) exclusively in the developing preoral organ, a putative homolog of the vertebrate adenohypophysis. Brain Res. Bull. 2008, 75, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Candiani, S.; Pestarino, M. Evidence for the presence of the tissue-specific transcription factor Pit-1 in lancelet larvae. J. Comp. Neurol. 1998, 400, 310–316. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Xing, C.; Zhang, H.; Shimeld, S.M.; Wang, Y. Cerberus–Nodal–Lefty–Pitx signaling cascade controls left–right asymmetry in amphioxus. Proc. Natl. Acad. Sci. USA 2017, 114, 3684–3689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kano, S. Genomics and Developmental Approaches to an Ascidian Adenohypophysis Primordium. Integr. Comp. Biol. 2010, 50, 35–52. [Google Scholar] [CrossRef] [Green Version]
- Christiaen, L.; Burighel, P.; Smith, W.C.; Vernier, P.; Bourrat, F.; Joly, J.-S. Pitx genes in Tunicates provide new molecular insight into the evolutionary origin of pituitary. Gene 2002, 287, 107–113. [Google Scholar] [CrossRef]
- Christiaen, L.; Bourrat, F.; Joly, J.-S. A modular cis-regulatory system controls isoform-specific pitx expression in ascidian stomodæum. Dev. Biol. 2005, 277, 557–566. [Google Scholar] [CrossRef]
- Di Fiore, M.M.; Rastogi, R.K.; Ceciliani, F.; Messi, E.; Botte, V.; Botte, L.; Pinelli, C.; D’Aniello, B.; D’Aniello, A. Mammalian and chicken I forms of gonadotropin-releasing hormone in the gonads of a protochordate, Ciona intestinalis. Proc. Natl. Acad. Sci. USA 2000, 97, 2343–2348. [Google Scholar] [CrossRef] [Green Version]
- Manni, L.; Lane, N.J.; Burighel, P.; Zaniolo, G. Are neural crest and placodes exclusive to vertebrates? Evol. Dev. 2001, 3, 297–298. [Google Scholar] [CrossRef]
- Pestarino, M. Immunocytochemical demonstration of prolactin-like activity in the neural gland of the ascidian Styela plicata. Gen. Comp. Endocrinol. 1984, 54, 444–449. [Google Scholar] [CrossRef]
- Boorman, C.J.; Shimeld, S.M. The evolution of left-right asymmetry in chordates. BioEssays 2002, 24, 1004–1011. [Google Scholar] [CrossRef]
- Poulin, G.; Lebel, M.; Chamberland, M.; Paradis, F.W.; Drouin, J.; Hall, C.; Nelson, D.M.; Ye, X.; Baker, K.; DeCaprio, J.A.; et al. Specific Protein-Protein Interaction between Basic Helix-Loop-Helix Transcription Factors and Homeoproteins of the Pitx Family. Mol. Cell. Biol. 2000, 20, 4826–4837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, M.; Goto, M.; Hojo, M.; Kita, A.; Kitagawa, M.; Ohtsuka, T.; Kageyama, R.; Miyamoto, S. The proneural bHLH genes Mash1, Math3 and NeuroD are required for pituitary development. J. Mol. Endocrinol. 2018, 61, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Joyce Tang, W.; Chen, J.S.; Zeller, R.W. Transcriptional regulation of the peripheral nervous system in Ciona intestinalis. Dev. Biol. 2013, 378, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Osugi, T.; Sasakura, Y.; Esatake, H. The nervous system of the adult ascidian Ciona intestinalis Type A (Ciona robusta): Insights from transgenic animal models. PLoS ONE 2017, 12, e0180227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, K.; Meinertzhagen, I.A. Neuronal identity: The neuron types of a simple chordate sibling, the tadpole larva of Ciona intestinalis. Curr. Opin. Neurobiol. 2019, 56, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Halluin, C.; Madelaine, R.; Naye, F.; Peers, B.; Roussigné, M.; Blader, P. Habenular Neurogenesis in Zebrafish Is Regulated by a Hedgehog, Pax6 Proneural Gene Cascade. PLoS ONE 2016, 11, e0158210. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, E.M.; Bailey, M.J.; Rath, M.F.; Shi, Q.; Morin, F.; Coon, S.L.; Møller, M.; Klein, D.C. NeuroD1: Developmental expression and regulated genes in the rodent pineal gland. J. Neurochem. 2007, 102, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Lacalli, T. Amphioxus, motion detection, and the evolutionary origin of the vertebrate retinotectal map. EvoDevo 2018, 9, 6. [Google Scholar] [CrossRef]
- Pergner, J.; Kozmik, Z. Amphioxus photoreceptors—Insights into the evolution of vertebrate opsins, vision and circadian rhythmicity. Int. J. Dev. Biol. 2017, 61, 665–681. [Google Scholar] [CrossRef] [Green Version]
- Cau, E.; Wilson, S.W. Ash1a and Neurogenin1 function downstream of Floating head to regulate epiphysial neurogenesis. Development 2003, 130, 2455–2466. [Google Scholar] [CrossRef] [Green Version]
- Utsumi, N.; Shimojima, Y.; Saiga, H. Analysis of ascidian Not genes highlights their evolutionarily conserved and derived features of structure and expression in development. Dev. Genes Evol. 2004, 214, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, T.; Tsuda, M. Photoreceptive Systems in Ascidians. Photochem. Photobiol. 2007, 83, 248–252. [Google Scholar] [CrossRef]
- Jamieson, D.; Roberts, A. Responses of young Xenopus laevis tadpoles to light dimming: Possible roles for the pineal eye. J. Exp. Biol. 2000, 203 Pt 12, 1857–1867. [Google Scholar] [PubMed]
- Kako, K.; Ishida, N. The role of transcription factors in circadian gene expression. Neurosci. Res. 1998, 31, 257–264. [Google Scholar] [CrossRef]
- Minamoto, T.; Hanai, S.; Kadota, K.; Oishi, K.; Matsumae, H.; Fujie, M.; Azumi, K.; Satoh, N.; Satake, M.; Ishida, N. Circadian clock in Ciona intestinalis revealed by microarray analysis and oxygen consumption. J. Biochem. 2010, 147, 175–184. [Google Scholar] [CrossRef]
- Zega, G.; Biggiogero, M.; Groppelli, S.; Candiani, S.; Oliveri, D.; Parodi, M.; Pestarino, M.; De Bernardi, F.; Pennati, R. Developmental expression ofglutamic acid decarboxylaseand ofγ-aminobutyricacid type B receptors in the ascidian Ciona intestinalis. J. Comp. Neurol. 2008, 506, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.; Dondorp, D.; Canon, L.; Tieo, S.; Chatzigeorgiou, M. Automated behavioural analysis reveals the basic behavioural repertoire of the urochordate Ciona intestinalis. Sci. Rep. 2019, 9, 2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, M.; Miyamoto, T.; Ohkuma, M.; Tsuda, M. Action spectrum for the photophobic response of Ciona intestinalis (Ascidieacea, Urochordata) larvae implicates retinal protein. Photochem. Photobiol. 1999, 70, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, I.; Shiraishi, S.; Tsuda, M. Photoresponse and Learning Behavior of Ascidian Larvae, a Primitive Chordate, to Repeated Stimuli of Step-Up and Step-Down of Light. J. Biol. Phys. 2002, 28, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Kusakabe, T.; Iwamoto, H.; Horie, T.; Nakashima, Y.; Nakagawa, M.; Okunou, K. Origin of the vertebrate visual cycle: II. Visual cycle proteins are localized in whole brain including photoreceptor cells of a primitive chordate. Vis. Res. 2003, 43, 3045–3053. [Google Scholar] [CrossRef] [Green Version]
- Bostwick, M.; Smith, E.L.; Borba, C.; Newman-Smith, E.; Guleria, I.; Kourakis, M.J.; Smith, W.C. Antagonistic Inhibitory Circuits Integrate Visual and Gravitactic Behaviors. Curr. Biol. 2020, 30, 600–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kratsios, P.; Stolfi, A.; Levine, M.; Hobert, O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 2011, 15, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaire, P.; Bertrand, V.; Hudson, C. Early steps in the formation of neural tissue in ascidian embryos. Dev. Biol. 2002, 252, 151–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolfi, A.; Ryan, K.; Meinertzhagen, I.A.; Christiaen, L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nat. Cell Biol. 2015, 527, 371–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, J.H.; Meinertzhagen, I.A. Neurons of the ascidian larval nervous system in Ciona intestinalis: I. Central nervous system. J. Comp. Neurol. 2007, 501, 316–334. [Google Scholar] [CrossRef]
- Ryan, K.; Lu, Z.; Meinertzhagen, I.A. The peripheral nervous system of the ascidian tadpole larva: Types of neurons and their synaptic networks. J. Comp. Neurol. 2018, 526, 583–608. [Google Scholar] [CrossRef]
- Kim, K.; Gibboney, S.; Razy-Krajka, F.; Lowe, E.K.; Wang, W.; Stolfi, A. Regulation of Neurogenesis by FGF Signaling and Neurogenin in the Invertebrate Chordate Ciona. Front. Cell Dev. Biol. 2020, 8, 477. [Google Scholar] [CrossRef]
- Horie, T.; Nakagawa, M.; Sasakura, Y.; Kusakabe, T. Cell type and function of neurons in the ascidian nervous system. Dev. Growth Differ. 2009, 51, 207–220. [Google Scholar] [CrossRef]
- Sorrentino, M.; Manni, L.; Lane, N.J.; Burighel, P. Evolution of cerebral vesicles and their sensory organs in an ascidian larva. Acta Zool. 2001, 81, 243–258. [Google Scholar] [CrossRef]
- Dilly, P.N. Studies on the receptors in the cerebral vesicle of the ascidian tadpole. 1. The Otolith. Q. J. Microsc. Sci. 1964, 103, 393–398. [Google Scholar]
- Hudson, C. The central nervous system of ascidian larvae. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 538–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilly, P.N. Studies on the receptors in the cerebral vesicle of the ascidian tadpole. 2. The Ocellus. Q. J. Microsc. Sci. 1964, 105, 13–20. [Google Scholar]
- Horie, T.; Sakurai, D.; Ohtsuki, H.; Terakita, A.; Shichida, Y.; Usukura, J.; Kusakabe, T.; Tsuda, M. Pigmented and nonpigmented ocelli in the brain vesicle of the ascidian larva. J. Comp. Neurol. 2008, 509, 88–102. [Google Scholar] [CrossRef]
- Oonuma, K.; Tanaka, M.; Nishitsuji, K.; Kato, Y.; Shimai, K.; Kusakabe, T.G. Revised lineage of larval photoreceptor cells in Ciona reveals archetypal collaboration between neural tube and neural crest in sensory organ formation. Dev. Biol. 2016, 420, 178–185. [Google Scholar] [CrossRef]
- D’Aniello, S.; D’Aniello, E.; Locascio, A.; Memoli, A.; Corrado, M.; Russo, M.T.; Aniello, F.; Fucci, L.; Brown, E.R.; Branno, M. The ascidian homolog of the vertebrate homeobox gene Rx is essential for ocellus development and function. Differentiation 2006, 74, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Oonuma, K.; Kusakabe, T.G. Spatio-temporal regulation of Rx and mitotic patterns shape the eye-cup of the photoreceptor cells in Ciona. Dev. Biol. 2019, 445, 245–255. [Google Scholar] [CrossRef]
- Hodgkinson, C.A.; Moore, K.J.; Nakayama, A.; Steingrimsson, E.; Copeland, N.G.; Jenkins, N.A.; Arnheiter, H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993, 74, 395–404. [Google Scholar] [CrossRef]
- Yajima, I.; Endo, K.; Sato, S.; Toyoda, R.; Wada, H.; Shibahara, S.; Numakunai, T.; Ikeo, K.; Gojobori, T.; Goding, C.R.; et al. Cloning and functional analysis of ascidian Mitf in vivo: Insights into the origin of vertebrate pigment cells. Mech. Dev. 2003, 120, 1489–1504. [Google Scholar] [CrossRef]
- Abitua, P.B.; Wagner, E.; Navarrete, I.A.; Levine, M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 2012, 492, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Curran, K.; Lister, J.A.; Kunkel, G.R.; Prendergast, A.; Parichy, D.M.; Raible, D.W. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010, 344, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Soo, K.; O’Rourke, M.P.; Khooa, P.L.; Steiner, K.A.; Wonga, N.; Behringer, R.R.; Tam, P.P. Twist Function Is Required for the Morphogenesis of the Cephalic Neural Tube and the Differentiation of the Cranial Neural Crest Cells in the Mouse Embryo. Dev. Biol. 2002, 247, 251–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanDusen, N.J.; Firulli, A.B. Twist factor regulation of non-cardiomyocyte cell lineages in the developing heart. Differentiation 2012, 84, 79–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokuoka, M.; Imai, K.S.; Satou, Y.; Satoh, N. Three distinct lineages of mesenchymal cells in Ciona intestinalis embryos demonstrated by specific gene expression. Dev. Biol. 2004, 274, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, H.; Satoh, N. Determination and regulation in the pigment cell lineage of the ascidian embryo. Dev. Biol. 1989, 132, 355–367. [Google Scholar] [CrossRef]
- Razy-Krajka, F.; Brown, E.R.; Horie, T.; Callebert, J.; Sasakura, Y.; Joly, J.S.; Kusakabe, T.G.; Vernier, P. Monoaminergic modulation of photoreception in ascidian: Evidence for a proto-hypothalamo-retinal territory. BMC Biol. 2012, 10, 45. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Saiga, H. Repression of Rx gene on the left side of the sensory vesicle by Nodal signaling is crucial for right-sided formation of the ocellus photoreceptor in the development of Ciona intestinalis. Dev. Biol. 2011, 354, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Takamura, K.; Minamida, N.; Okabe, S. Neural Map of the Larval Central Nervous System in the Ascidian Ciona intestinalis. Zool. Sci. 2010, 27, 191–203; PMID: 20141424. [Google Scholar] [CrossRef]
- Sasakura, Y.; Hozumi, A. Formation of adult organs through metamorphosis in ascidians. Wiley Interdiscip. Rev. Dev. Biol. 2017, 7. [Google Scholar] [CrossRef]
- Satoh, N. An Advanced Filter-Feeder Hypothesis for Urochordate Evolution. Zool. Sci. 2009, 26, 97–111. [Google Scholar] [CrossRef]
- Horie, T.; Shinki, R.; Ogura, Y.; Kusakabe, T.; Satoh, N.; Sasakura, Y. Ependymal cells of chordate larvae are stem-like cells that form the adult nervous system. Nature 2011, 469, 525–528. [Google Scholar] [CrossRef]
- Lambert, C.C.; Brandt, C.L. The Effect of Light on the Spawning of Ciona intestinalis. Biol. Bull. 1967, 132, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Auger, H.; Sasakura, Y.; Joly, J.-S.; Jeffery, W.R. Regeneration of oral siphon pigment organs in the ascidian Ciona intestinalis. Dev. Biol. 2010, 339, 374–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkuma, M.; Katagiri, Y.; Nakagawa, M.; Tsuda, M. Possible involvement of light regulated gonadotropin-releasing hormone neurons in biological clock for reproduction in the cerebral ganglion of the ascidian, Halocynthia roretzi. Neurosci. Lett. 2000, 293, 5–8. [Google Scholar] [CrossRef]
- Tsutsui, H.; Oka, Y. Light-sensitive voltage responses in the neurons of the cerebral ganglion of Ciona savignyi (Chordata: Ascidiacea). Biol. Bull. 2000, 198, 26–28. [Google Scholar] [CrossRef]
- Ogasawara, M.; Sasaki, A.; Metoki, H.; Shin-I, T.; Kohara, Y.; Satoh, N.; Satou, Y. Gene expression profiles in young adult Ciona intestinalis. Dev. Genes Evol. 2002, 212, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Gorički, Š.; Byerly, M.S.; Satoh, N.; Jeffery, W.R. Evolution of the chordate regeneration blastema: Differential gene expression and conserved role of notch signaling during siphon regeneration in the ascidian Ciona. Dev. Biol. 2015, 405, 304–315. [Google Scholar] [CrossRef] [Green Version]
- Pennati, R.; Ficetola, G.F.; Brunetti, R.; Caicci, F.; Gasparini, F.; Griggio, F.; Sato, A.; Stach, T.; Kaul-Strehlow, S.; Gissi, C.; et al. Morphological Differences between Larvae of the Ciona intestinalis Species Complex: Hints for a Valid Taxonomic Definition of Distinct Species. PLoS ONE 2015, 10, e0122879. [Google Scholar] [CrossRef] [Green Version]
- Dehal, P.; Satou, Y.; Campbell, R.K.; Chapman, J.; Degnan, B.; De Tomaso, A.; Davidson, B.; Di Gregorio, A.; Gelpke, M.; Goodstein, D.M.; et al. The Draft Genome of Ciona intestinalis: Insights into Chordate and Vertebrate Origins. Science 2002, 298, 2157–2167. [Google Scholar] [CrossRef] [Green Version]
- Satou, Y.; Nakamura, R.; Yu, D.; Yoshida, R.; Hamada, M.; Fujie, M.; Hisata, K.; Takeda, H.; Satoh, N.; Nakmura, R.; et al. A Nearly Complete Genome of Ciona intestinalis Type A (C. robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol. Evol. 2019, 11, 3144–3157. [Google Scholar] [CrossRef] [Green Version]
- Kusakabe, T.; Yoshida, R.; Kawakami, I.; Kusakabe, R.; Mochizuki, Y.; Yamada, L.; Shin-I, T.; Kohara, Y.; Satoh, N.; Tsuda, M.; et al. Gene Expression Profiles in Tadpole Larvae of Ciona intestinalis. Dev. Biol. 2002, 242, 188–203. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, S.; Maeda, Y.; Shin-I, T.; Kohara, Y.; Takatori, N.; Satou, Y.; Satoh, N. Gene expression profiles in Ciona intestinalis cleavage-stage embryos. Mech. Dev. 2002, 112, 115–127. [Google Scholar] [CrossRef]
- Miwata, K.; Chiba, T.; Horii, R.; Yamada, L.; Kubo, A.; Miyamura, D.; Satoh, N.; Satou, Y. Systematic analysis of embryonic expression profiles of zinc finger genes in Ciona intestinalis. Dev. Biol. 2006, 292, 546–554. [Google Scholar] [CrossRef]
- Yamada, L.; Shoguchi, E.; Wada, S.; Kobayashi, K.; Mochizuki, Y.; Satou, Y.; Satoh, N. Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development 2003, 130, 6485–6495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamada, M.; Wada, S.; Kobayashi, K.; Satoh, N. Novel genes involved in Ciona intestinalis embryogenesis: Characterization of gene knockdown embryos. Dev. Dyn. 2007, 236, 1820–1831. [Google Scholar] [CrossRef] [PubMed]
- Nitta, K.R.; Vincentelli, R.; Jacox, E.; Cimino, A.; Ohtsuka, Y.; Sobral, D.; Satou, Y.; Cambillau, C.; Lemaire, P. High-Throughput Protein Production Combined with High- Throughput SELEX Identifies an Extensive Atlas of Ciona robusta Transcription Factor DNA-Binding Specificities. Methods Mol. Biol. 2019, 2025, 487–517. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Imai, K.S.; Levine, M.; Kohara, Y.; Rokhsar, D.; Satoh, N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. Dev. Genes Evol. 2003, 213, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004, 5, 226. [Google Scholar] [CrossRef] [Green Version]
- Menon, S.; Lawrence, C. Helix-Turn-Helix Motif. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier BV: Amsterdam, The Netherlands, 2013; pp. 412–415. [Google Scholar]
- Murre, C. Helix–loop–helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev. 2019, 33, 6–25. [Google Scholar] [CrossRef] [Green Version]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Sato, F.; Bhawal, U.K. The basic helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates Twist1 through an E-box element. Biochem. Biophys. Res. Commun. 2014, 455, 390–395. [Google Scholar] [CrossRef]
- Di Gregorio, A.; Spagnuolo, A.; Ristoratore, F.; Pischetola, M.; Aniello, F.; Branno, M.; Cariello, L.; Di Lauro, R. Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene 1995, 156, 253–257. [Google Scholar] [CrossRef]
- Wada, S.; Tokuoka, M.; Shoguchi, E.; Kobayashi, K.; Di Gregorio, A.; Spagnuolo, A.; Branno, M.; Kohara, Y.; Rokhsar, D.S.; Levine, M.; et al. A genomewide survey of developmentally relevant genes in Ciona intestinalis. Dev. Genes Evol. 2003, 213, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, A. T-Box Genes and Developmental Gene Regulatory Networks in Ascidians. Curr. Top. Dev. Biol. 2017, 122, 55–91. [Google Scholar] [CrossRef]
- Meedel, T.H.; Farmer, S.C.; Lee, J.J. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: Implications for the evolution of chordate muscle gene regulation. Development 1997, 124, 1711–1721. [Google Scholar] [PubMed]
- Ratcliffe, L.E.; Asiedu, E.K.; Pickett, C.J.; Warburton, M.A.; Izzi, S.A.; Meedel, T.H. The Ciona myogenic regulatory factor functions as a typical MRF but possesses a novel N-terminus that is essential for activity. Dev. Biol. 2019, 448, 210–225. [Google Scholar] [CrossRef]
- Aase-Remedios, M.E.; Coll-Lladó, C.; Ferrier, D.E.K. More than one-to-four via 2R: Evidence of an independent amphioxus expansion and two-gene ancestral vertebrate state for MyoD-related Muscle Regulatory Factors (MRFs). Mol. Biol. Evol. 2020, 147. [Google Scholar] [CrossRef]
- Davidson, B.; Levine, M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc. Natl. Acad. Sci. USA 2003, 100, 11469–11473. [Google Scholar] [CrossRef] [Green Version]
- Satou, Y.; Imai, K.S.; Satoh, N. The ascidian Mesp gene specifies heart precursor cells. Development 2004, 131, 2533–2541. [Google Scholar] [CrossRef] [Green Version]
- Waldrop, L.D.; Miller, L.A. The role of the pericardium in the valveless, tubular heart of the tunicate Ciona savignyi. J. Exp. Biol. 2015, 218 Pt 17, 2753–2763. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Xu, C.; Chen, X.; Li, X.; Lu, C.; Zhou, P.; Yin, L.; Qian, R.; Chen, S.; Ling, Z.; et al. The roles of Mesp family proteins: Functional diversity and redundancy in differentiation of pluripotent stem cells and mammalian mesodermal development. Protein Cell 2015, 6, 553–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waki, K.; Imai, K.S.; Satou, Y. Genetic pathways for differentiation of the peripheral nervous system in ascidians. Nat. Commun. 2015, 6, 8719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.S.; Levine, M.; Satoh, N.; Satou, Y. Regulatory Blueprint for a Chordate Embryo. Science 2006, 312, 1183–1187. [Google Scholar] [CrossRef]
- Horie, T.; Horie, R.; Chen, K.; Cao, C.; Nakagawa, M.; Kusakabe, T.G.; Satoh, N.; Sasakura, Y.; Levine, M. Regulatory cocktail for dopaminergic neurons in a protovertebrate identified by whole-embryo single-cell transcriptomics. Genes Dev. 2018, 32, 1297–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kugler, J.E.; Wu, Y.; Katikala, L.; Passamaneck, Y.J.; Addy, J.; Caballero, N.; Oda-Ishii, I.; Maguire, J.E.; Li, R.; Di Gregorio, A. Positioning a multifunctional basic helix-loop-helix transcription factor within the Ciona notochord gene regulatory network. Dev. Biol. 2019, 448, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Adell, T.; Gómez-Cuadrado, A.; Skoudy, A.; Pettengill, O.S.; Longnecker, D.S.; Real, F.X. Role of the basic helix-loop-helix transcription factor p48 in the differentiation phenotype of exocrine pancreas cancer cells. Cell Growth Differ. 2000, 11, 137–147. [Google Scholar] [PubMed]
- Fujitani, Y.; Fujitani, S.; Luo, H.; Qiu, F.; Burlison, J.; Long, Q.; Kawaguchi, Y.; Edlund, H.; Macdonald, R.J.; Furukawa, T.; et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 2006, 133, 4439–4450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, M.; Nakamura, S.; Mori, K.; Kawauchi, T.; Terao, M.; Nishimura, Y.V.; Fukuda, A.; Fuse, T.; Matsuo, N.; Sone, M.; et al. Ptf1a, a bHLH Transcriptional Gene, Defines GABAergic Neuronal Fates in Cerebellum. Neuron 2005, 47, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burlison, J.S.; Long, Q.-M.; Fujitani, Y.; Wright, C.V.; Magnuson, M.A. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev. Biol. 2008, 316, 74–86. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, Y.; Cooper, B.; Gannon, M.; Ray, M.; Macdonald, R.J.; Wright, C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002, 32, 128–134. [Google Scholar] [CrossRef]
- Seo, S.; Lim, J.W.; Yellajoshyula, D.; Chang, L.W.; Kroll, K.L. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007, 26, 5093–5108. [Google Scholar] [CrossRef] [Green Version]
- Sommer, L.; Ma, Q.; Anderson, D.J. neurogenins, a Novel Family ofatonal-Related bHLH Transcription Factors, Are Putative Mammalian Neuronal Determination Genes That Reveal Progenitor Cell Heterogeneity in the Developing CNS and PNS. Mol. Cell. Neurosci. 1996, 8, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Jensen, J.B.; Parmar, M.; Guillemot, F.; Björklund, A. Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development 2006, 133, 507–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kele, J.; Simplicio, N.; Ferri, A.L.; Mira, H.; Guillemot, F.; Arenas, E.; Ang, S.L. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 2006, 133, 495–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [Green Version]
- Coppola, U.; Kamal, A.K.; Stolfi, A.; Ristoratore, F. The Cis-Regulatory Code for Kelch-like 21/30 Specific Expression in Ciona robusta Sensory Organs. Front. Collect. 2020, 8. [Google Scholar] [CrossRef]
- Hotta, K.; Takahashi, H.; Asakura, T.; Saitoh, B.; Takatori, N.; Satou, Y.; Satoh, N. Characterization of Brachyury-Downstream Notochord Genes in the Ciona intestinalis Embryo. Dev. Biol. 2000, 224, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Hotta, K.; Takahashi, H.; Satoh, N.; Gojobori, T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: Integrating notochord specification, morphogenesis and chordate evolution. Evol. Dev. 2008, 10, 37–51. [Google Scholar] [CrossRef]
- Soyal, S.M.; Amleh, A.; Dean, J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development 2000, 127, 4645–4654. [Google Scholar] [PubMed]
- Pangas, S.A.; Rajkovic, A. Transcriptional regulation of early oogenesis: In search of masters. Hum. Reprod. Update 2006, 12, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Furness, S.G.B.; Lees, M.J.; Whitelaw, M.L. The dioxin (aryl hydrocarbon) receptor as a model for adaptive responses of bHLH/PAS transcription factors. FEBS Lett. 2007, 581, 3616–3625. [Google Scholar] [CrossRef]
- Taylor, B.L.; Zhulin, I.B. PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [CrossRef] [Green Version]
- Möglich, A.; Ayers, R.A.; Moffat, K. Structure and Signaling Mechanism of Per-ARNT-Sim Domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaud, J.L.; DeRossi, C.; May, N.R.; Holdener, B.C.; Fan, C.M. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 2000, 90, 253–261. [Google Scholar] [CrossRef]
- Pasini, A.; Amiel, A.; Rothbächer, U.; Roure, A.; Lemaire, P.; Darras, S. Formation of the ascidian epidermal sensory neurons: Insights into the origin of the chordate peripheral nervous system. PLoS Biol. 2006, 4, e225. [Google Scholar] [CrossRef]
- Daburon, V.; Mella, S.; Plouhinec, J.-L.; Mazan, S.; Crozatier, M.; Vincent, A. The metazoan history of the COE transcription factors. Selection of a variant HLH motif by mandatory inclusion of a duplicated exon in vertebrates. BMC Evol. Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razy-Krajka, F.; Lam, K.; Wang, W.; Stolfi, A.; Joly, M.; Bonneau, R.; Christiaen, L. Collier/OLF/EBF-Dependent Transcriptional Dynamics Control Pharyngeal Muscle Specification from Primed Cardiopharyngeal Progenitors. Dev. Cell 2014, 29, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Stolfi, A.; Sasakura, Y.; Chalopin, M.; Satou, Y.; Christiaen, L.; Dantec, C.; Endo, T.; Naville, M.; Nishida, H.; Swalla, B.J.; et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis 2014, 53, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Roure, A.; Darras, S. Msxb is a core component of the genetic circuitry specifying the dorsal and ventral neurogenic midlines in the ascidian embryo. Dev. Biol. 2016, 409, 277–287. [Google Scholar] [CrossRef] [Green Version]
- Tolkin, T.; Christiaen, L. Rewiring of an ancestral Tbx1/10-Ebf-Mrf network for pharyngeal muscle specification in distinct embryonic lineages. Development 2016, 143, 3852–3862. [Google Scholar] [CrossRef] [Green Version]
- Bonfig, W.; Krude, H.; Schmidt, H. A novel mutation of LHX3 is associated with combined pituitary hormone deficiency including ACTH deficiency, sensorineural hearing loss, and short neck—A case report and review of the literature. Eur. J. Pediatr. 2011, 170, 1017–1021. [Google Scholar] [CrossRef]
- Carreno, G.; Apps, J.R.; Lodge, E.J.; Panousopoulos, L.; Haston, S.; Gonzalez-Meljem, J.M.; Hahn, H.; Andoniadou, C.L.; Martinez-Barbera, J.P. Hypothalamic sonic hedgehog is required for cell specification and proliferation of LHX3/LHX4 pituitary embryonic precursors. Development 2017, 144, 3289–3302. [Google Scholar] [CrossRef] [Green Version]
- Sheng, H.Z.; Moriyama, K.; Yamashita, T.; Li, H.; Potter, S.S.; Mahon, K.A.; Westphal, H. Multistep Control of Pituitary Organogenesis. Science 1997, 278, 1809–1812. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Ensini, M.; Morton, S.B.; Baldassare, M.; Edlund, T.; Jessell, T.M.; Pfaff, S.L. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 1994, 79, 957–970. [Google Scholar] [CrossRef]
- Stern, J.E. Nitric oxide and homeostatic control: An intercellular signalling molecule contributing to autonomic and neuroendocrine integration? Prog. Biophys. Mol. Biol. 2004, 84, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Castellano, I.; Ercolesi, E.; Palumbo, A. Nitric Oxide Affects ERK Signaling through Down-Regulation of MAP Kinase Phosphatase Levels during Larval Development of the Ascidian Ciona intestinalis. PLoS ONE 2014, 9, e102907. [Google Scholar] [CrossRef] [Green Version]
- Ledent, V.; Paquet, O.; Vervoort, M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Patoori, S.; Jean-Charles, N.; Gopal, A.; Sulaiman, S.; Gopal, S.; Wang, B.; Souferi, B.; Emerson, M.M. Cis-regulatory analysis of Onecut1 expression in fate-restricted retinal progenitor cells. Neural Dev. 2020, 15, 1–20. [Google Scholar] [CrossRef] [Green Version]
- D’Aniello, E.; Pezzotti, M.R.; Locascio, A.; Branno, M. Onecut is a direct neural-specific transcriptional activator of Rx in Ciona intestinalis. Dev. Biol. 2011, 355, 358–371. [Google Scholar] [CrossRef] [Green Version]
- Pezzotti, M.R.; Locascio, A.; Racioppi, C.; Fucci, L.; Branno, M. Auto and cross regulatory elements control Onecut expression in the ascidian nervous system. Dev. Biol. 2014, 390, 273–287. [Google Scholar] [CrossRef] [Green Version]
- Song, H.W.; Wilkinson, M.F. Transcriptional control of spermatogonial maintenance and differentiation. Semin. Cell Dev. Biol. 2014, 30, 14–26. [Google Scholar] [CrossRef] [Green Version]
- Pangas, S.A.; Choi, Y.; Ballow, D.J.; Zhao, Y.; Westphal, H.; Matzuk, M.M.; Rajkovic, A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc. Natl. Acad. Sci. USA 2006, 103, 8090–8095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter, I.S. Regulatory states in the developmental control of gene expression. Brief. Funct. Genom. 2017, 16, 281–287. [Google Scholar] [CrossRef]
- Yan, R.T.; Ma, W.; Liang, L.; Wang, S.Z. bHLH Genes and Retinal Cell Fate Specification. Mol. Neurobiol. 2005, 32, 157–172. [Google Scholar] [CrossRef]
- Jahan, I.; Pan, N.; Kersigo, J.; Fritzsch, B. Neurod1 Suppresses Hair Cell Differentiation in Ear Ganglia and Regulates Hair Cell Subtype Development in the Cochlea. PLoS ONE 2010, 5, e11661. [Google Scholar] [CrossRef] [Green Version]
- Mazet, F.; Hutt, J.A.; Milloz, J.; Millard, J.; Graham, A.; Shimeld, S.M. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 2005, 282, 494–508. [Google Scholar] [CrossRef] [Green Version]
- Brozovic, M.; Martin, C.; Dantec, C.; Dauga, D.; Mendez, M.; Simion, P.; Percher, M.; Laporte, B.; Scornavacca, C.; Di Gregorio, A.; et al. ANISEED 2015: A digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 2016, 44, D808–D818. [Google Scholar] [CrossRef]
- Satou, Y.; Kawashima, T.; Shoguchi, E.; Nakayama, A.; Satoh, N. An Integrated Database of the Ascidian, Ciona intestinalis: Towards Functional Genomics. Zool. Sci. 2005, 22, 837–843. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Negrón-Piñeiro, L.J.; Wu, Y.; Di Gregorio, A. Transcription Factors of the bHLH Family Delineate Vertebrate Landmarks in the Nervous System of a Simple Chordate. Genes 2020, 11, 1262. https://doi.org/10.3390/genes11111262
Negrón-Piñeiro LJ, Wu Y, Di Gregorio A. Transcription Factors of the bHLH Family Delineate Vertebrate Landmarks in the Nervous System of a Simple Chordate. Genes. 2020; 11(11):1262. https://doi.org/10.3390/genes11111262
Chicago/Turabian StyleNegrón-Piñeiro, Lenny J., Yushi Wu, and Anna Di Gregorio. 2020. "Transcription Factors of the bHLH Family Delineate Vertebrate Landmarks in the Nervous System of a Simple Chordate" Genes 11, no. 11: 1262. https://doi.org/10.3390/genes11111262
APA StyleNegrón-Piñeiro, L. J., Wu, Y., & Di Gregorio, A. (2020). Transcription Factors of the bHLH Family Delineate Vertebrate Landmarks in the Nervous System of a Simple Chordate. Genes, 11(11), 1262. https://doi.org/10.3390/genes11111262





