Molecular Evolution of Maize Ascorbate Peroxidase Genes and Their Functional Divergence
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening of APX Protein Sequences
2.2. Analysis of Phylogeny, Splice Site, and Duplication Events of APX Genes
2.3. Analysis of Subcellular and Tissue Locations and Expression Pattern of APX Genes in Maize
3. Results
3.1. Identification of APX Genes
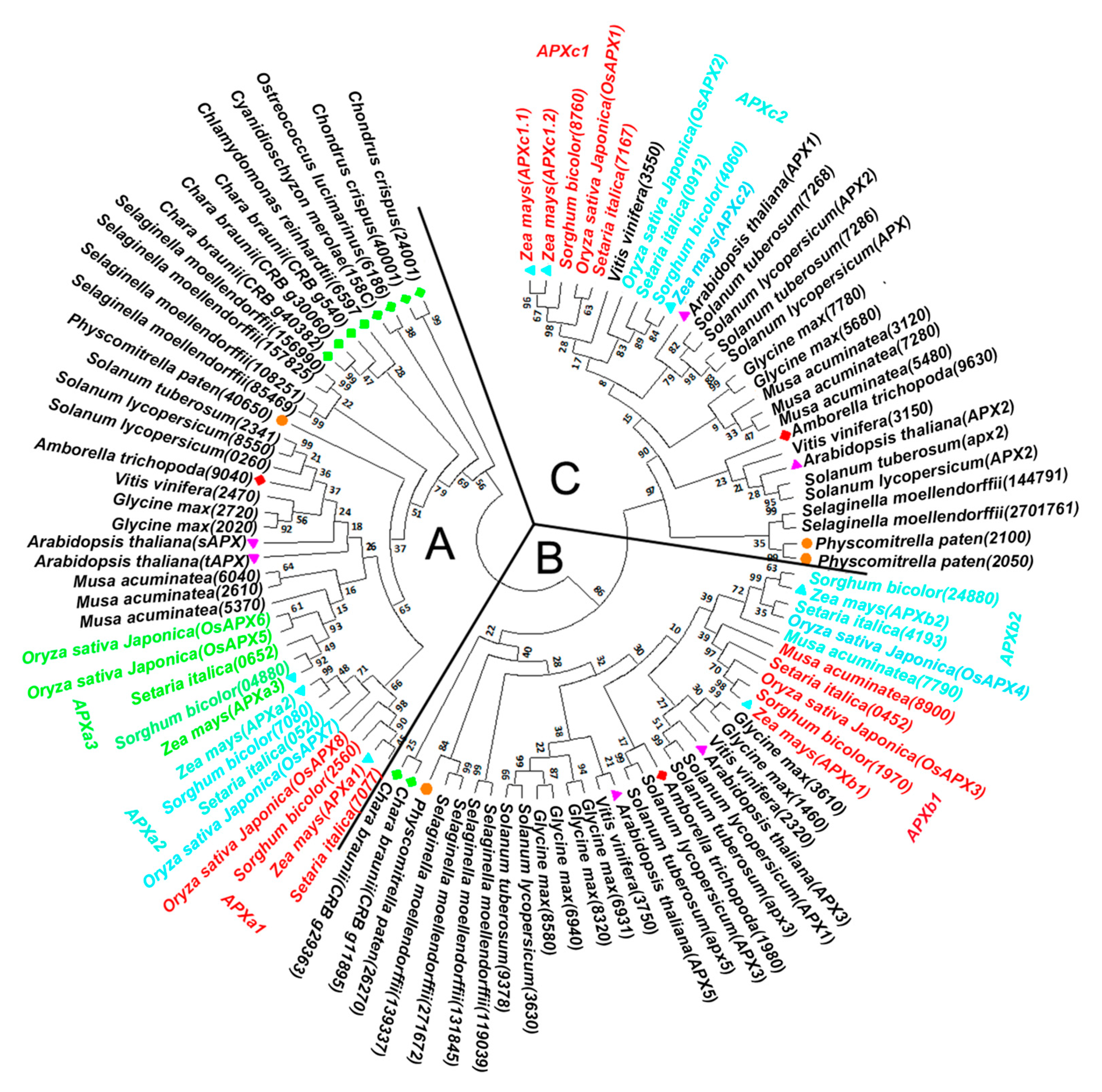
3.2. Phylogenetic Analysis of APX Genes
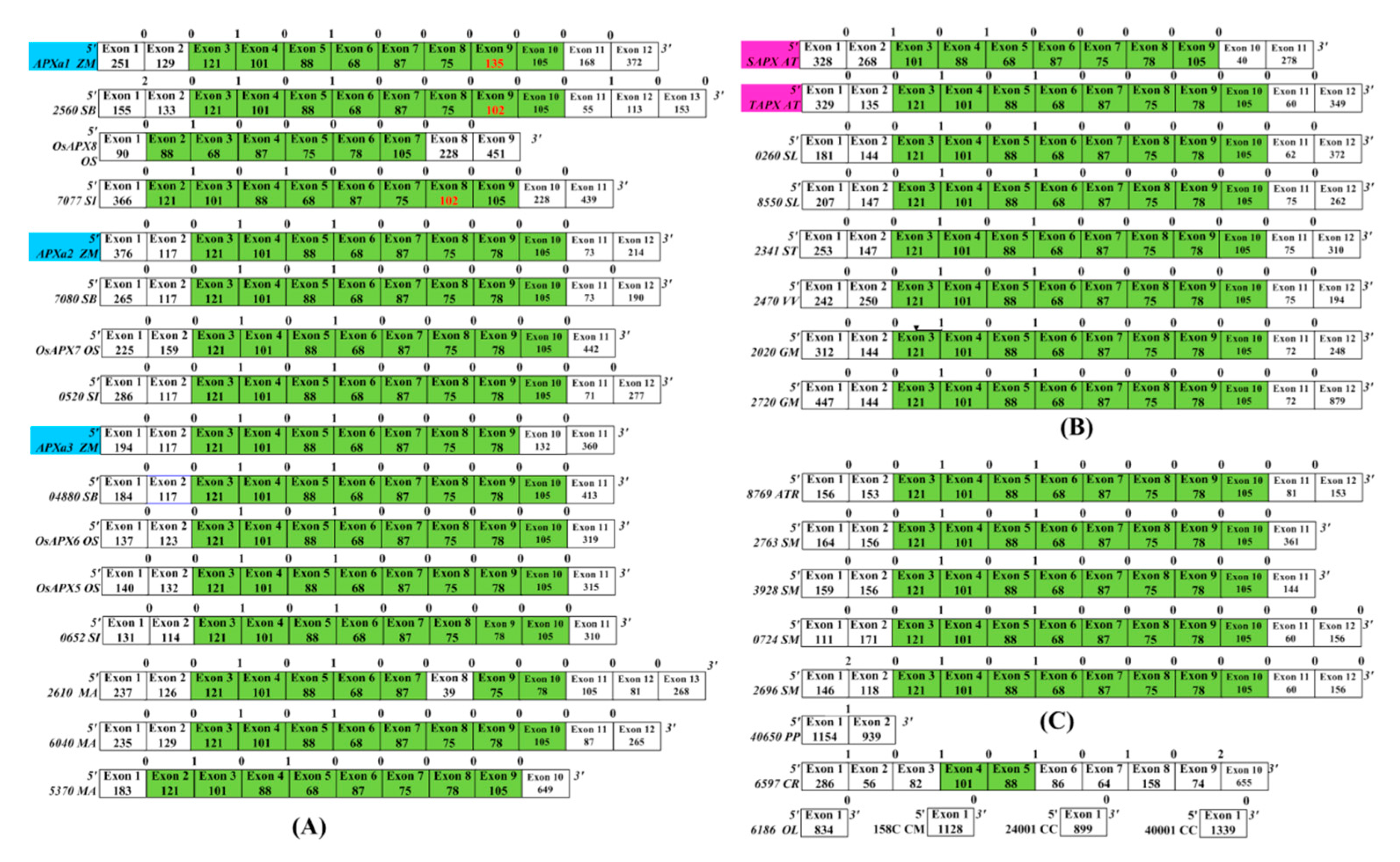
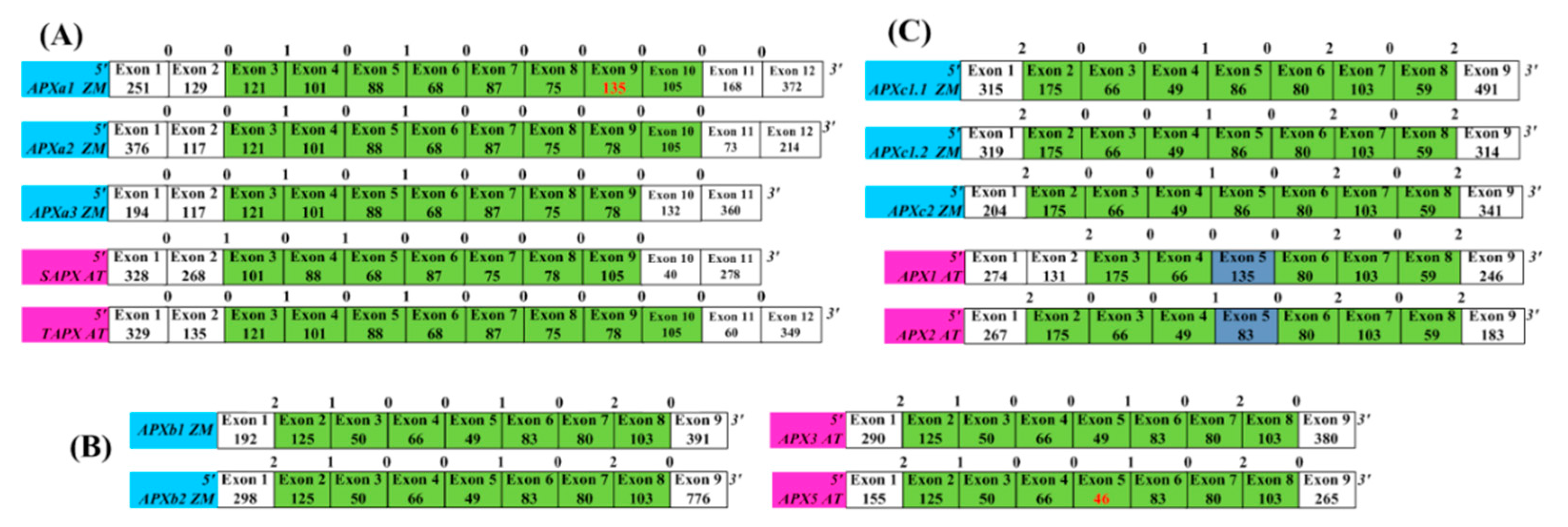
3.3. Splice Sites of APX Genes
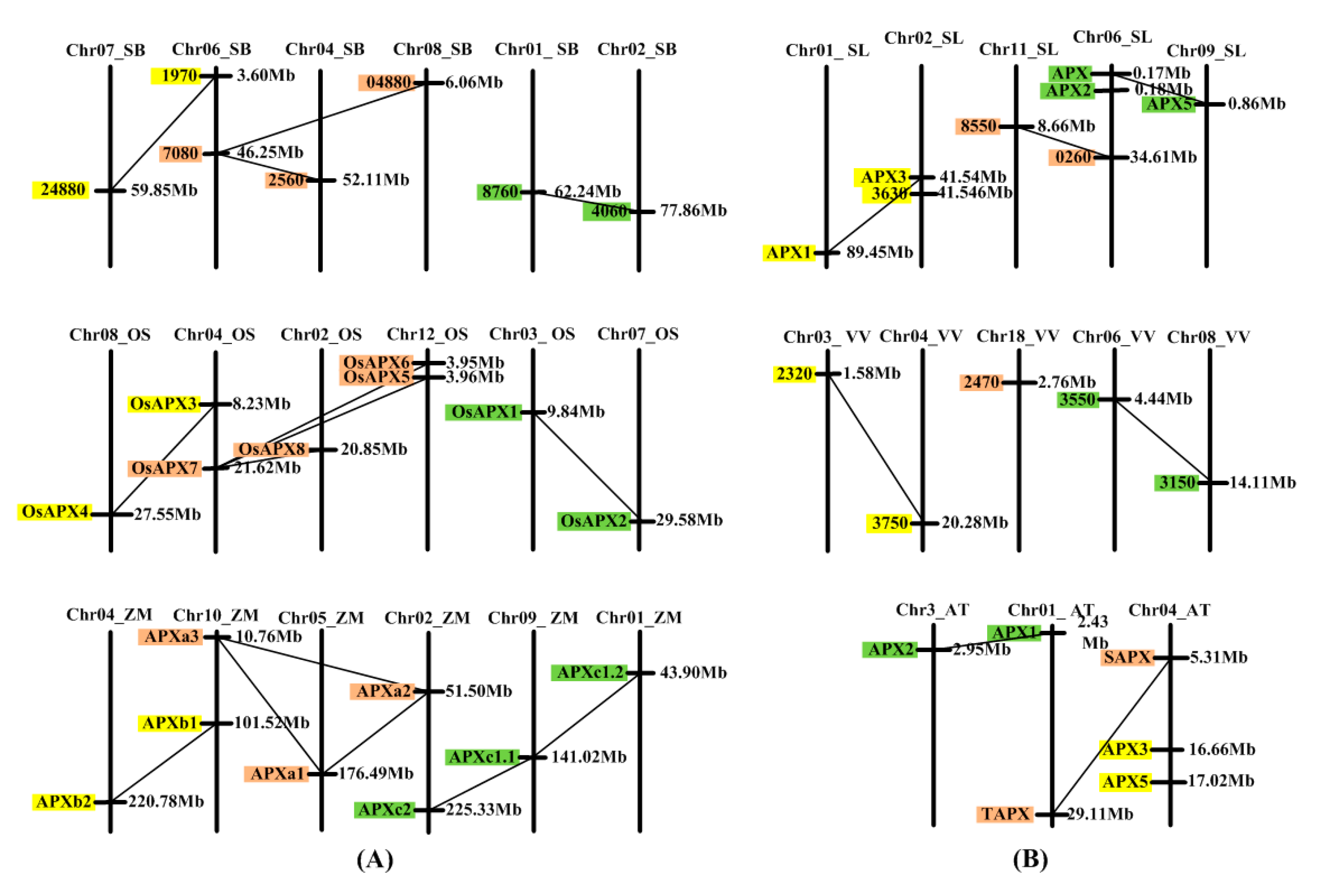
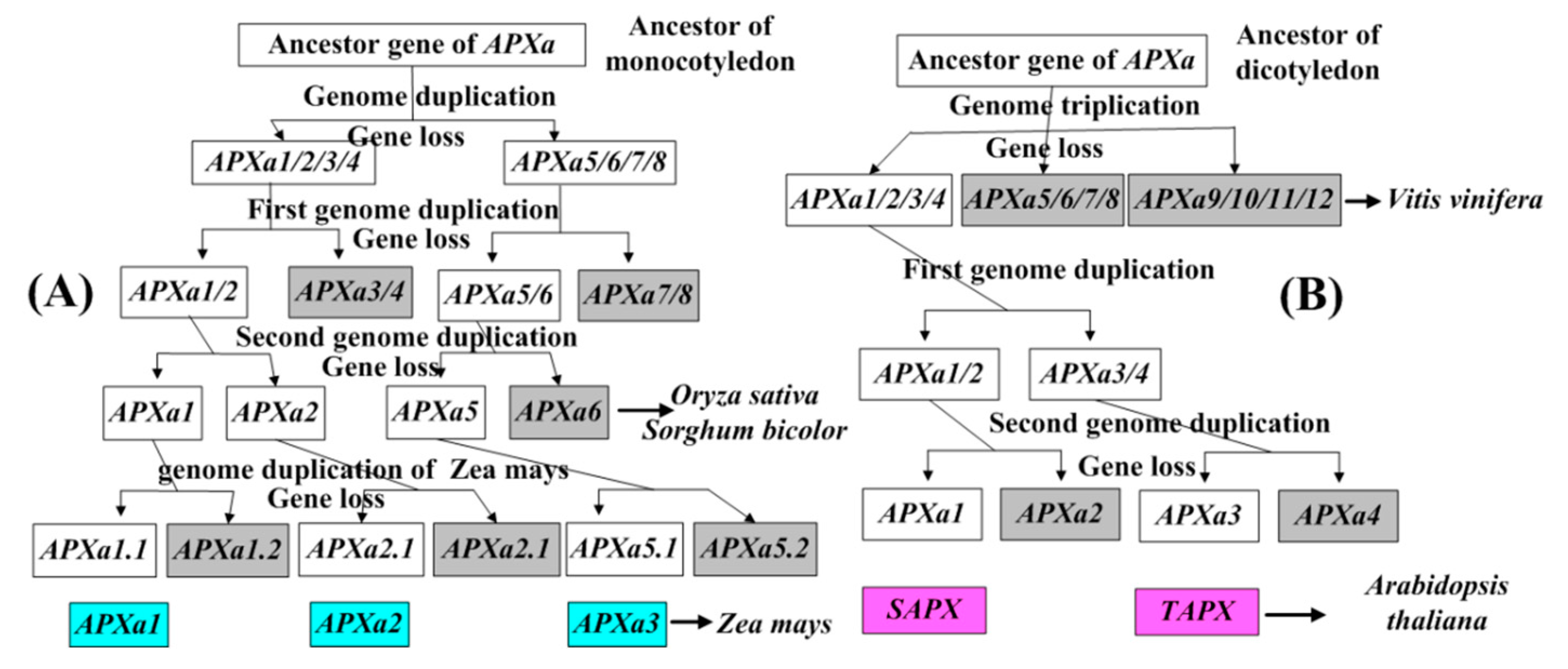
3.4. Analysis of Chromosome Distribution and Duplication Events of APX Genes of Angiosperms
3.5. Subcellular Localization of APX Isoenzymes in Maize
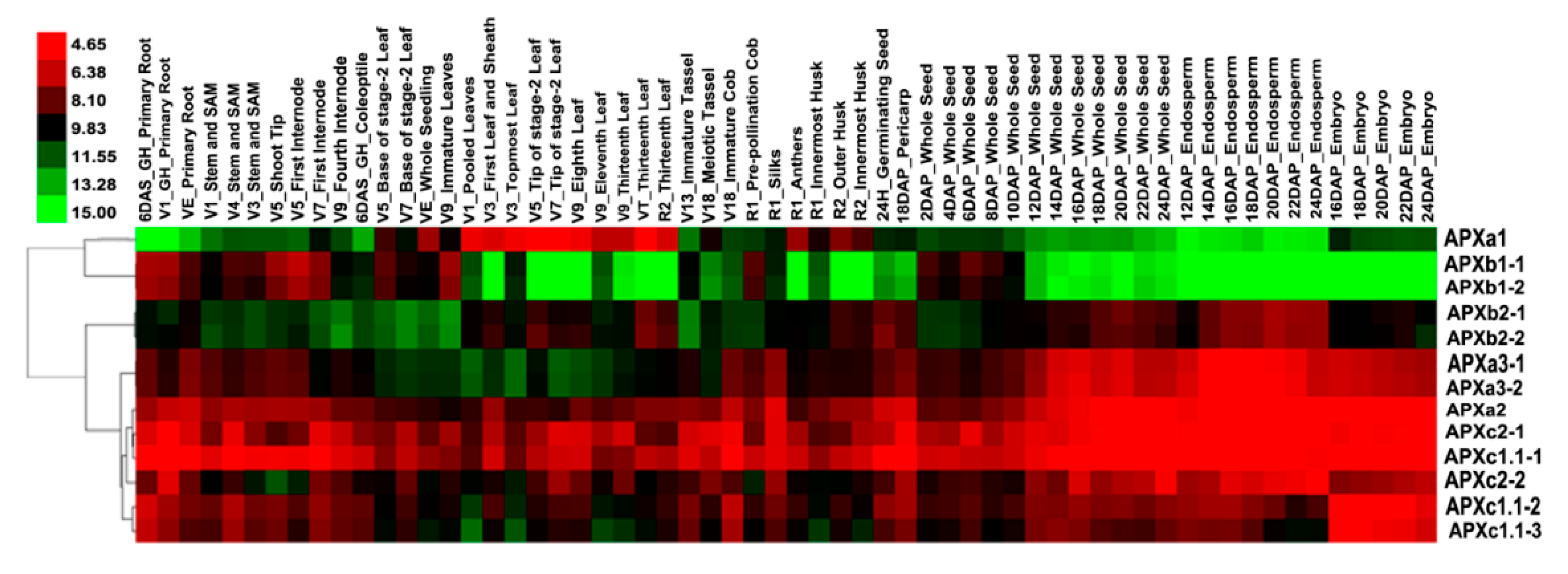
3.6. Expression Pattern Analysis of Maize APX Genes in Different Tissues
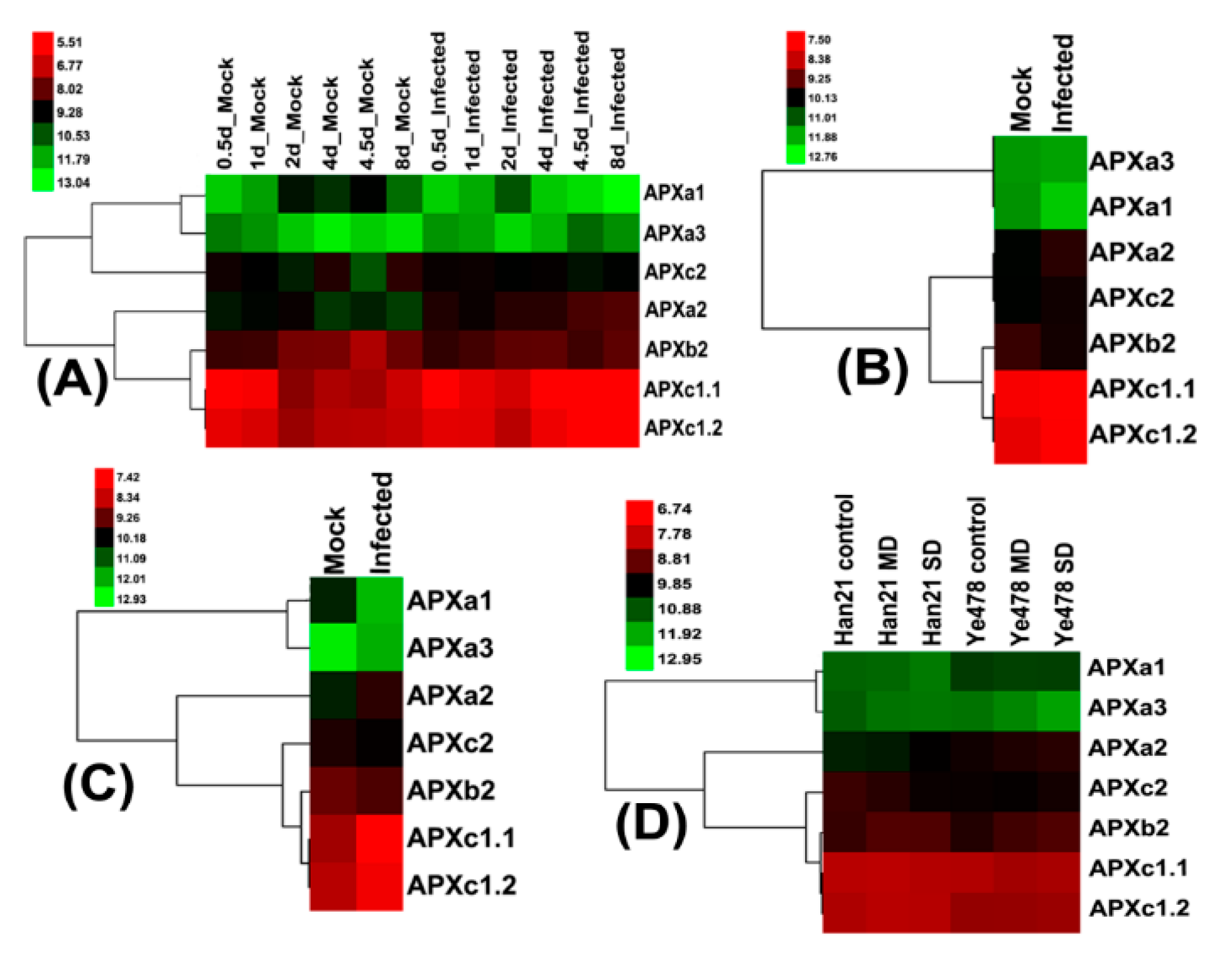
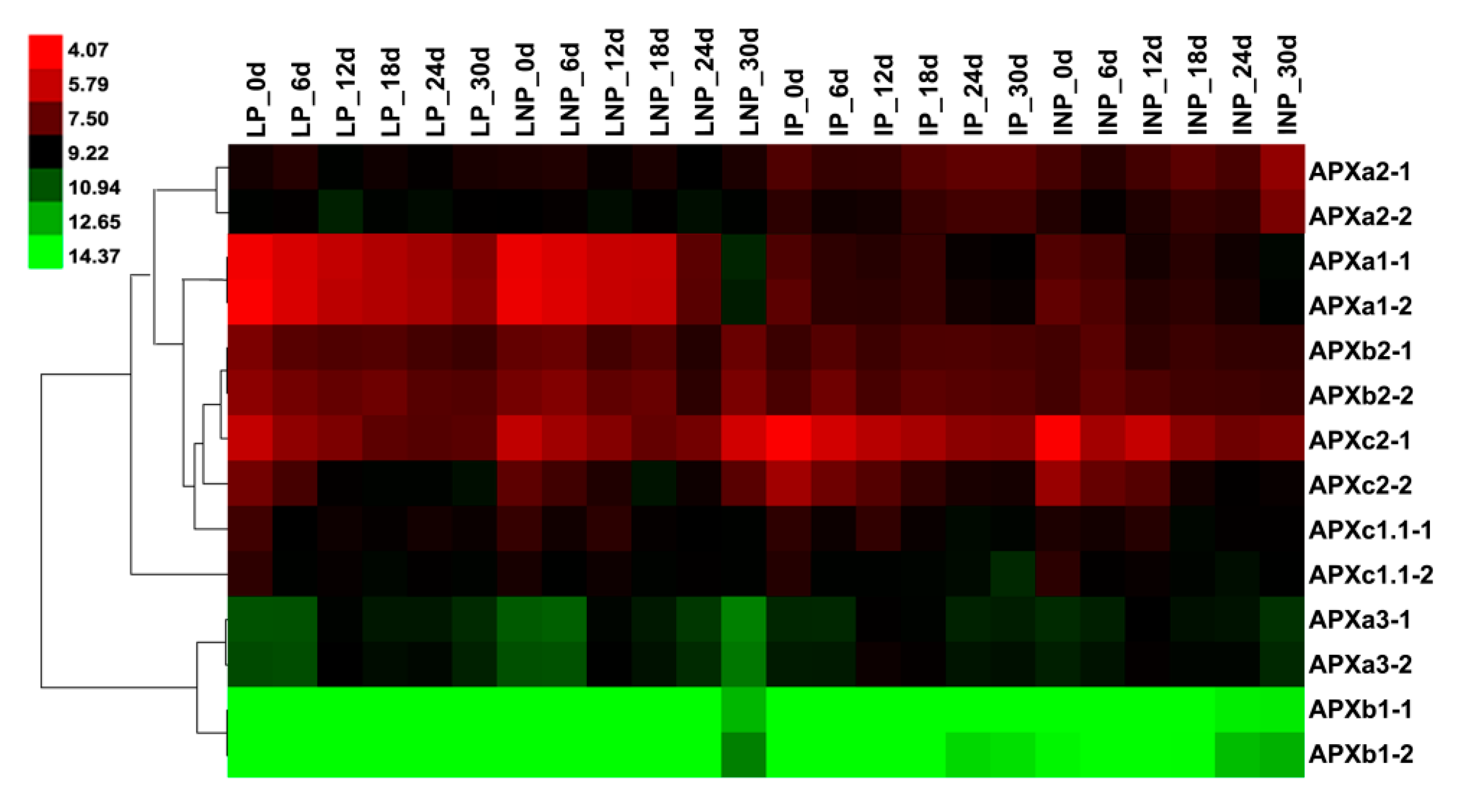
3.7. Expression Pattern of Maize Leaf APX Genes in Response to Infection with Ustilago Maydis and Drought Stress
3.8. Expression Patterns of Induced Senescence of APXs in Maize Leaves and Internodes
4. Discussion
4.1. Evolution of Maize APX Genes
4.2. Subcellular Localization of APX Isozymes and Differences in Tissue Expression of Maize APX Genes
4.3. The Difference of Maize APX Gene Expression Level in Response to Stress and Induced Senescence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Quan, L.-J.; Zhang, B.; Shi, W.-W.; Li, H.-Y. Hydrogen Peroxide in Plants: A Versatile Molecule of the Reactive Oxygen Species Network. J. Integr. Plant Biol. 2008, 50, 2–18. [Google Scholar] [CrossRef]
- Ribeiro, C.W.; Korbes, A.P.; Garighan, J.A.; Jardim-Messeder, D.; Carvalho, F.E.; Sousa, R.H.; Caverzan, A.; Teixeira, F.K.; Silveira, J.A.G.; Margis, R. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017, 263, 55–65. [Google Scholar] [CrossRef]
- Petrov, V.D.; Van Breusegem, F. Hydrogen peroxide—A central hub for information flow in plant cells. AoB Plants 2012, 2012, pls014. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Wilkinson, S.R.; Obado, S.O.; Maurício, I.L.; Kelly, J.M. Trypanosoma cruzi expresses a plant-like ascorbate-dependent hemoperoxidase localized to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2002, 99, 13453–13458. [Google Scholar] [CrossRef]
- Takeda, T.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Purification and characterization of ascorbate peroxidase in Chlorella vulgaris. Biochimie 1998, 80, 295–301. [Google Scholar] [CrossRef]
- Takeda, T.; Yoshimura, K.; Yoshii, M.; Kanahoshi, H.; Miyasaka, H.; Shigeoka, S. Molecular Characterization and Physiological Role of Ascorbate Peroxidase from Halotolerant Chlamydomonas sp. W80 Strain. Arch. Biochem. Biophys. 2000, 376, 82–90. [Google Scholar] [CrossRef]
- Sano, S.; Ueda, M.; Kitajima, S.; Takeda, T.; Shigeoka, S.; Kurano, N.; Miyachi, S.; Miyake, C.; Yokota, A. Characterization of Ascorbate Peroxidases from Unicellular Red Alga Galdieria partita. Plant Cell Physiol. 2001, 42, 433–440. [Google Scholar] [CrossRef]
- Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Diversity and Evolution of Ascorbate Peroxidase Functions in Chloroplasts: More Than Just a Classical Antioxidant Enzyme? Plant Cell Physiol. 2016, 57, 1377–1386. [Google Scholar] [CrossRef]
- Özyiğit, I.I.; Filiz, E.; Vatansever, R.; Kurtoglu, K.Y.; Koç, I.; Öztürk, M.X.; Anjum, N.A. Identification and Comparative Analysis of H2O2-Scavenging Enzymes (Ascorbate Peroxidase and Glutathione Peroxidase) in Selected Plants Employing Bioinformatics Approaches. Front. Plant Sci. 2016, 7, 301. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L. Analysis of the Molecular Evolutionary History of the Ascorbate Peroxidase Gene Family: Inferences from the Rice Genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, Y.; Liu, Y.-Y.; Liu, Y.; Fu, J.; Zheng, J.; Wang, G. Gene families of maize glutathione–ascorbate redox cycle respond differently to abiotic stresses. J. Plant Physiol. 2012, 169, 183–192. [Google Scholar] [CrossRef]
- Granlund, I.; Storm, P.; Schubert, M.; García-Cerdán, J.G.; Funk, C.; Schröder, W.P. The TL29 Protein is Lumen Located, Associated with PSII and Not an Ascorbate Peroxidase. Plant Cell Physiol. 2009, 50, 1898–1910. [Google Scholar] [CrossRef]
- Morita, S.; Kaminaka, H.; Yokoi, H.; Masumura, T.; Tanaka, K. Cloning and characterization of cytosolic ascorbate peroxidase cDNA from rice (PGR97-012). Plant Physiol. 1997, 113, 306. [Google Scholar]
- Morita, S.; Kaminaka, H.; Masumura, T.; Tanaka, K. Induction of Rice Cytosolic Ascorbate Peroxidase mRNA by Oxidative Stress; the Involvement of Hydrogen Peroxide in Oxidative Stress Signalling. Plant Cell Physiol. 1999, 40, 417–422. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Galvão, V.C.; Margis, R.; Margis-Pinheiro, M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 2006, 224, 300–314. [Google Scholar] [CrossRef]
- Hong, C.-Y.; Hsu, Y.T.; Tsai, Y.-C.; Kao, C.H. Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 2007, 58, 3273–3283. [Google Scholar] [CrossRef]
- Xu, L.; Carrie, C.; Law, S.R.; Murcha, M.W.; Whelan, J. Acquisition, Conservation, and Loss of Dual-Targeted Proteins in Land Plants. Plant Physiol. 2012, 161, 644–662. [Google Scholar] [CrossRef]
- Jespersen, H.M.; Kjærsgård, I.V.H.; Østergaard, L.; Welinder, K.G. From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem. J. 1997, 326, 305–310. [Google Scholar] [CrossRef]
- Panchuk, I.; Volkov, R.; Schöffl, F. Heat Stress- and Heat Shock Transcription Factor-Dependent Expression and Activity of Ascorbate Peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, I.; Ezentgraf, U.; Volkov, R. Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 2005, 222, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic Ascorbate Peroxidase 1 Is a Central Component of the Reactive Oxygen Gene Network of Arabidopsis. Plant Cell 2004, 17, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Koussevitzky, S.; Suzuki, N.; Huntington, S.; Armijo, L.; Sha, W.; Cortes, D.; Shulaev, V.; Mittler, R. Ascorbate Peroxidase 1 Plays a Key Role in the Response of Arabidopsis thaliana to Stress Combination. J. Biol. Chem. 2008, 283, 34197–34203. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.-H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2012, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Zuccolo, A.; Bowers, J.E.; Estill, J.C.; Xiong, Z.; Luo, M.; Sebastian, A.; Goicoechea, J.-L.; Collura, K.; Yu, Y.; Jiao, Y.; et al. A physical map for the Amborella trichopoda genome sheds light on the evolution of angiosperm genome structure. Genome Biol. 2011, 12, R48. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated Syntenic and Phylogenomic Analyses Reveal an Ancient Genome Duplication in Monocots. Plant Cell 2014, 26, 2792–2802. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-N.; Chen, C.-T.; Sung, T.-Y.; Ho, S.-Y.; Hsu, W.-L. Protein subcellular localization prediction of eukaryotes using a knowledge-based approach. BMC Bioinform. 2009, 10, S8. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting Subcellular Localization of Proteins Based on their N-terminal Amino Acid Sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Dash, S.; Van Hemert, J.; Hong, L.; Wise, R.P.; Dickerson, J. PLEXdb: Gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2011, 40, D1194–D1201. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; Mcgettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Zheng, J.; Fu, J.; Gou, M.; Huai, J.; Liu, Y.; Jian, M.; Huang, Q.; Guo, X.; Dong, Z.; Wang, H.; et al. Genome-wide transcriptome analysis of two maize inbred lines under drought stress. Plant Mol. Biol. 2009, 72, 407–421. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Childs, K.L.; Santoro, N.; Foster, C.E.; Buell, C.R.; De Leon, N.; Kaeppler, S.M. Transcriptional and Metabolic Analysis of Senescence Induced by Preventing Pollination in Maize. Plant Physiol. 2012, 159, 1730–1744. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; De Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Doehlemann, G.; Van Der Linde, K.; Assmann, D.; Schwammbach, D.; Hof, A.; Mohanty, A.; Jackson, D.; Kahmann, R. Pep1, a Secreted Effector Protein of Ustilago maydis, Is Required for Successful Invasion of Plant Cells. PLoS Pathog. 2009, 5, e1000290. [Google Scholar] [CrossRef]
- Doehlemann, G.; Wahl, R.; Horst, R.J.; Voll, L.M.; Usadel, B.; Poree, F.; Stitt, M.; Pons-Kühnemann, J.; Sonnewald, U.; Kahmann, R.; et al. Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008, 56, 181–195. [Google Scholar] [CrossRef]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Gouy, M.; Yang, Y.W.; Sharp, P.M.; Li, W.H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed]
- Wikström, N.; Savolainen, V.; Chase, M.W. Evolution of the angiosperms: Calibrating the family tree. Proc. R. Soc. B Boil. Sci. 2001, 268, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hu, J.; Qu, C.; Wang, L.; Huang, G.; Niu, P.; Zhong, Z.; Hong, F.; Wang, G.; Postlethwait, J.H.; et al. Molecular evolution and functional divergence of zebrafish (Danio rerio) cryptochrome genes. Sci. Rep. 2015, 5, srep08113. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Paterson, A.H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 2009, 107, 472–477. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nat. Cell Biol. 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Initiative, T.A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nat. Cell Biol. 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Nickel, U.; Allen, R.D.; Goodman, H.M. Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol. Biol. 1997, 34, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Narendra, S.; Venkataramani, S.; Shen, G.; Wang, J.; Pasapula, V.; Lin, Y.; Kornyeyev, D.; Holaday, A.S.; Zhang, H. The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 2006, 57, 3033–3042. [Google Scholar] [CrossRef]
- Shen, G.; Kuppu, S.; Venkataramani, S.; Wang, J.; Yan, J.; Qiu, X.; Zhang, H. ANKYRIN REPEAT-CONTAINING PROTEIN 2A Is an Essential Molecular Chaperone for Peroxisomal Membrane-Bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell 2010, 22, 811–831. [Google Scholar] [CrossRef]
- Tiwari, V.; Chaturvedi, A.K.; Mishra, A.; Jha, B. The Transcriptional Regulatory Mechanism of the Peroxisomal Ascorbate Peroxidase (pAPX) Gene Cloned from an Extreme Halophyte, Salicornia brachiata. Plant Cell Physiol. 2013, 55, 201–217. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Charlton, W.L.; Johnson, B.; Graham, I.A.; Baker, A. Non-coordinate expression of peroxisome biogenesis, β-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 2004, 23, 647–653. [Google Scholar] [CrossRef]
- Baker, A.; Graham, I.A.; Holdsworth, M.J.; Smith, A.M.; Theodoulou, F.L. Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 2006, 11, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.A. Seed Storage Oil Mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef]
- Karpinska, G.W.B.; Wingsle, G.; Karpiński, S.; Karpińska, B. Antagonistic Effects of Hydrogen Peroxide and Glutathione on Acclimation to Excess Excitation Energy in Arabidopsis. IUBMB Life 2000, 50, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Elaloi, C.; Przybyla, D.; Apel, K. A genetic approach towards elucidating the biological activity of different reactive oxygen species in Arabidopsis thaliana. J. Exp. Bot. 2006, 57, 1719–1724. [Google Scholar] [CrossRef]
- Del Río, L.A.; Sandalio, L.M.; Corpas, F.J.; Palma, J.M.; Barroso, J.B. Reactive Oxygen Species and Reactive Nitrogen Species in Peroxisomes. Production, Scavenging, and Role in Cell Signaling. Plant Physiol. 2006, 141, 330–335. [Google Scholar] [CrossRef]
- Noctor, G.; Veljovic-Jovanovic, S.; Driscoll, S.; Novitskaya, L.; Foyer, C.H. Drought and Oxidative Load in the Leaves of C3 Plants: A Predominant Role for Photorespiration? Ann. Bot. 2002, 89, 841–850. [Google Scholar] [CrossRef]

| Gene Name | KnowPredsite II | Target P 1.1 | CELLO v.2.5 | WoLF PSORT | Result |
|---|---|---|---|---|---|
| APXa1 | Plastid (100) Chloroplast (100) Plasma membrane (40) Mitochondrion (20) | Chloroplast (1) | Chloroplast (2.2) | Chloroplast (13) | Thylakoid membrane of the chloroplast |
| APXa2 | Plastid (100) Chloroplast (100) Plasma membrane (40) Mitochondrion (20) | Chloroplast (4) | Mitochondrion (2.3) Chloroplast (1.4) | Chloroplast (11) Mitochondrion (3) | Matrix of chloroplast (matrix of plastid) |
| APXa3 | Plastid (100) Chloroplast (100) Plasma membrane (40) Mitochondrion (20) | Mitochondrion (1) | Mitochondrion (2.6) Chloroplast (1.2) | Chloroplast (9) Mitochondrion (5) | Mitochondrion |
| APXb1 | Plasma membrane (100) Peroxisome (40) | Other (3) | Cytoplasm (2.8) Mitochondrion (0.9) | Cytoplasm (7.5) | Peroxisome |
| APXb2 | Plasma membrane (100) Peroxisome (40) | Mitochondrion (5) Other (1) | Cytoplasm (1.9) Mitochondrion (1.2) Chloroplast (1.0) | Chloroplast (13) | Peroxisome |
| APXc1.1 | Cytoplasm (100) | Other (5) | Cytoplasm (2.4) | Mitochondrion (6) Cytoplasm (4) | Cytoplasm |
| APXc1.2 | Cytoplasm (100) | Other (5) | Cytoplasm (2.6) | Cytoplasm (4) Chloroplast (3) | Cytoplasm |
| APXc2 | Cytoplasm (100) | Other (3) | Cytoplasm (2.6) | Cytoplasm (4) | Cytoplasm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, C.; Wang, L.; Zhao, Y.; Liu, C. Molecular Evolution of Maize Ascorbate Peroxidase Genes and Their Functional Divergence. Genes 2020, 11, 1204. https://doi.org/10.3390/genes11101204
Qu C, Wang L, Zhao Y, Liu C. Molecular Evolution of Maize Ascorbate Peroxidase Genes and Their Functional Divergence. Genes. 2020; 11(10):1204. https://doi.org/10.3390/genes11101204
Chicago/Turabian StyleQu, Chunxiang, Lin Wang, Yingwei Zhao, and Chao Liu. 2020. "Molecular Evolution of Maize Ascorbate Peroxidase Genes and Their Functional Divergence" Genes 11, no. 10: 1204. https://doi.org/10.3390/genes11101204
APA StyleQu, C., Wang, L., Zhao, Y., & Liu, C. (2020). Molecular Evolution of Maize Ascorbate Peroxidase Genes and Their Functional Divergence. Genes, 11(10), 1204. https://doi.org/10.3390/genes11101204




