R2 and Non-Site-Specific R2-Like Retrotransposons of the German Cockroach, Blattella germanica
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Isolation, Library Construction, and TE Amplification/Cloning/Sequencing
2.2. Identification of Sequences of Retrotransposons with Maximum Similarity to the R2 Retrotransposon of B. germanica
2.3. Domain Architecture Analysis
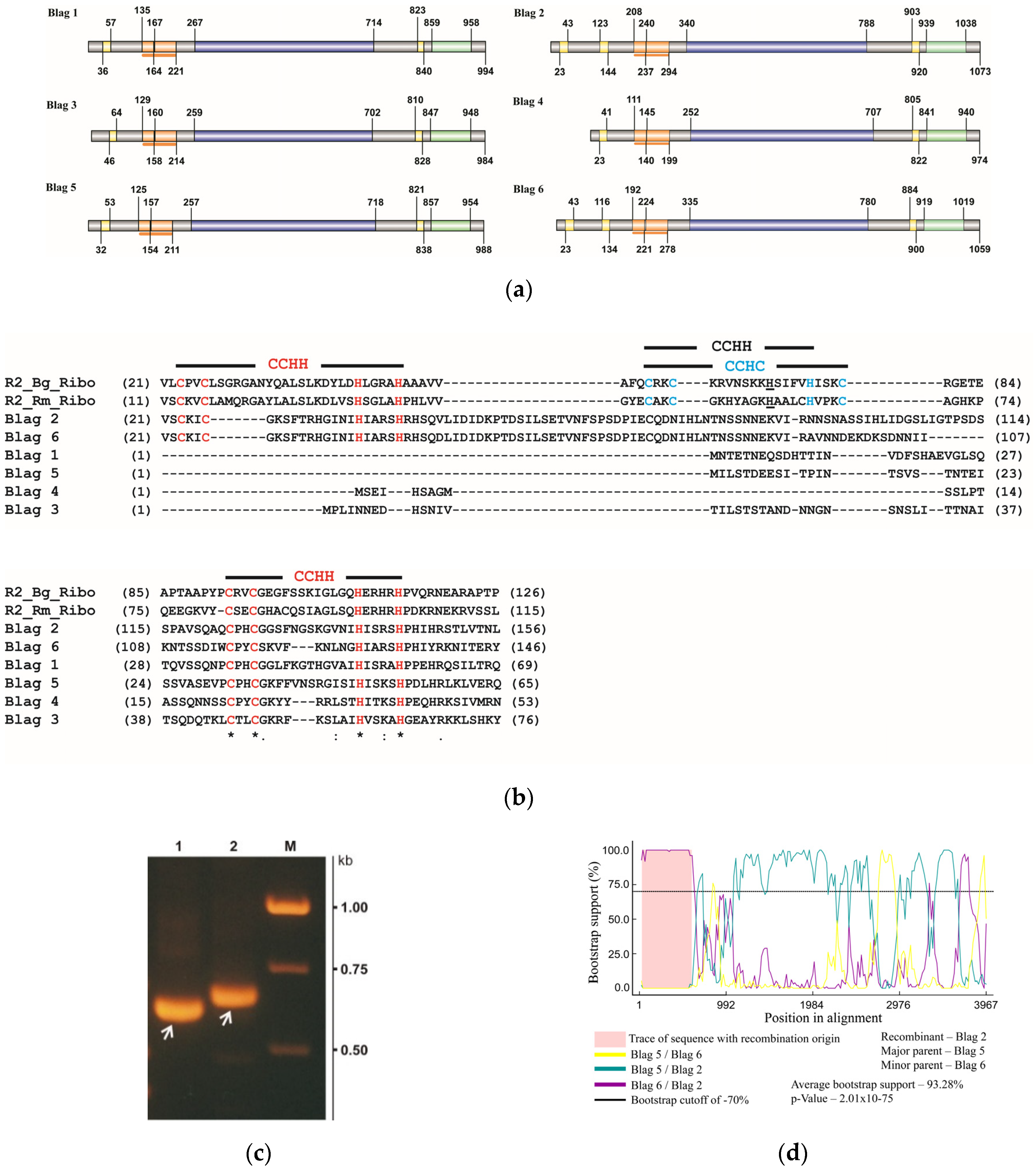
2.4. Recombination Analysis
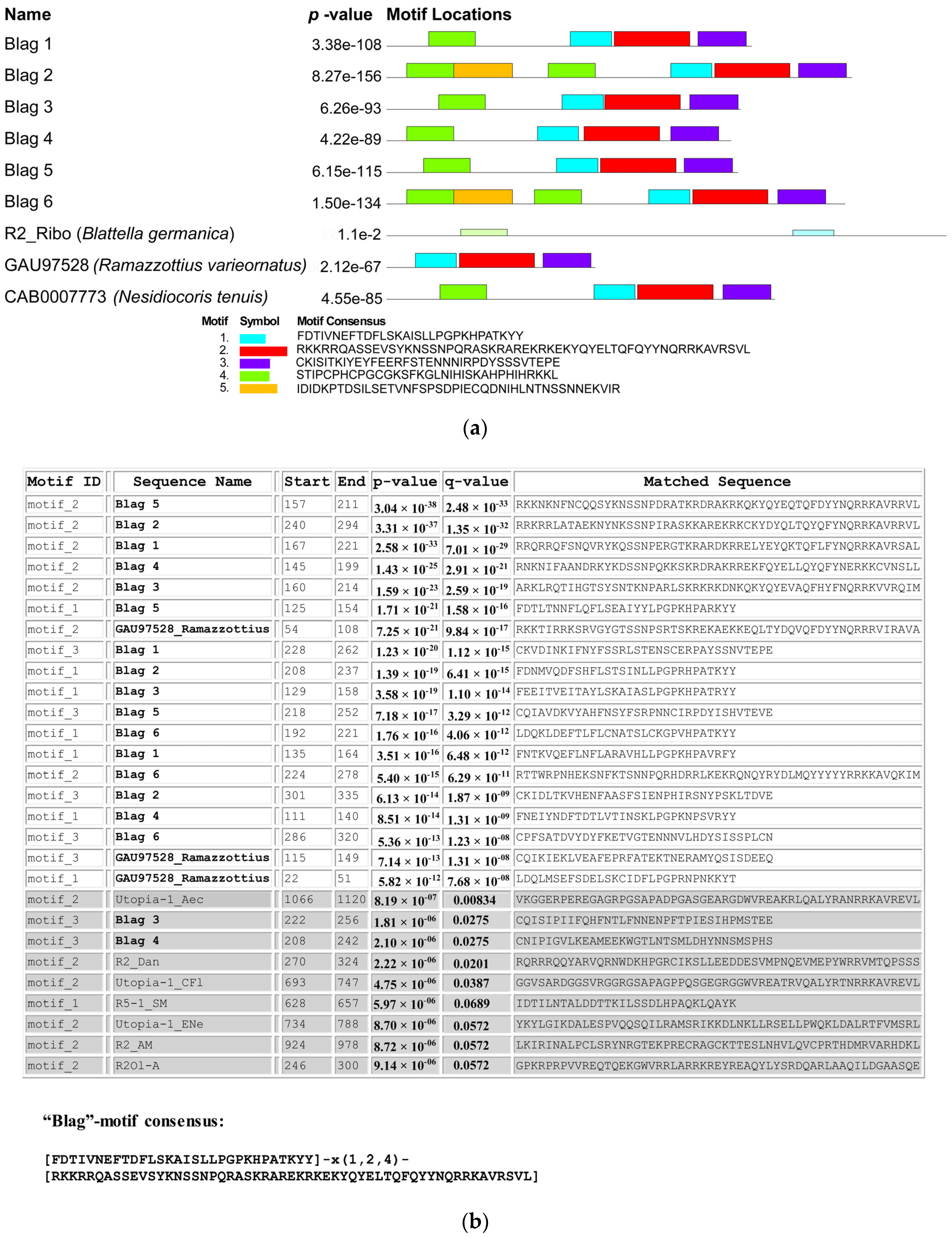
2.5. Conserved Motif Analyses
2.6. Sequence Alignment and Phylogenetic Analysis
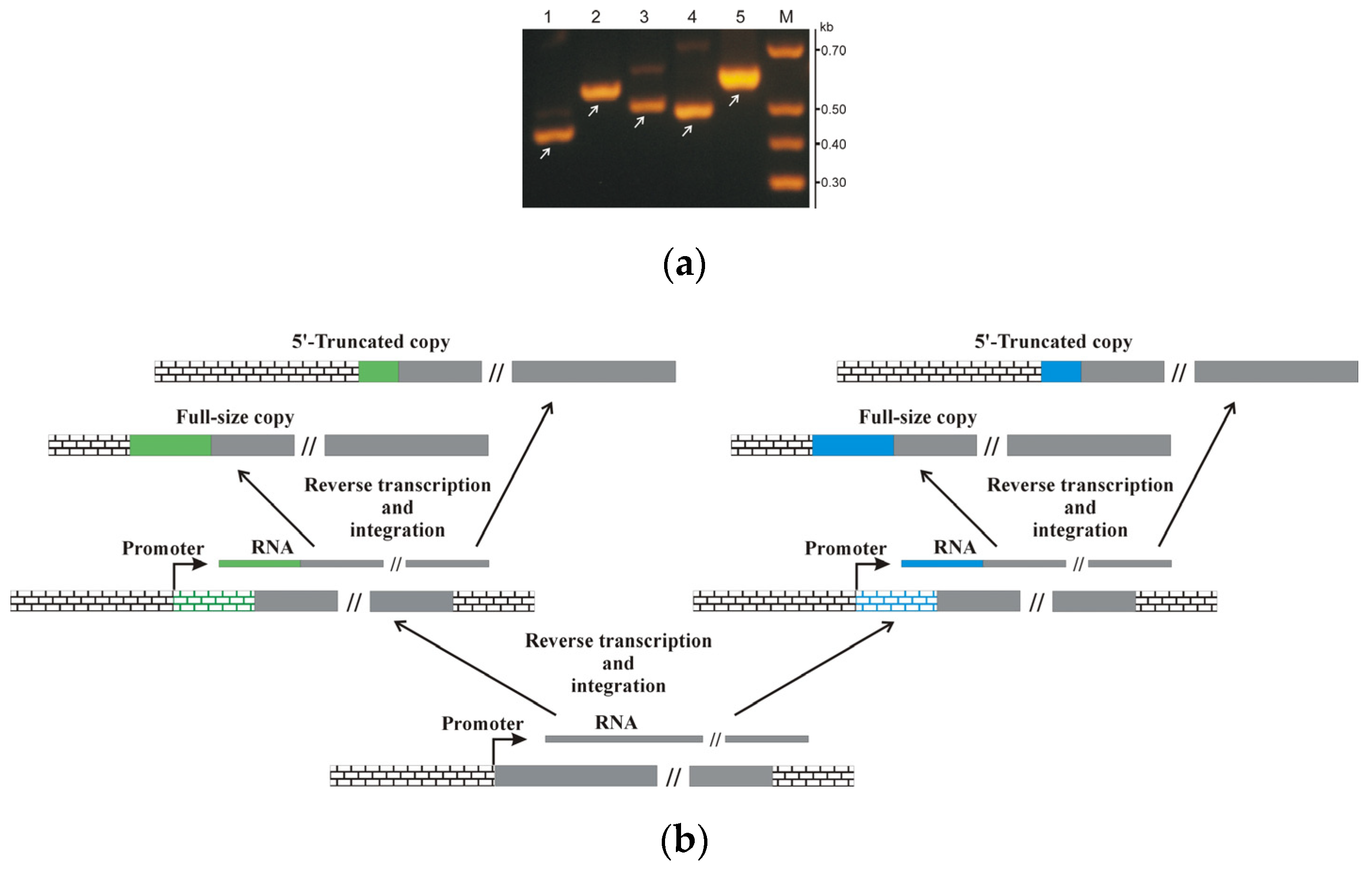
2.7. R2 Retrotransposon 5′-truncated Fragments Analysis
2.8. Bayesian Clustering and Principal Coordinate Analysis
2.9. Data Deposition
3. Results and Discussion
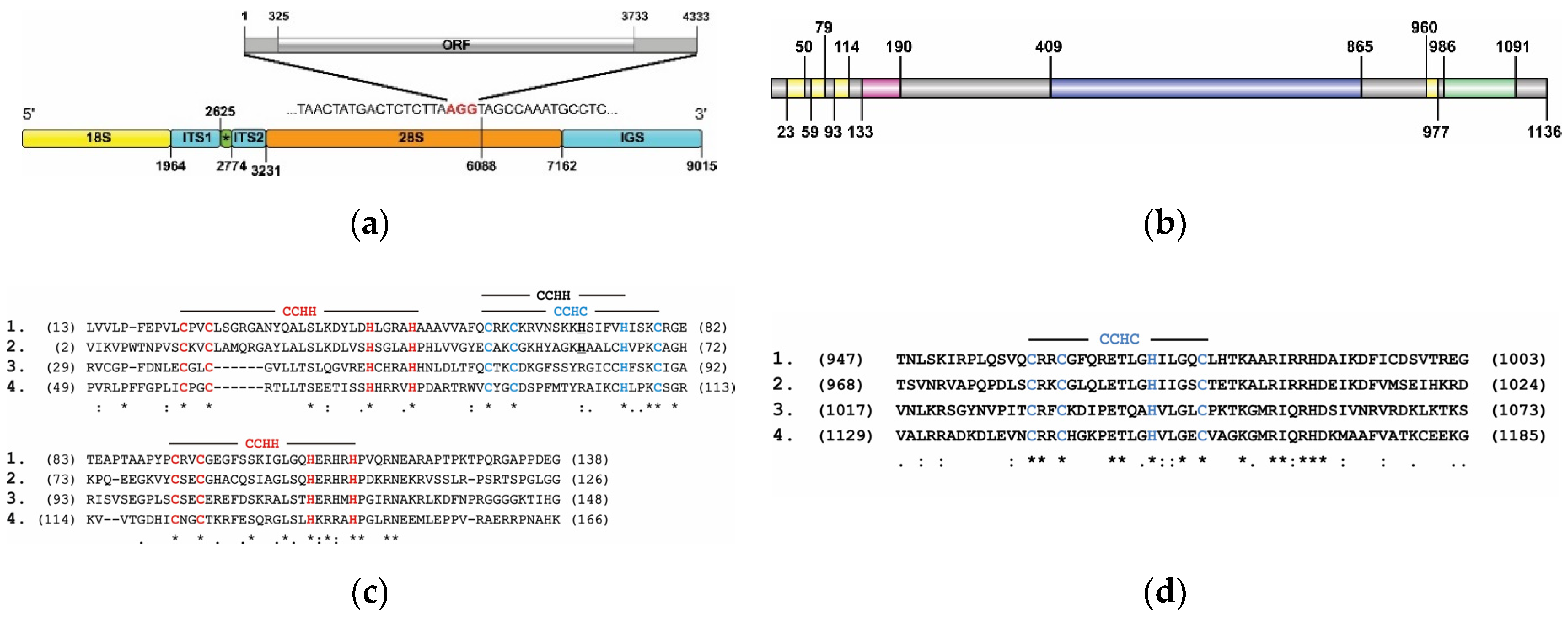
3.1. Complete rDNA Repeat Sequence of B. germanica
3.2. Structural and Functional Organization of Full-Length Copies of the R2 Retrotransposon of B. germanica
3.3. Detection and Analysis of the Non-Site-Specific R2-like Retrotransposon Distribution in the Genome of B. germanica
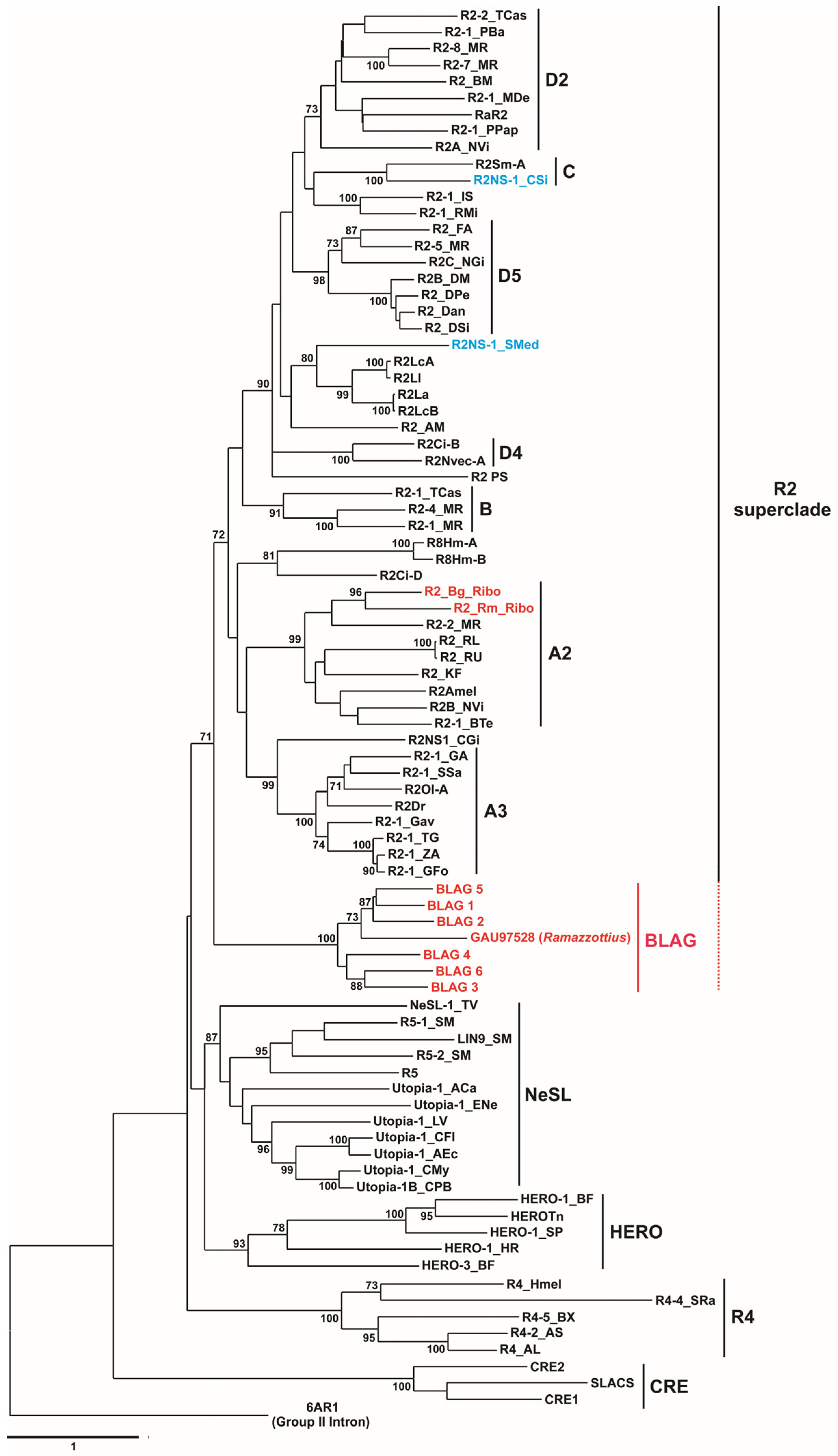
3.4. Structural and Functional Organization of BLAG Retrotransposons
- #1—FDTIVNEFTDFLSKAISLLPGPKHPATKYY,
- #2—RKKRRQASSEVSYKNSSNPQRASKRAREKRKEKYQYELTQFQYYNQRRKAVRSVL,
- #3—CKISITKIYEYFEERFSTENNNIRPDYSSSVTEPE.
3.5. Retrotransposon Phylogeny
3.6. R2 Retrotransposon Dynamics and Population Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oliver, K.R.; Greene, W.K. Transposable elements: Powerful facilitators of evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Bire, S.; Rouleux-Bonnin, F. Transposable Elements as Tools for Reshaping the Genome: It Is a Huge World after All! In Methods in Molecular Biology; Bigot, Y., Ed.; Humana Press: New York, NY, USA, 2012; Volume 859, pp. 1–28. [Google Scholar]
- Kim, Y.J.; Lee, J.; Han, K. Transposable elements: No more ‘JunkDNA’. Genom. Inform. 2012, 10, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, E.; González, J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Chénais, B. Transposable elements and human cancer: A causal relationship? Biochim. Biophys. Acta 2013, 1835, 28–35. [Google Scholar] [CrossRef]
- Trizzino, M.; Kapusta, A.; Brown, C.D. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genom. 2018, 19, 468. [Google Scholar] [CrossRef]
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of Transposable Elements in the Different Layers of Gene Expression Regulation. Int. J. Mol. Sci. 2019, 20, 5755. [Google Scholar] [CrossRef]
- Craig, N.L.; Craigie, R.; Gellert, M.; Lambowitz, A.M. Mobile DNA II; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Kapitonov, V.V.; Tempel, S.; Jurka, J. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene 2009, 448, 207–213. [Google Scholar] [CrossRef]
- Makałowski, W.; Gotea, V.; Pande, A.; Makałowska, I. Transposable Elements: Classification, Identification, and Their Use as a Tool For Comparative Genomics. Methods Mol. Biol. 2019, 1910, 177–207. [Google Scholar]
- Kojima, K.K.; Fujiwara, H. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol. Biol. Evol. 2005, 22, 2157–2165. [Google Scholar] [CrossRef]
- Kojima, K.K.; Seto, Y.; Fujiwara, H. The Wide Distribution and Change of Target Specificity of R2 Non-LTR Retrotransposons in Animals. PLoS ONE 2016, 11, e0163496. [Google Scholar] [CrossRef]
- Kojima, K.K.; Kuma, K.; Toh, H.; Fujiwara, H. Identification of rDNA-Specific Non-LTR Retrotransposons in Cnidaria. Mol. Biol. Evol. 2006, 23, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.; Arkhipova, I. Rotifer rDNA-specific R9 retrotransposable elements generate an exceptionally long target site duplication upon insertion. Gene 2009, 448, 145–150. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burke, W.D.; Malik, H.S.; Jones, J.P.; Eickbush, T.H. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 1999, 16, 502–511. [Google Scholar] [CrossRef]
- Yang, J.; Malik, H.S.; Eickbush, T.H. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 1999, 96, 7847–7852. [Google Scholar] [CrossRef]
- Luan, D.D.; Eickbush, T.H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell Biol. 1995, 15, 3882–3891. [Google Scholar] [CrossRef]
- Bibillo, A.; Eickbush, T.H. The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 2002, 316, 459–473. [Google Scholar] [CrossRef]
- Bibillo, A.; Eickbush, T.H. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J. Biol. Chem. 2004, 279, 14945–14953. [Google Scholar] [CrossRef]
- Christensen, S.M.; Bibillo, A.; Eickbush, T.H. Role of the Bombyx mori R2 element N-terminal domain in the target- primed reverse transcription (TPRT) reaction. Nucleic Acids Res. 2005, 33, 6461–6468. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.M.; Eickbush, T.H. R2 Target-primed reverse transcription: Ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell. Biol. 2005, 25, 6617–6628. [Google Scholar] [CrossRef]
- Christensen, S.M.; Ye, J.; Eickbush, T.H. RNA from the 5′-end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc. Natl. Acad. Sci. USA 2006, 103, 17602–17607. [Google Scholar] [CrossRef]
- Lathe, W.C.; Eickbush, T.H. A single lineage of R2 retrotransposable elements is an active, evolutionary stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 1997, 14, 1232–1241. [Google Scholar] [CrossRef][Green Version]
- Pérez-González, C.E.; Eickbush, T.H. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 2001, 158, 1557–1567. [Google Scholar] [PubMed]
- Zhang, X.; Eickbush, T.H. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics 2005, 170, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Eickbush, D.G.; Ye, J.; Zhang, X.; Burke, W.D.; Eickbush, T.H. Epigenetic regulation of retrotransposons within the nucleolus of Drosophila. Mol. Cell Biol. 2008, 28, 6452–6461. [Google Scholar] [CrossRef]
- Zhou, J.; Eickbush, T.H. The Pattern of R2 Retrotransposon Activity in Natural Populations of Drosophila simulans Reflects the Dynamic Nature of the rDNA Locus. PLoS Genet. 2009, 5, e1000386. [Google Scholar] [CrossRef] [PubMed]
- Mingazzini, V.; Luchetti, A.; Mantovani, B. R2 dynamics in Triops cancriformis (Bosc, 1801) (Crustacea, Branchiopoda, Notostraca): Turnover rate and 28S concerted evolution. Heredity 2011, 106, 567–575. [Google Scholar] [CrossRef][Green Version]
- Ghesini, S.; Luchetti, A.; Marini, M.; Mantovani, B. The Non-LTR Retrotransposon R2 in Termites (Insecta, Isoptera): Characterization and Dynamics. J. Mol. Evol. 2011, 72, 296–305. [Google Scholar] [CrossRef]
- Roiha, H.; Glover, D.M. Duplicated rDNA sequences of variable lengths flanking the short type I insertions in the rDNA of Drosophila melanogaster. Nucleic Acids Res. 1981, 9, 5521–5532. [Google Scholar] [CrossRef][Green Version]
- Fujiwara, H.; Ogura, T.; Takada, N.; Miyajima, N.; Ishikawa, H.; Maekawa, H. Introns and their flanking sequences of Bombyx mori rDNA. Nucleic Acids Res. 1984, 12, 6861–6869. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.K.; Fujiwara, H. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: Various multicopy RNA genes and microsatellites are selected as targets. Mol. Biol. Evol. 2004, 21, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.D.; Malik, H.S.; Lathe, W.C., 3rd; Eickbush, T.H. Are retrotransposons long-term hitchhikers? Nature 1998, 392, 141–142. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Jurka, J. R2 non-LTR retrotransposons in the bird genome. Repbase Rep. 2009, 9, 1329. [Google Scholar]
- Schal, C.; Hamilton, R.L. Integrated suppression of synanthropic cockroaches. Annu. Rev. Entomol. 1990, 35, 521–551. [Google Scholar] [CrossRef] [PubMed]
- Brenner, R.J. Economics and medical importance of German cockroaches. In Understanding and Controlling the German Cockroach; Rust, M.K., Owens, J.M., Reierson, D.A., Eds.; Oxford University Press: New York, NY, USA, 1995; pp. 72–92. [Google Scholar]
- Rosenstreich, D.L.; Eggleston, P.; Kattan, M.; Baker, D.; Slavin, R.G.; Gergen, P.; Mitchell, H.; McNiffMortimer, K.; Lynn, H.; Ownby, D.; et al. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. N. Eng. J. Med. 1997, 336, 1356–1363. [Google Scholar] [CrossRef]
- Gore, J.C.; Schal, C. Cockroach allergen biology and mitigation in the indoor environment. Annu. Rev. Entonol. 2007, 52, 439–463. [Google Scholar] [CrossRef] [PubMed]
- Mukha, D.V.; Kagramanova, A.S.; Lazebnaya, I.V.; Lazebnyl, O.E.; Vargo, E.L.; Schal, C. Intraspecific variation and population structure of the German cockroach, Blattella germanica, revealed with RFLP analysis of the non-transcribed spacer region of ribosomal DNA. Med. Vet. Entomol. 2007, 21, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Crissman, J.R.; Booth, W.; Santangelo, R.G.; Mukha, D.V.; Vargo, E.L.; Schal, C. Population genetic structure of the German cockroach (Blattodea: Blattellidae) in apartment buildings. J. Med. Entomol. 2010, 47, 553–564. [Google Scholar] [CrossRef]
- Booth, W.; Santangelo, R.G.; Vargo, E.L.; Mukha, D.V.; Schal, C. Population genetic structure in German cockroaches, Blattella germanica: Differentiated islands in an agricultural landscape. J. Hered. 2011, 102, 175–183. [Google Scholar] [CrossRef][Green Version]
- Vargo, E.L.; Crissman, J.R.; Booth, W.; Santangelo, R.G.; Mukha, D.V.; Schal, C. Hierarchical genetic analysis of German cockroach (Blattella germanica) populations from within buildings to across continents. PLoS ONE 2014, 9, e102321. [Google Scholar] [CrossRef]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, N.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; Volume 1–3. [Google Scholar]
- Kagramanova, A.S.; Kapelinskaya, T.V.; Korolev, A.L.; Mukha, D.V. R1 and R2 Retrotransposons of German Cockroach Blatella germanica: A Comparative Study of 5′-Truncated Copies Integrated into the Genome. Mol. Biol. 2007, 41, 608–615. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Kelley, L.; Mezulis, S.; Yates, C.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Söding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple sequence and structure alignment. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Martínez, A.M.; Gama, L.T.; Cañón, J.; Ginja, C.; Delgado, J.V.; Dunner, S.; Landi, V.; Martín-Burriel, I.; Penedo, M.C.; Rodellar, C.; et al. Genetic footprints of Iberian cattle in America 500 years after the arrival of Columbus. PLoS ONE 2012, 7, e49066. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Mukha, D.V.; Sidorenko, A.P.; Lazebnaya, I.V.; Wiegmann, B.M.; Schal, C. Analysis of intraspecies polymorphism in the ribosomal DNA cluster of the cockroach Blattella germanica. Insect Mol. Biol. 2000, 9, 217–222. [Google Scholar] [CrossRef]
- Mukha, D.V.; Wiegmann, B.M.; Schal, C. Evolution and phylogenetic information content of the ribosomal DNA repeat unit in the Blattoidea (Insecta). Insect Mol. Biol. 2002, 32, 951–960. [Google Scholar] [CrossRef]
- Mukha, D.V.; Mysina, V.; Mavropulo, V.; Schal, C. Structure and molecular evolution of the ribosomal DNA external transcribed spacer in the cockroach genus Blattella. Genome 2011, 54, 222–234. [Google Scholar] [CrossRef]
- Beauregard, A.; Curcio, M.J.; Belfort, M. The take and give between retrotransposable elements and their hosts. Annu Rev. Genet. 2008, 42, 587–617. [Google Scholar] [CrossRef] [PubMed]
- Belfort, M.; Curcio, M.J.; Lue, N.F. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 20304–20310. [Google Scholar] [CrossRef] [PubMed]
- Mukha, D.V.; Pasyukova, E.G.; Kapelinskaya, T.V.; Kagramanova, A.S. Endonuclease domain of the Drosophila melanogaster R2 non-LTR retrotransposon and related retroelements: A new model for transposition. Front. Genet. 2013, 4, 63. [Google Scholar] [CrossRef]
- Ruminski, D.J.; Webb, C.H.; Riccitelli, N.J.; Lupták, A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 2011, 286, 41286–41295. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Eickbush, T.H. NeSL-1, an ancient lineage of site-specific non-LTR retrotransposons from Caenorhabditis elegans. Genetics 2000, 154, 193–203. [Google Scholar]
- Furano, A.V. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acids Res. Mol. Biol. 2000, 64, 255–294. [Google Scholar]
- Fedorov, A. Regulation of mammalian LINE1 retrotransposon transcription. Cell Tissue Biol. 2009, 3, 1. [Google Scholar] [CrossRef]
- Alexandrova, E.A.; Olovnikov, I.A.; Malakhova, G.V.; Zabolotneva, A.A.; Suntsova, M.V.; Dmitriev, S.E.; Buzdina, A.A. Sense transcripts originated from an internal part of the human retrotransposon LINE-1 5′ UTR. Gene 2012, 511, 46–53. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Novikova, O.S.; Blinov, A.G. Origin, evolution, and distribution of different groups of non-LTR retrotransposons among eukaryotes. Russ. J. Genet. 2009, 45, 129–138. [Google Scholar] [CrossRef]
- Kagramanova, A.S.; Korolev, A.L.; Schal, C.; Mukha, D.V. Length Polymorphism of Integrated Copies of R1 and R2 Retrotransposons in the German Cockroach (Blattella germanica) as a Potential Marker for Population and Phylogenetic Studies. Russ. J. Genet. 2006, 42, 397–401. [Google Scholar] [CrossRef]
- Kagramanova, A.S.; Korolev, A.L.; Mukha, D.V. Analysis of the Inheritance Patterns of 5′-Truncated Copies of the German Cockroach R2 Retroposons in Individual Crosses. Russ. J. Genet. 2010, 46, 1290–1294. [Google Scholar] [CrossRef]
- Oyun, N.Y.; Zagoskina, A.S.; Mukha, D.V. Inheritance of 5′-Truncated Copies of R2 Retrotransposon in a Series of Generations of German Cockroach, Blattella germanica. Russ. J. Genet. 2018, 54, 1438–1444. [Google Scholar] [CrossRef]
- Cochran, D.G.; Ross, M.H. Preliminary studies of the chromosomes of twelve cockroach species (Blattaria: Blattidae, Blatellidae, Blaberidae). Ann. Entomol. Am. 1967, 60, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Roth, L.M. Chromosome numbers of the Blattaria. Ann. Entomol. Soc. Am. 1970, 63, 1520–1547. [Google Scholar] [CrossRef]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef]
- Nei, M.; Gu, X.; Sitnikova, T. Evolution by the birthand-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 1997, 94, 7799–7806. [Google Scholar] [CrossRef]
- Gao, X.; Hou, Y.; Ebina, H.; Levin, H.L.; Voytas, D.F. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008, 18, 359–369. [Google Scholar] [CrossRef]
- Linheiro, R.S.; Bergman, C.M. Whole genome resequencing reveals natural target site preferences of transposable elements in Drosophila melanogaster. PLoS ONE 2012, 7, e30008. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.G.; Esnault, C.; Guo, Y.; Hung, S.; McQueen, P.G.; Levin, H.L. Serial number tagging reveals a prominent sequence preference of retrotransposon integration. Nucleic Acids Res. 2014, 42, 8449–8460. [Google Scholar] [CrossRef] [PubMed]
- Kent, T.V.; Uzunovic, J.; Wright, S.I. Coevolution between transposable elements and recombination. Phil. Trans. R. Soc. B 2017, 372, 20160458. [Google Scholar] [CrossRef] [PubMed]
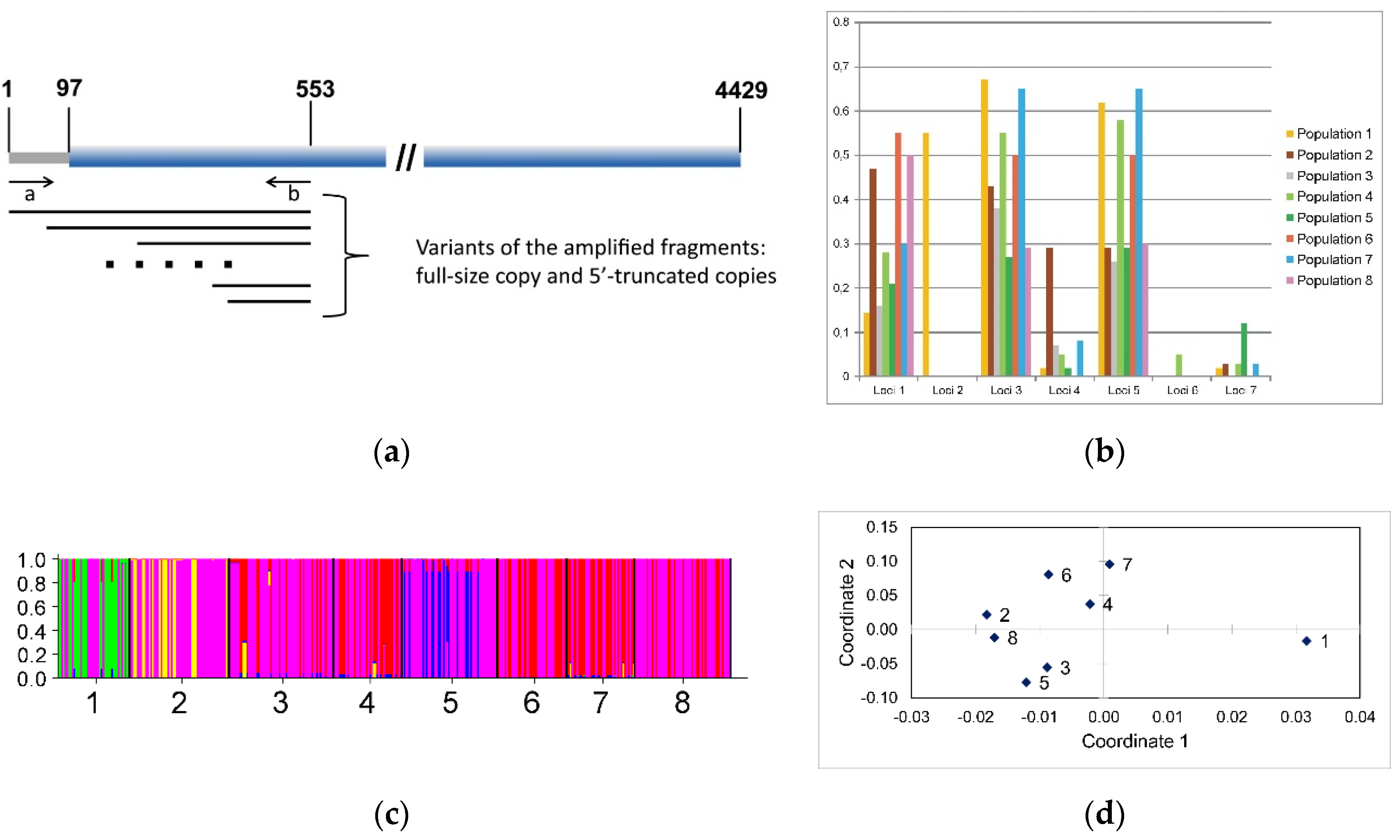
| Population | Number of Indiviuals * | Loci ** | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 1 | 42/9 | 6/0.143 | 23/0.55 | 28/0.67 | 1/0.02 | 26/0.62 | 0/0 | 1/0.02 |
| 2 | 58/16 | 27/0.47 | 0/0 | 25/0.43 | 17/0.29 | 17/0.29 | 0/0 | 2/0.03 |
| 3 | 61/31 | 10/0.16 | 0/0 | 23/0.38 | 4/0.07 | 16/0.26 | 0/0 | 0/0 |
| 4 | 40/10 | 11/0.28 | 0/0 | 22/0.55 | 2/0.05 | 23/0.58 | 2/0.05 | 1/0.03 |
| 5 | 56/23 | 12/0.21 | 0/0 | 15/0.27 | 1/0.02 | 16/0.29 | 0/0 | 6/0.12 |
| 6 | 40/8 | 22/0.55 | 0/0 | 20/0.50 | 0/0 | 20/0.50 | 0/0 | 0/0 |
| 7 | 40/10 | 12/0.30 | 0/0 | 26/0.65 | 3/0.08 | 26/0.65 | 0/0 | 1/0.03 |
| 8 | 56/20 | 28/0.50 | 0/0 | 17/0.29 | 0/0 | 17/0.30 | 0/0 | 0/0 |
| Population | Cluster | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 1 | 0.014 | 0.600 | 0.006 | 0.000 | 0.379 |
| 2 | 0.001 | 0.000 | 0.000 | 0.293 | 0.706 |
| 3 | 0.246 | 0.001 | 0.009 | 0.013 | 0.731 |
| 4 | 0.548 | 0.001 | 0.012 | 0.007 | 0.432 |
| 5 | 0.023 | 0.001 | 0.257 | 0.000 | 0.719 |
| 6 | 0.497 | 0.000 | 0.003 | 0.000 | 0.500 |
| 7 | 0.631 | 0.000 | 0.009 | 0.010 | 0.350 |
| 8 | 0.303 | 0.000 | 0.000 | 0.000 | 0.697 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zagoskina, A.; Firsov, S.; Lazebnaya, I.; Lazebny, O.; Mukha, D.V. R2 and Non-Site-Specific R2-Like Retrotransposons of the German Cockroach, Blattella germanica. Genes 2020, 11, 1202. https://doi.org/10.3390/genes11101202
Zagoskina A, Firsov S, Lazebnaya I, Lazebny O, Mukha DV. R2 and Non-Site-Specific R2-Like Retrotransposons of the German Cockroach, Blattella germanica. Genes. 2020; 11(10):1202. https://doi.org/10.3390/genes11101202
Chicago/Turabian StyleZagoskina, Arina, Sergei Firsov, Irina Lazebnaya, Oleg Lazebny, and Dmitry V. Mukha. 2020. "R2 and Non-Site-Specific R2-Like Retrotransposons of the German Cockroach, Blattella germanica" Genes 11, no. 10: 1202. https://doi.org/10.3390/genes11101202
APA StyleZagoskina, A., Firsov, S., Lazebnaya, I., Lazebny, O., & Mukha, D. V. (2020). R2 and Non-Site-Specific R2-Like Retrotransposons of the German Cockroach, Blattella germanica. Genes, 11(10), 1202. https://doi.org/10.3390/genes11101202





