Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis
Abstract
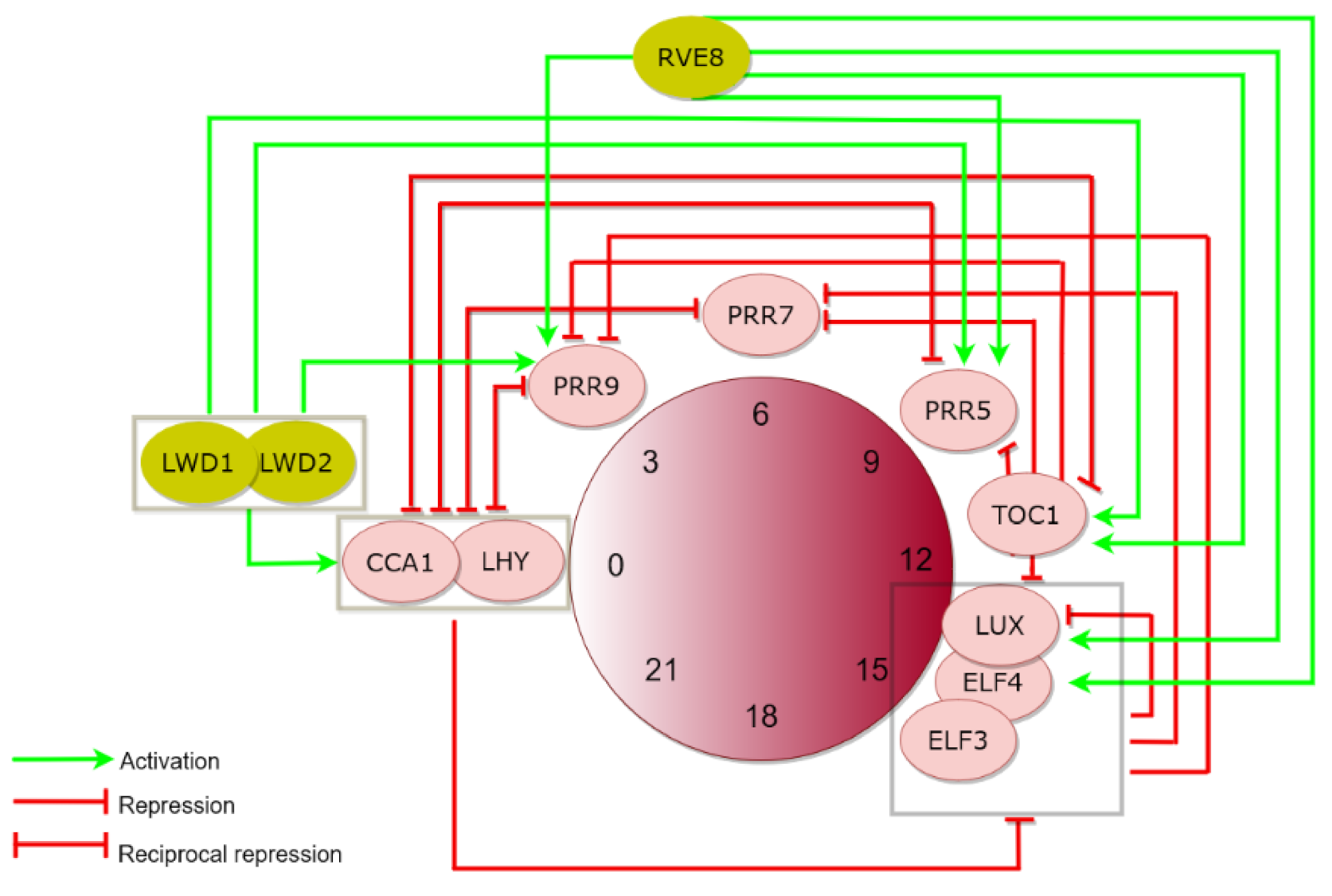
1. The Plant Circadian Clock
2. Transcriptional Dynamics and Chromatin Status
3. Functional Link between Chromatin and Circadian Transcription
4. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gachon, F.; Nagoshi, E.; Brown, S.A.; Ripperger, J.; Schibler, U. The mammalian circadian timing sytem: From gene expression to physiology. Chromosoma 2004, 113, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends Endocrinol. Metab. 2016, 27, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.H.; Takahashi, J.S. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef] [PubMed]
- Doherty, C.J.; Kay, S.A. Circadian surprise—It’s not all about transcription. Science 2012, 338, 338–400. [Google Scholar] [CrossRef] [PubMed]
- Young, M.W.; Kay, S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Gen. 2001, 2, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tingay, S.; Wang, Z.Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Resco, V.; Hartwell, J.; Hall, A. Ecological implications of plants ability to tell the time. Ecol. Lett. 2009, 12, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, S.; Green, R.M. Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 2009, 12, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef] [PubMed]
- Más, P. Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008, 18, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Mas, P. Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell. 2014, 26, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; de Leone, M.J.; Torchio, J.; Reichel, M.; Staiger, D. Beyond transcription: Fine-tuning of circadian timekeeping by post-transcriptional regulation. Genes 2018, 9, 616. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, J.; Huang, F.Y.; Yang, S.; Wu, K. The plant circadian clock and chromatin modifications. Genes 2018, 9, 561. [Google Scholar] [CrossRef] [PubMed]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef]
- Millar, A.J.; Carré, I.A.; Strayer, C.A.; Chua, N.H.; Kay, S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 1995, 267, 1161–1163. [Google Scholar] [CrossRef]
- DSomers, E.; Webb, A.A.R.; Pearson, M.; Kay, S.A. The short-period mutant toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998, 125, 485–494. [Google Scholar]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Mas, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Más, P.; Alabadí, D.; Yanovsky, M.J.; Oyama, T.; Kay, S.A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 2003, 15, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Nakamichi, N. Pseudo-Response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 2005, 46, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Makino, S.; Kiba, T.; Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 791–803. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Manfield, I.; Stockley, P.; Carré, I.A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 2015, 10, e0143943. [Google Scholar] [CrossRef] [PubMed]
- Hicks, K.A.; Albertson, T.M.; Wagner, D.R. EARLY FLOWERING3 Encodes a Novel Protein That Regulates Circadian Clock Function and Flowering in Arabidopsis. Plant Cell 2001, 13, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognár, L.; Nagy, F.; Millar, A.J. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef]
- Onai, K.; Ishiura, M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005, 10, 963–972. [Google Scholar] [CrossRef]
- McWatters, H.G.; Kolmos, E.; Hall, A.; Doyle, M.R.; Amasino, R.M.; Gyula, P.; Nagy, F.; Millar, A.J.; Davis, S.J. ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 2007, 144, 391–401. [Google Scholar] [CrossRef]
- Kolmos, E.; Nowak, M.; Werner, M.; Fischer, K.; Schwarz, G.; Mathews, S.; Schoof, H.; Nagy, F.; Bujnicki, J.M.; Davis, S.J. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009, 3, 350–366. [Google Scholar] [CrossRef]
- McWatters, H.G.; Bastow, R.M.; Hall, A.; Millar, A.J. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 2000, 408, 716–720. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed]
- Ezer, D.; Jung, J.H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stockle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 Regulates GIGANTEA Chromatin Access through Subnuclear Sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; López-Salmerón, V.; Davière, J.-M.; Prat, S. ELF3-PIF4 Interaction Regulates Plant Growth Independently of the Evening Complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyam, T.; Kinoshita, T.; Nakamichi, N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef]
- Pokhilko, A.; Fernandez, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1,is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 335, 75–79. [Google Scholar] [CrossRef]
- Portolés, S.; Más, P. The functional interplay between protein kinase CK2 and cca1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010, 6, e1001201. [Google Scholar] [CrossRef]
- Li, G.; Siddiqui, H.; Teng, Y.; Lin, R.; Wan, X.; Li, J.; Lau, O.-S.; Ouyang, X.; Dai, M.; Wan, J.; et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 2011, 13, 616–622. [Google Scholar] [CrossRef]
- SLu, X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.J.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 Interact in the Control of Hypocotyl Length and Flowering Time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar]
- Dixon, L.E.; Knox, K.; Kozma-Bognar, L.; Southern, M.M.; Pokhilko, A.; Millar, A.J. Temporal Repression of Core Circadian Genes Is Mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011, 21, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.Y.; Helfer, A.; Nusinow, D.A.; Kay, S.A. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav. 2012, 7, 170–173. [Google Scholar] [CrossRef]
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976. [Google Scholar] [CrossRef]
- Wu, J.F.; Wang, Y.; Wu, S.H. Two new clock proteins, LWD1 and LWD2, regulate arabidopsis photoperiodic flowering. Plant Physiol. 2008, 148, 948–959. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.F.; Nakamichi, N.; Sakakibara, H.; Nam, H.G.; Wu, S.H. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 Form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011, 23, 486–498. [Google Scholar] [CrossRef]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef]
- Xie, Q.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.; Zhang, C.; Li, Y.; Xing, H.; Zhi, L.; Yue, Z.; et al. LNK1 and LNK2 Are Transcriptional Coactivators in the Arabidopsis Circadian Oscillator. Plant Cell Online 2014, 26, 2843–2857. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Mas, P. Interactive roles of chromatin regulation and circadian clock function in plants. Genome Biol. 2019, 20, 62. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Ronald, J. Making the clock tick: The transcriptional landscape of the plant circadian clock. F1000Research 2017, 6. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Pfluger, J.; Wagner, D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007, 10, 645–652. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Scheid, O.M. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Probst, A.V.; Desvoyes, B.; Gutierrez, C. Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 2020, 71, 5191–5204. [Google Scholar] [CrossRef]
- Dong, P.; Tu, X.; Liang, Z.; Kang, B.-H.; Zhong, S. Plant and animal chromatin three-dimensional organization: Similar structures but different functions. J. Exp. Bot. 2020, 71, 5119–5128. [Google Scholar] [CrossRef]
- Chen, Z.J.; Tian, L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 2007, 1769, 295–307. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef]
- Servet, C.; Silva, N.C.E.; Zhou, D.X. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Singh, R.; Lodhi, N. Histone demethylases and control of gene expression in plants. Cell. Mol. Biol. 2014, 60, 97–105. [Google Scholar]
- Perales, M.; Más, P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 2007, 19, 2111–2123. [Google Scholar] [CrossRef]
- Song, H.R.; Noh, Y.S. Rhythmic Oscillation of Histone Acetylation and Methylation at the Arabidopsis Central Clock Loci. Mol. Cells 2012, 34, 279–287. [Google Scholar] [CrossRef]
- Malapeira, J.; Khaitova, L.C.; Mas, P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2012, 109, 21540–21545. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. The HAF2 protein shapes histone acetylation levels of PRR5 and LUX loci in Arabidopsis. Planta 2018, 248, 513–518. [Google Scholar] [CrossRef]
- Lee, H.G.; Hong, C.; Seo, P.J. The Arabidopsis sin3-hdac complex facilitates temporal histone deacetylation at the Cca1 and Prr9 loci for robust circadian oscillation. Front. Plant Sci. 2019, 10, 171. [Google Scholar] [CrossRef]
- Lee, K.; Mas, P.; Seo, P.J. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2019, 2, 143. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Baek, D.; Cha, J.Y.; Liao, X.; Kang, S.H.; McClung, C.R.; Lee, S.Y.; Yun, D.J.; Kim, W.Y. Hos15 interacts with the histone deacetylase hda9 and the evening complex to epigenetically regulate the floral activator gigantea. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.-Y.; Chen, F.-F.; Li, C.; Chen, C.; Lai, Y.-C.; Chen, J.-H.; Cui, Y.; Wu, K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018, 46, 10669–10681. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Chen, J.H.; Cui, Y.; Wu, K. The LDL1/2-HDA6 histone modification complex interacts with TOC1 and regulates the core circadian clock components in Arabidopsis. Front. Plant Sci. 2019, 10, 233. [Google Scholar] [CrossRef]
- Song, Q.; Huang, T.-Y.; Yu, H.H.; Ando, A.; Mas, P.; Ha, M.; Chen, Z.J. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 2019, 20, 170. [Google Scholar] [CrossRef]
- Ma, Y.; Gil, S.; Grasser, K.D.; Mas, P. Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in arabidopsis. Plant Cell 2018, 30, 907–924. [Google Scholar] [CrossRef]
- Zheng, S.; Hu, H.; Ren, H.; Yang, Z.; Qiu, Q.; Qi, W.; Liu, X.; Chen, X.; Cui, X.; Li, S.; et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 2019, 10, 1303. [Google Scholar] [CrossRef]
- Woloszynska, M.; le Gall, S.; Neyt, P.; Boccardi, T.M.; Grasser, M.; Längst, G.; Aesaert, S.; Coussens, G.; Dhondt, S.; van de Slijke, E.; et al. Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc. Natl. Acad. Sci. USA 2019, 116, 8060–8069. [Google Scholar] [CrossRef]
- Tong, M.; Lee, K.; Ezer, D.; Cortijo, S.; Jung, J.; Charoensawan, V.; Box, M.S.; Jaeger, K.E.; Takahashi, N.; Mas, P.; et al. The evening complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. 2020, 182, 612–625. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 2013, 110, 761–766. [Google Scholar] [CrossRef]
- Raisner, R.M.; Madhani, H.D. Patterning chromatin: Form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 2006, 16, 119–124. [Google Scholar] [CrossRef] [PubMed]
- 92. Raisner, R.M.; Hartley, P.D.; Meneghini, M.D.; Bao, M.Z.; Liu, C.L.; Schreiber, S.L.; Rando, O.J.; Madhani, H.D. Histone variant H2A.Z marks the 5’ ends of both active and inactive genes in euchromatin. Cell 2005, 123, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Rugnone, M.L.; Soverna, A.F.; Sanchez, S.E.; Schlaen, R.G.; Hernando, C.E.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Mas, P.; et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, P.; Ma, Y.; Yanovsky, M.J.; Mas, P. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 5249–5253. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Takeuchi, A.; Nomoto, Y.; Nakamichi, N.; Yamashino, T. The LNK1 night light-inducible and clock-regulated gene is induced also in response to warm-night through the circadian clock nighttime repressor in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e28505. [Google Scholar] [CrossRef] [PubMed]
- Nishio, H.; Nagano, A.J.; Ito, T.; Suzuki, Y.; Kudoh, H. Seasonal plasticity and diel stability of H3K27me3 in natural fluctuating environments. Nat. Plants 2020, 1091–1097. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Johansson, M.; Staiger, D. Time to flower: Interplay between photoperiod and the circadian clock. J. Exp. Bot. 2015, 66, 719–730. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef]
- Duren, Z.; Wang, Y.; Wang, J.; Zhao, X.-M.; Lv, L.; Li, X.; Liu, J.; Zhu, X.-G.; Chen, L.; Wang, Y. Hierarchical graphical model reveals HFR1 bridging circadian rhythm and flower development in Arabidopsis thaliana. NPJ Syst. Biol. Appl. 2019, 5, 28. [Google Scholar] [CrossRef]
- You, Y.; Sawikowska, A.; Neumann, M.; Posé, D.; Capovilla, G.; Langenecker, T.; Neher, R.A.; Krajewski, P.; Schmid, M. Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nat. Commun. 2017, 8, 15120. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, Z.; Liu, Y.; An, Z.; Peng, M.; Shen, W.H.; Dong, A.; Yu, Y. MRG1/2 histone methylation readers and HD2C histone deacetylase associate in repression of the florigen gene FT to set a proper flowering time in response to day-length changes. New Phytol. 2020, 227, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Wagner, D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 2015, 23, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Gao, Z.; Wang, Y.; Chen, Z.; Zhang, W.; Huang, J.; Yu, H.; He, Y. The NUCLEAR FACTOR-CONSTANS complex antagonizes Polycomb repression to de-repress FLOWERING LOCUS T expression in response to inductive long days in Arabidopsis. Plant J. 2018, 95, 17–29. [Google Scholar] [CrossRef]
- Jones, M.A.; Morohashi, K.; Grotewold, E.; Harmer, S.L. Arabidopsis JMJD5/JMJ30 Acts Independently of LUX ARRHYTHMO Within the Plant Circadian Clock to Enable Temperature Compensation. Front. Plant Sci. 2019, 10, 57. [Google Scholar] [CrossRef]
- Suzuki, M.; McCarty, D.R. Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 2008, 11, 548–553. [Google Scholar] [CrossRef]
- Zha, P.; Liu, S.; Li, Y.; Ma, T.; Yang, L.; Jing, Y.; Lin, R. The Evening Complex and the Chromatin-Remodeling Factor PICKLE Coordinately Control Seed Dormancy by Directly Repressing DOG1 in Arabidopsis. Plant Commun. 2020, 1, 100011. [Google Scholar] [CrossRef]
- Nonogaki, H. The Long-Standing Paradox of Seed Dormancy Unfolded? Trends Plant Sci. 2019, 24, 989–998. [Google Scholar] [CrossRef]
- Sai, J.; Johnson, C.H. Different circadian oscillators control Ca(2+) fluxes and lhcb gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 11659–11663. [Google Scholar] [CrossRef]
- Thain, S.C.; Murtas, G.; Lynn, J.R.; McGrath, R.B.; Millar, A.J. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002, 130, 102–110. [Google Scholar] [CrossRef]
- Michael, T.P.; Salome, P.A.; McClung, C.R. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci. USA 2003, 100, 6878–6883. [Google Scholar] [CrossRef] [PubMed]
- Muranaka, T.; Oyama, T. Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci. Adv. 2016, 2, e1600500. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Takahashi, N.; Hirata, Y.; Ronald, J.; Porco, S.; Davis, S.J.; Nusinow, D.A.; Kay, S.A.; Mas, P. Amobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 2020, 6, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Yakir, E.; Hassidim, M.; Melamed-Book, N.; Hilman, D.; Kron, I.; Green, R.M. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011, 68, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Ukai, K.; Oyama, T. Self-arrangement of cellular circadian rhythms through phase-resetting in plant roots. Phys. Rev. E 2012, 86, 41917. [Google Scholar] [CrossRef] [PubMed]
- Thain, S.C.; Hall, A.; Millar, A.J. Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 2000, 10, 951–956. [Google Scholar] [CrossRef]
- Fukuda, H.; Nakamichi, N.; Hisatsune, M.; Murase, H.; Mizuno, T. Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Phys. Rev. Lett. 2007, 99, 98102. [Google Scholar] [CrossRef]
- Wenden, B.; Toner, D.L.K.; Hodge, S.K.; Grima, R.; Millar, A.J. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc. Natl. Acad. Sci. USA 2012, 109, 6757–6762. [Google Scholar] [CrossRef]
- Greenwood, M.; Domijan, M.; Gould, P.D.; Hall, A.J.W.; Locke, J.C.W. Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana. PLOS Biol. 2019, 17, e3000407. [Google Scholar] [CrossRef]
- Gould, P.D.; Domijan, M.; Greenwood, M.; Tokuda, I.T.; Rees, H.; Kozma-Bognar, L.; Hall, A.J.; Locke, J.C. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 2018, 7, e31700. [Google Scholar] [CrossRef]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A Hierarchical Multi-oscillator Network Orchestrates the Arabidopsis Circadian System. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [PubMed]
- James, A.B.; Monreal, J.A.; Nimmo, G.A.; Kelly, C.L.; Herzyk, P.; Jenkins, G.I.; Nimmo, H.G. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 2008, 322, 1832–1835. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, H.G. Entrainment of Arabidopsis roots to the light:dark cycle by light piping. Plant Cell Environ. 2018, 41, 1742–1748. [Google Scholar] [CrossRef]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef]
- Yu, M.; Ren, B. The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol. 2017, 33, 265–289. [Google Scholar] [CrossRef]
| Histone Mark | Clock/Chromatin-Related Factor | Regulated Clock Component | Reference |
|---|---|---|---|
| Acetylation | HAF2 | PRR5, LUX | [79] |
| Deacetylation | Sin3-HDAC | CCA1, PRR9 | [80] |
| ELF3-HDA9 | TOC1 | [81] | |
| EC-HDA9-HOS15 | GI | [82] | |
| HDA6-CCA1/LHY | TOC1 | [83] | |
| HDA6-TOC1 | CCA1, LHY, other clock genes | [84] | |
| Methylation | SDG2 (ATXR3) | CCA1, LHY | [78,85] |
| RVE8/LNKs | PRR5, TOC1 nascent RNAs | [86] | |
| Demethylation | JMJ14 | CCA1, LHY | [85] |
| CCA1/LHY-LDL1/2 | TOC1 | [83] | |
| TOC1-LDL1/2 | CCA1, LHY, other clock genes | [84] | |
| JMJ13 | CCA1, LHY | [87] | |
| Monoubiquitination | HUB1/HUB2 | CCA1 | [88] |
| Histone Variant H2A.Z | ELF3-SWR1 | PRR7, PRR9 | [89] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maric, A.; Mas, P. Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis. Genes 2020, 11, 1170. https://doi.org/10.3390/genes11101170
Maric A, Mas P. Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis. Genes. 2020; 11(10):1170. https://doi.org/10.3390/genes11101170
Chicago/Turabian StyleMaric, Aida, and Paloma Mas. 2020. "Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis" Genes 11, no. 10: 1170. https://doi.org/10.3390/genes11101170
APA StyleMaric, A., & Mas, P. (2020). Chromatin Dynamics and Transcriptional Control of Circadian Rhythms in Arabidopsis. Genes, 11(10), 1170. https://doi.org/10.3390/genes11101170




