Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23
Abstract
1. Introduction
2. Materials and Methods
2.1. Leishmania Strains and Culture
2.2. Promastigote Cultivation
2.3. Transfections, Selection, and Cell Cloning
2.4. Construction and Preparation of Recombinant DNA
2.5. PCR-Amplification of Targeting Constructs
2.6. Analytical PCR
2.7. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qRT-PCR)
2.8. Next Generation Sequencing
2.9. Western Blotting
2.10. Immunofluorescence Assays
2.11. Flow Cytometry Cell Analysis
2.12. In Vitro Infection of Murine Bone Marrow-Derived Macrophages
2.13. In Silico Procedures
3. Results
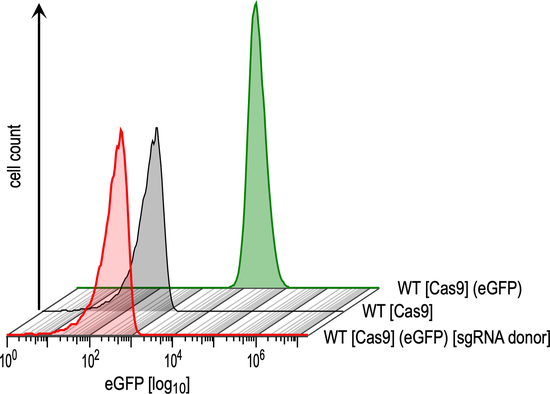
3.1. Optimisation and Validation of the CRISPR-Cas9 System in L. braziliensis
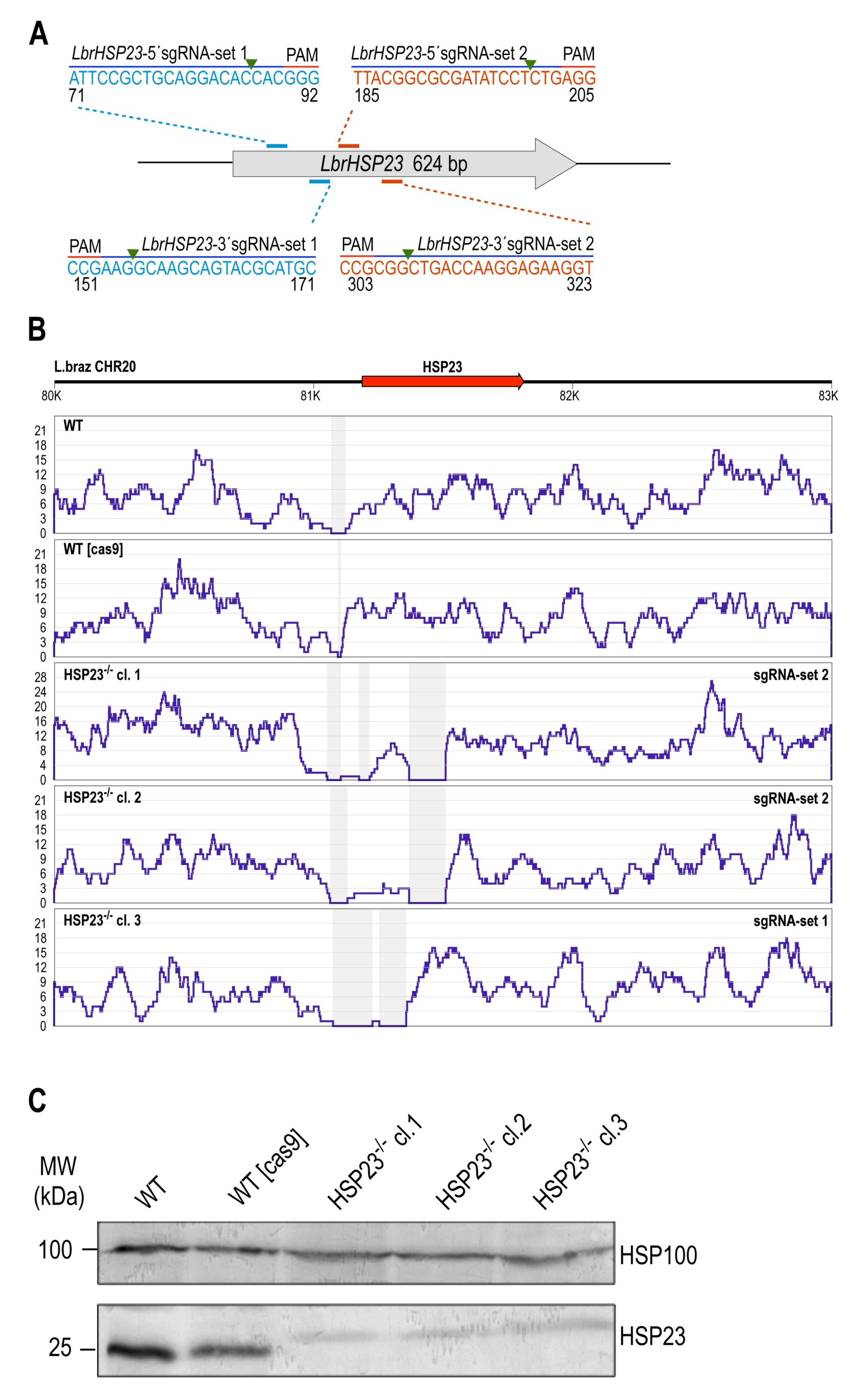
3.2. CRISPR–Cas9-Mediated Disruption of Endogenous HSP23 and HSP100 Genes in L. braziliensis
3.2.1. LbrHSP23 Gene Replacement
3.2.2. LbrHSP100 Gene Replacement
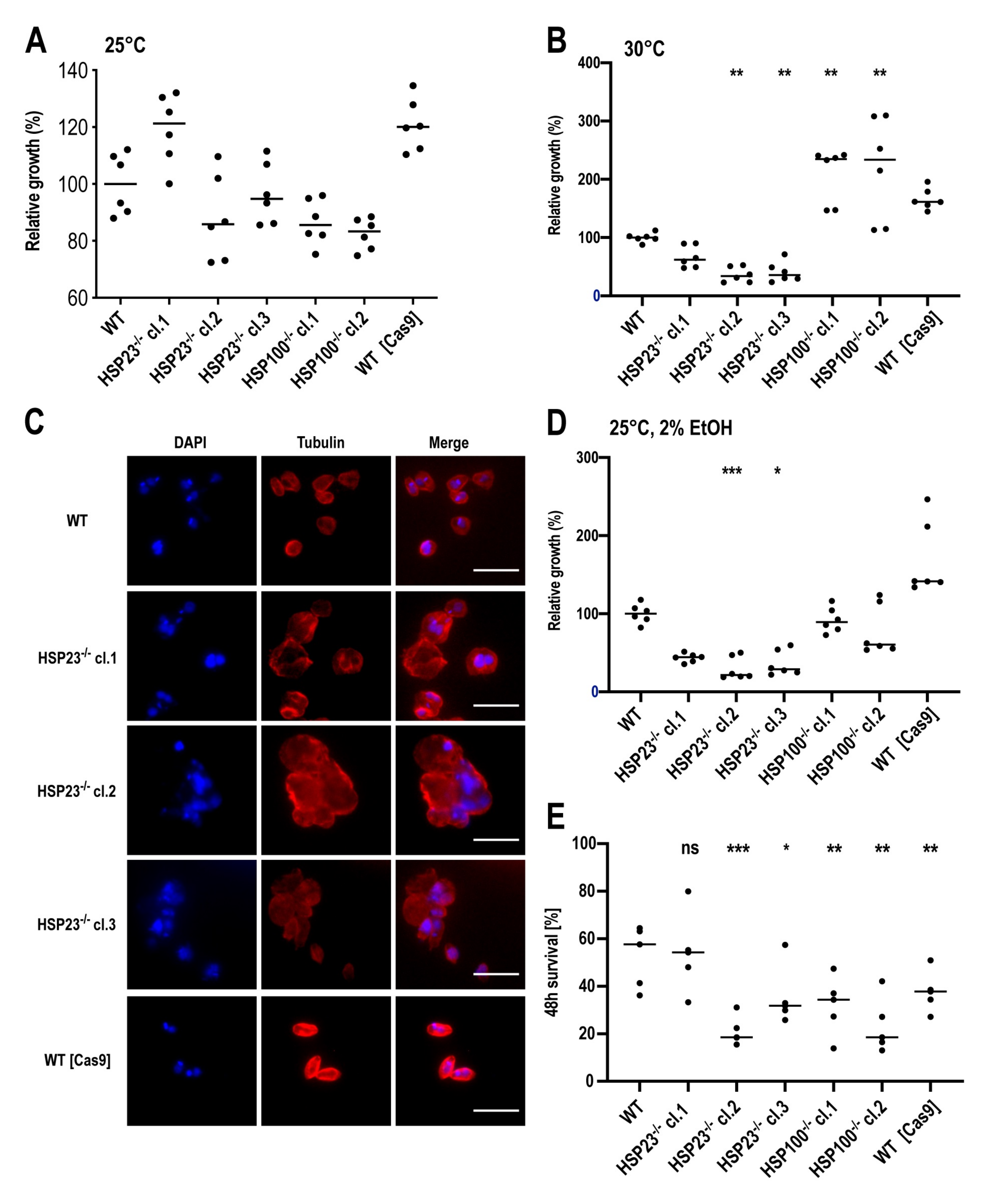
3.3. L. braziliensis HSP23- and HSP100-Null Mutant Phenotypes Resemble Those Described for Old World Leishmania
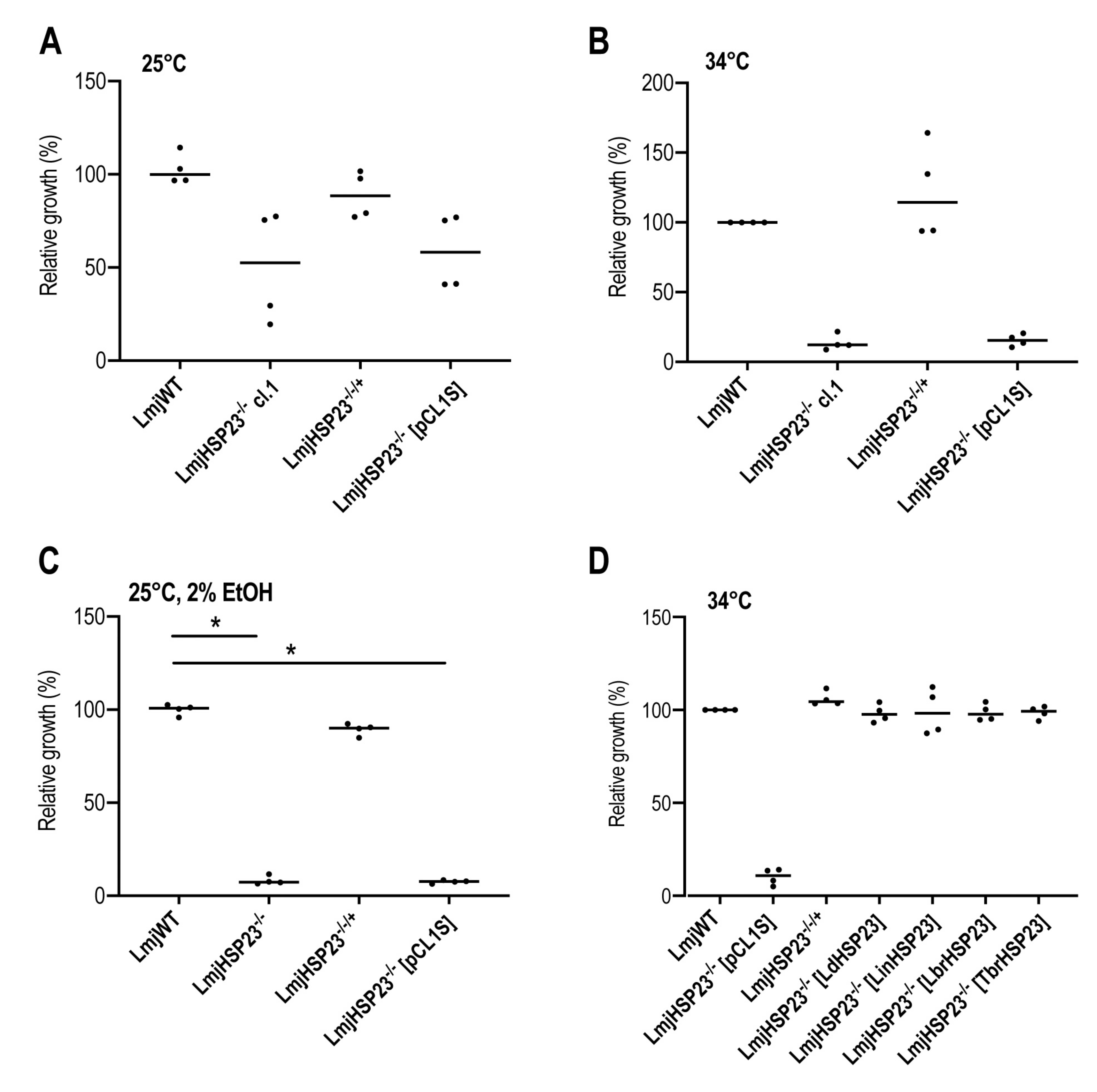
3.4. Complementation Studies in L. major HSP23-Null Mutants Indicate a Conserved Function in Thermotolerance for Trypanosomatid HSP23
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marsden, P.D. Mucosal leishmaniasis ("espundia" Escomel, 1911). Trans R Soc. Trop. Med. Hyg. 1986, 80, 859–876. [Google Scholar] [CrossRef]
- Amato, V.S.; Tuon, F.F.; Siqueira, A.M.; Nicodemo, A.C.; Neto, V.A. Treatment of mucosal leishmaniasis in Latin America: systematic review. Am. J. Trop. Med. Hyg. 2007, 77, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, J.; Ramirez, L.; Adaui, V.; Zimic, M.; Tulliano, G.; Miranda-Verastegui, C.; Lazo, M.; Loayza-Muro, R.; De Doncker, S.; Maurer, A.; et al. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J. Infect. Dis. 2007, 195, 1846–1851. [Google Scholar] [CrossRef] [PubMed]
- Reithinger, R.; Dujardin, J.C.; Louzir, H.; Pirmez, C.; Alexander, B.; Brooker, S. Cutaneous leishmaniasis. Lancet Infect. Dis. 2007, 7, 581–596. [Google Scholar] [CrossRef]
- Cruz, A.; Beverley, S.M. Gene replacement in parasitic protozoa. Nature 1990, 348, 171–173. [Google Scholar] [CrossRef]
- Cruz, A.; Coburn, C.M.; Beverley, S.M. Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. USA 1991, 88, 7170–7174. [Google Scholar] [CrossRef]
- Zirpel, H.; Clos, J. Gene Replacement by Homologous Recombination. Methods Mol. Biol. 2019, 1971, 169–188. [Google Scholar]
- Peacock, C.S.; Seeger, K.; Harris, D.; Murphy, L.; Ruiz, J.C.; Quail, M.A.; Peters, N.; Adlem, E.; Tivey, A.; Aslett, M.; et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007, 39, 839–847. [Google Scholar] [CrossRef]
- Lye, L.F.; Owens, K.; Shi, H.; Murta, S.M.; Vieira, A.C.; Turco, S.J.; Tschudi, C.; Ullu, E.; Beverley, S.M. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010, 6, e1001161. [Google Scholar] [CrossRef]
- De Paiva, R.M.; Grazielle-Silva, V.; Cardoso, M.S.; Nakagaki, B.N.; Mendonca-Neto, R.P.; Canavaci, A.M.; Souza Melo, N.; Martinelli, P.M.; Fernandes, A.P.; daRocha, W.D.; et al. Amastin Knockdown in Leishmania braziliensis Affects Parasite–Macrophage Interaction and Results in Impaired Viability of Intracellular Amastigotes. PLoS Pathog. 2015, 11, e1005296. [Google Scholar] [CrossRef]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off–target gene regulation by RNAi. Nat Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.J.; Doudna, J.A. CRISPR–Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual–RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double–Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Bibikova, M.; Beumer, K.; Trautman, J.K.; Carroll, D. Enhancing gene targeting with designed zinc finger nucleases. Science 2003, 300, 764. [Google Scholar] [CrossRef]
- Peng, D.; Kurup, S.P.; Yao, P.Y.; Minning, T.A.; Tarleton, R.L. CRISPR–Cas9–mediated single–gene and gene family disruption in Trypanosoma cruzi. mBio 2014, 6, e02097-14. [Google Scholar] [CrossRef]
- Beneke, T.; Madden, R.; Makin, L.; Valli, J.; Sunter, J.; Gluenz, E. A CRISPR Cas9 high–throughput genome editing toolkit for kinetoplastids. R Soc. Open Sci. 2017, 4, 170095. [Google Scholar] [CrossRef]
- Vasquez, J.J.; Wedel, C.; Cosentino, R.O.; Siegel, T.N. Exploiting CRISPR–Cas9 technology to investigate individual histone modifications. Nucleic Acids Res. 2018, 46, e106. [Google Scholar] [CrossRef]
- Sollelis, L.; Ghorbal, M.; MacPherson, C.R.; Martins, R.M.; Kuk, N.; Crobu, L.; Bastien, P.; Scherf, A.; Lopez-Rubio, J.-J.; Sterkers, Y. First efficient CRISPR–Cas9–mediated genome editing in Leishmania parasites. Cell. Microbiol. 2015, 17, 1405–1412. [Google Scholar] [CrossRef]
- Zhang, W.W.; Matlashewski, G. CRISPR–Cas9–Mediated Genome Editing in Leishmania donovani. MBio 2015, 6, e00861. [Google Scholar] [CrossRef]
- Martel, D.; Beneke, T.; Gluenz, E.; Spath, G.F.; Rachidi, N. Characterisation of Casein Kinase 1.1 in Leishmania donovani Using the CRISPR Cas9 Toolkit. Biomed. Res. Int. 2017, 2017, 4635605. [Google Scholar] [CrossRef] [PubMed]
- Soares Medeiros, L.C.; South, L.; Peng, D.; Bustamante, J.M.; Wang, W.; Bunkofske, M.; Perumal, N.; Sanchez-Valdez, F.; Tarleton, R.L. Rapid, Selection–Free, High–Efficiency Genome Editing in Protozoan Parasites Using CRISPR–Cas9 Ribonucleoproteins. mBio 2017, 8. [Google Scholar] [CrossRef]
- Fernandez-Prada, C.; Sharma, M.; Plourde, M.; Bresson, E.; Roy, G.; Leprohon, P.; Ouellette, M. High–throughput Cos–Seq screen with intracellular Leishmania infantum for the discovery of novel drug–resistance mechanisms. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Ishemgulova, A.; Hlavacova, J.; Majerova, K.; Butenko, A.; Lukes, J.; Votypka, J.; Volf, P.; Yurchenko, V. CRISPR/Cas9 in Leishmania mexicana: A case study of LmxBTN1. PloS ONE 2018, 13, e0192723. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.M.; Baumgarten, S.; Glover, L.; Hutchinson, S.; Rachidi, N. CRISPR in Parasitology: Not Exactly Cut and Dried! Trends Parasitol 2019, 35, 409–422. [Google Scholar] [CrossRef]
- Cruz, A.K.; Titus, R.; Beverley, S.M. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc. Natl. Acad. Sci. USA 1993, 90, 1599–1603. [Google Scholar] [CrossRef]
- Sterkers, Y.; Crobu, L.; Lachaud, L.; Pages, M.; Bastien, P. Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014, 30, 429–435. [Google Scholar] [CrossRef]
- Dumetz, F.; Imamura, H.; Sanders, M.; Seblova, V.; Myskova, J.; Pescher, P.; Vanaerschot, M.; Meehan, C.J.; Cuypers, B.; De Muylder, G.; et al. Modulation of Aneuploidy in Leishmania donovani during Adaptation to Different In Vitro and In Vivo Environments and Its Impact on Gene Expression. MBio 2017, 8. [Google Scholar] [CrossRef]
- Duncan, S.M.; Jones, N.G.; Mottram, J.C. Recent advances in Leishmania reverse genetics: Manipulating a manipulative parasite. Mol. Biochem. Parasitol. 2017, 216, 30–38. [Google Scholar] [CrossRef]
- Zhang, W.W.; Lypaczewski, P.; Matlashewski, G. Optimized CRISPR–Cas9 Genome Editing for Leishmania and Its Use To Target a Multigene Family, Induce Chromosomal Translocation, and Study DNA Break Repair Mechanisms. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Zhang, W.W.; Matlashewski, G. Single–Strand Annealing Plays a Major Role in Double–Strand DNA Break Repair following CRISPR–Cas9 Cleavage in Leishmania. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Damasceno, J.D.; Reis-Cunha, J.; Crouch, K.; Beraldi, D.; Lapsley, C.; Tosi, L.R.O.; Bartholomeu, D.; McCulloch, R. Conditional knockout of RAD51–related genes in Leishmania major reveals a critical role for homologous recombination during genome replication. PLoS Genet. 2020, 16, e1008828. [Google Scholar] [CrossRef] [PubMed]
- Yagoubat, A.; Crobu, L.; Berry, L.; Kuk, N.; Lefebvre, M.; Sarrazin, A.; Bastien, P.; Sterkers, Y. Universal highly efficient conditional knockout system in Leishmania, with a focus on untranscribed region preservation. Cell. Microbiol. 2020, 22, e13159. [Google Scholar] [CrossRef] [PubMed]
- Yardley, V.; Ortuno, N.; Llanos-Cuentas, A.; Chappuis, F.; Doncker, S.D.; Ramirez, L.; Croft, S.; Arevalo, J.; Adaui, V.; Bermudez, H.; et al. American tegumentary leishmaniasis: Is antimonial treatment outcome related to parasite drug susceptibility? J. Infect. Dis. 2006, 194, 1168–1175. [Google Scholar] [CrossRef]
- Rosenzweig, D.; Smith, D.; Opperdoes, F.; Stern, S.; Olafson, R.W.; Zilberstein, D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J 2008, 22, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Al-Jawabreh, A.; Diezmann, S.; Muller, M.; Wirth, T.; Schnur, L.F.; Strelkova, M.V.; Kovalenko, D.A.; Razakov, S.A.; Schwenkenbecher, J.; Kuhls, K.; et al. Identification of geographically distributed sub–populations of Leishmania (Leishmania) major by microsatellite analysis. BMC Evol. Biol. 2008, 8, 183. [Google Scholar] [CrossRef]
- Kapler, G.M.; Coburn, C.M.; Beverley, S.M. Stable transfection of the human parasite Leishmania major delineates a 30–kilobase region sufficient for extrachromosomal replication and expression. Mol. Cell. Biol. 1990, 10, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Krobitsch, S.; Brandau, S.; Hoyer, C.; Schmetz, C.; Hübel, A.; Clos, J. Leishmania donovani heat shock protein 100: characterization and function in amastigote stage differentiation. J. Biol. Chem. 1998, 273, 6488–6494. [Google Scholar] [CrossRef]
- Ommen, G.; Lorenz, S.; Clos, J. One–step generation of double–allele gene replacement mutants in Leishmania donovani. Int. J. Parasitol. 2009, 39, 541–546. [Google Scholar] [CrossRef]
- Beneke, T.; Gluenz, E. LeishGEdit: A Method for Rapid Gene Knockout and Tagging Using CRISPR–Cas9. Methods Mol. Biol. 2019, 1971, 189–210. [Google Scholar]
- Schumann Burkard, G.; Jutzi, P.; Roditi, I. Genome–wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol. Biochem. Parasitol. 2011, 175, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, K.; Hombach-Barrigah, A.; Clos, J. Hsp90 inhibitors radicicol and geldanamycin have opposing effects on Leishmania Aha1–dependent proliferation. Cell Stress Chaperones 2017, 22, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, K.; Zander, D.; Kube, M.; Reinhardt, R.; Clos, J. Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int. J. Parasitol. 2008, 38, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real–time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kröber-Boncardo, C.; Lorenzen, S.; Brinker, C.; Clos, J. Casein Kinase 1.2 Over Expression Restores Stress Resistance toLeishmania donovaniHSP23 Null Mutants. Sci. Rep. 2020, 10. in press. [Google Scholar]
- Van den Broeck, F.; Savill, N.J.; Imamura, H.; Sanders, M.; Maes, I.; Cooper, S.; Mateus, D.; Jara, M.; Adaui, V.; Arevalo, J.; et al. Ecological divergence and hybridization of Neotropical Leishmania parasites. Proc. Natl. Acad. Sci. USA 2020, 10, 210. [Google Scholar] [CrossRef]
- Hombach, A.; Ommen, G.; MacDonald, A.; Clos, J. A small heat shock protein is essential for thermotolerance and intracellular survival of Leishmania donovani. J. Cell Sci. 2014, 127, 4762–4773. [Google Scholar] [CrossRef]
- Hombach-Barrigah, A.; Bartsch, K.; Smirlis, D.; Rosenqvist, H.; MacDonald, A.; Dingli, F.; Loew, D.; Spath, G.F.; Rachidi, N.; Wiese, M.; et al. Leishmania donovani 90 kD Heat Shock Protein – Impact of Phosphosites on Parasite Fitness, Infectivity and Casein Kinase Affinity. Sci. Rep. 2019, 9, 5074. [Google Scholar] [CrossRef]
- Bifeld, E.; Tejera Nevado, P.; Bartsch, J.; Eick, J.; Clos, J. A versatile qPCR assay to quantify trypanosomatidic infections of host cells and tissues. Med. Microbiol. Immunol. 2016, 205, 449–458. [Google Scholar] [CrossRef]
- Bifeld, E. Quantification of Intracellular Leishmania spp. Using Real–Time Quantitative PCR (qPCR). Methods Mol. Biol. 2019, 1971, 249–263. [Google Scholar]
- Bifeld, E. Generation of Bone Marrow–Derived Macrophages for In Vitro Infection Experiments. Methods Mol. Biol. 2019, 1971, 237–247. [Google Scholar] [PubMed]
- Peng, D.; Tarleton, R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 2015, 1, e000033. [Google Scholar] [CrossRef]
- Hoyer, C.; Zander, D.; Fleischer, S.; Schilhabel, M.; Kroener, M.; Platzer, M.; Clos, J. A Leishmania donovani gene that confers accelerated recovery from stationary phase growth arrest. Int. J. Parasitol. 2004, 34, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Ward, J.D.; Reiner, D.J.; Goldstein, B. Engineering the Caenorhabditis elegans genome using Cas9–triggered homologous recombination. Nat. Methods 2013, 10, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, R.; Hollmann, M.; Merk, K.; Nitschko, V.; Obermaier, C.; Philippou-Massier, J.; Wieland, I.; Gaul, U.; Forstemann, K. Efficient chromosomal gene modification with CRISPR/cas9 and PCR–based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 2014, 42, e89. [Google Scholar] [CrossRef]
- Hübel, A.; Krobitsch, S.; Horauf, A.; Clos, J. Leishmania major Hsp100 is required chiefly in the mammalian stage of the parasite. Mol. Cell Biol. 1997, 17, 5987–5995. [Google Scholar] [CrossRef]
- Krobitsch, S.; Clos, J. A novel role for 100 kD heat shock proteins in the parasite Leishmania donovani. Cell Stress Chaperones 1999, 4, 191–198. [Google Scholar] [CrossRef]
- Van Montfort, R.L.; Basha, E.; Friedrich, K.L.; Slingsby, C.; Vierling, E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 2001, 8, 1025–1030. [Google Scholar] [CrossRef]
- Nuhs, A.; Schafer, C.; Zander, D.; Trube, L.; Tejera Nevado, P.; Schmidt, S.; Arevalo, J.; Adaui, V.; Maes, L.; Dujardin, J.C.; et al. A novel marker, ARM58, confers antimony resistance to Leishmania spp. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 37–47. [Google Scholar] [CrossRef]
- Callahan, H.L.; Portal, I.F.; Bensinger, S.J.; Grogl, M. Leishmania spp: temperature sensitivity of promastigotes in vitro as a model for tropism in vivo. Exp. Parasitol. 1996, 84, 400–409. [Google Scholar] [CrossRef]
- Piper, P.W. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 1995, 134, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Barak, E.; Amin-Spector, S.; Gerliak, E.; Goyard, S.; Holland, N.; Zilberstein, D. Differentiation of Leishmania donovani in host–free system: analysis of signal perception and response. Mol. Biochem. Parasitol. 2005, 141, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Cupolillo, E.; Brahim, L.R.; Toaldo, C.B.; de Oliveira-Neto, M.P.; de Brito, M.E.; Falqueto, A.; de Farias Naiff, M.; Grimaldi, G., Jr. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J. Clin. Microbiol. 2003, 41, 3126–3132. [Google Scholar] [CrossRef] [PubMed]
- Tobin, J.F.; Laban, A.; Wirth, D.F. Homologous recombination in Leishmania enriettii. Proc. Natl. Acad. Sci. USA 1991, 88, 864–868. [Google Scholar] [CrossRef]
- Beverley, S.M. Protozomics: trypanosomatid parasite genetics comes of age. Nat. Rev. Genet. 2003, 4, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wiese, M. A mitogen–activated protein (MAP) kinase homologue of Leishmania mexicana is essential for parasite survival in the infected host. Embo. J. 1998, 17, 2619–2628. [Google Scholar] [CrossRef]
- Coelho, A.C.; Oliveira, J.C.; Espada, C.R.; Reimao, J.Q.; Trinconi, C.T.; Uliana, S.R. A Luciferase–Expressing Leishmania braziliensis Line That Leads to Sustained Skin Lesions in BALB/c Mice and Allows Monitoring of Miltefosine Treatment Outcome. PLoS Negl. Trop. Dis. 2016, 10, e0004660. [Google Scholar] [CrossRef]
- Bastos, M.S.; Souza, L.A.; Onofre, T.S.; Silva, A.J.; Almeida, M.R.; Bressan, G.C.; Fietto, J.L. Achievement of constitutive fluorescent pLEXSY–egfp Leishmania braziliensis and its application as an alternative method for drug screening in vitro. Mem. Inst. Oswaldo. Cruz. 2017, 112, 155–159. [Google Scholar] [CrossRef]
- Sharma, R.; Silveira-Mattos, P.S.; Ferreira, V.C.; Rangel, F.A.; Oliveira, L.B.; Celes, F.S.; Viana, S.M.; Wilson, M.E.; de Oliveira, C.I. Generation and Characterization of a Dual–Reporter Transgenic Leishmania braziliensis Line Expressing eGFP and Luciferase. Front. Cell. Infect. Microbiol. 2019, 9, 468. [Google Scholar] [CrossRef]
- Andrade, J.M.; Murta, S.M. Functional analysis of cytosolic tryparedoxin peroxidase in antimony–resistant and –susceptible Leishmania braziliensis and Leishmania infantum lines. Parasites Vectors 2014, 7, 406. [Google Scholar] [CrossRef]
- Andrade, J.M.; Baba, E.H.; Machado-de-Avila, R.A.; Chavez-Olortegui, C.; Demicheli, C.P.; Frezard, F.; Monte-Neto, R.L.; Murta, S.M. Silver and Nitrate Oppositely Modulate Antimony Susceptibility through Aquaglyceroporin 1 in Leishmania (Viannia) Species. Antimicrob. Agents Chemother 2016, 60, 4482–4489. [Google Scholar] [CrossRef] [PubMed]
- Moreira, D.S.; Xavier, M.V.; Murta, S.M.F. Ascorbate peroxidase overexpression protects Leishmania braziliensis against trivalent antimony effects. Mem. Inst. Oswaldo Cruz 2018, 113, e180377. [Google Scholar] [CrossRef] [PubMed]
- De Toledo, J.S.; Junqueira dos Santos, A.F.; Rodrigues de Moura, T.; Antoniazi, S.A.; Brodskyn, C.; Indiani de Oliveira, C.; Barral, A.; Cruz, A.K. Leishmania (Viannia) braziliensis transfectants overexpressing the miniexon gene lose virulence in vivo. Parasitol. Int. 2009, 58, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Greenside, P.G.; Natoli, T.; Lahr, D.L.; Wadden, D.; Tirosh, I.; Narayan, R.; Root, D.E.; Golub, T.R.; Subramanian, A.; et al. Evaluation of RNAi and CRISPR technologies by large–scale gene expression profiling in the Connectivity Map. PLoS Biol. 2017, 15, e2003213. [Google Scholar] [CrossRef] [PubMed]
- Adaui, V.; Lye, L.F.; Akopyants, N.S.; Zimic, M.; Llanos-Cuentas, A.; Garcia, L.; Maes, I.; De Doncker, S.; Dobson, D.E.; Arevalo, J.; et al. Association of the Endobiont Double–Stranded RNA Virus LRV1 With Treatment Failure for Human Leishmaniasis Caused by Leishmania braziliensis in Peru and Bolivia. J. Infect. Dis. 2016, 213, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Bourreau, E.; Ginouves, M.; Prevot, G.; Hartley, M.A.; Gangneux, J.P.; Robert-Gangneux, F.; Dufour, J.; Sainte-Marie, D.; Bertolotti, A.; Pratlong, F.; et al. Presence of Leishmania RNA Virus 1 in Leishmania guyanensis Increases the Risk of First–Line Treatment Failure and Symptomatic Relapse. J. Infect. Dis. 2016, 213, 105–111. [Google Scholar] [CrossRef]
- Cantanhede, L.M.; Fernandes, F.G.; Ferreira, G.E.M.; Porrozzi, R.; Ferreira, R.G.M.; Cupolillo, E. New insights into the genetic diversity of Leishmania RNA Virus 1 and its species–specific relationship with Leishmania parasites. PloS ONE 2018, 13, e0198727. [Google Scholar] [CrossRef]
- Ives, A.; Ronet, C.; Prevel, F.; Ruzzante, G.; Fuertes-Marraco, S.; Schutz, F.; Zangger, H.; Revaz-Breton, M.; Lye, L.F.; Hickerson, S.M.; et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 2011, 331, 775–778. [Google Scholar] [CrossRef]
- Eren, R.O.; Reverte, M.; Rossi, M.; Hartley, M.A.; Castiglioni, P.; Prevel, F.; Martin, R.; Desponds, C.; Lye, L.F.; Drexler, S.K.; et al. Mammalian Innate Immune Response to a Leishmania–Resident RNA Virus Increases Macrophage Survival to Promote Parasite Persistence. Cell. Host Microbe. 2016, 20, 318–328. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR–Cas9–mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Wong, N.; Liu, W.; Wang, X. WU–CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Xiao, T.; Chen, C.H.; Li, W.; Meyer, C.A.; Wu, Q.; Wu, D.; Cong, L.; Zhang, F.; Liu, J.S.; et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015, 25, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Labuhn, M.; Adams, F.F.; Ng, M.; Knoess, S.; Schambach, A.; Charpentier, E.M.; Schwarzer, A.; Mateo, J.L.; Klusmann, J.H.; Heckl, D. Refined sgRNA efficacy prediction improves large– and small–scale CRISPR–Cas9 applications. Nucleic Acids Res. 2018, 46, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Graf, R.; Li, X.; Chu, V.T.; Rajewsky, K. sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9–Mediated Gene Editing. Cell Rep. 2019, 26, 1098–1103.e3. [Google Scholar] [CrossRef]
- Yuen, G.; Khan, F.J.; Gao, S.; Stommel, J.M.; Batchelor, E.; Wu, X.; Luo, J. CRISPR/Cas9–mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res. 2017, 45, 12039–12053. [Google Scholar] [CrossRef]
- Ng, H.; Dean, N. Dramatic Improvement of CRISPR/Cas9 Editing in Candida albicans by Increased Single Guide RNA Expression. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Jara, M.; Maes, I.; Imamura, H.; Domagalska, M.A.; Dujardin, J.C.; Arevalo, J. Tracking of quiescence in Leishmania by quantifying the expression of GFP in the ribosomal DNA locus. Sci. Rep. 2019, 9, 18951. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adaui, V.; Kröber-Boncardo, C.; Brinker, C.; Zirpel, H.; Sellau, J.; Arévalo, J.; Dujardin, J.-C.; Clos, J. Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23. Genes 2020, 11, 1159. https://doi.org/10.3390/genes11101159
Adaui V, Kröber-Boncardo C, Brinker C, Zirpel H, Sellau J, Arévalo J, Dujardin J-C, Clos J. Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23. Genes. 2020; 11(10):1159. https://doi.org/10.3390/genes11101159
Chicago/Turabian StyleAdaui, Vanessa, Constanze Kröber-Boncardo, Christine Brinker, Henner Zirpel, Julie Sellau, Jorge Arévalo, Jean-Claude Dujardin, and Joachim Clos. 2020. "Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23" Genes 11, no. 10: 1159. https://doi.org/10.3390/genes11101159
APA StyleAdaui, V., Kröber-Boncardo, C., Brinker, C., Zirpel, H., Sellau, J., Arévalo, J., Dujardin, J.-C., & Clos, J. (2020). Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23. Genes, 11(10), 1159. https://doi.org/10.3390/genes11101159