Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala dermatitidis
Abstract
1. Introduction
2. Materials and Methods
2.1. Development and Growth of Fungal Strains Used E. dermatitidis
2.2. Collection and Analysis of Genome Sequence of Evolved Strain
2.3. Irradiation of E. dermatitidis
2.4. Cell Lysis and Protein Extraction
2.5. Proteolysis
2.6. Liquid Chromatography and Tandem Mass Spectrometry
2.7. Data Analysis
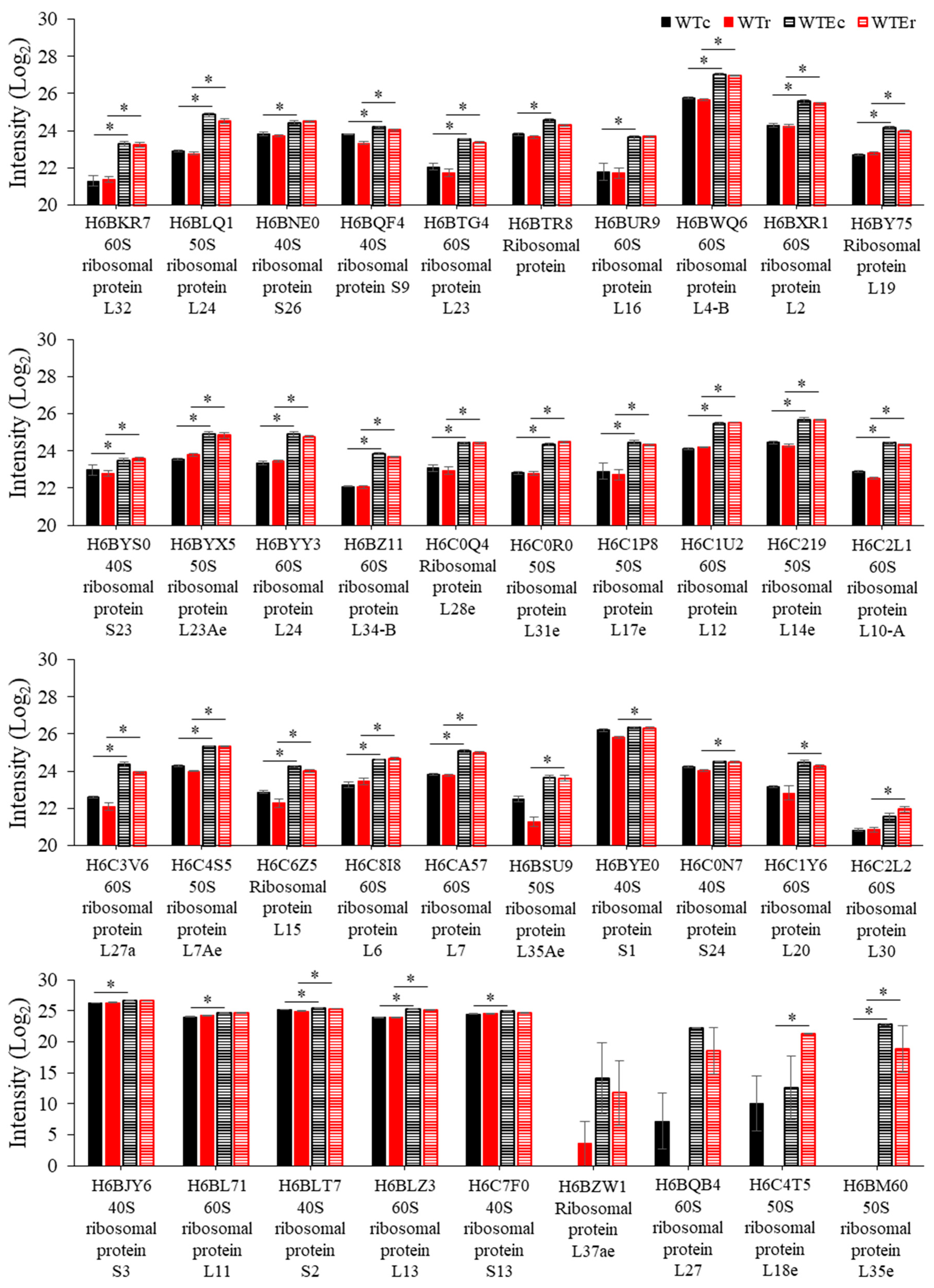
3. Results
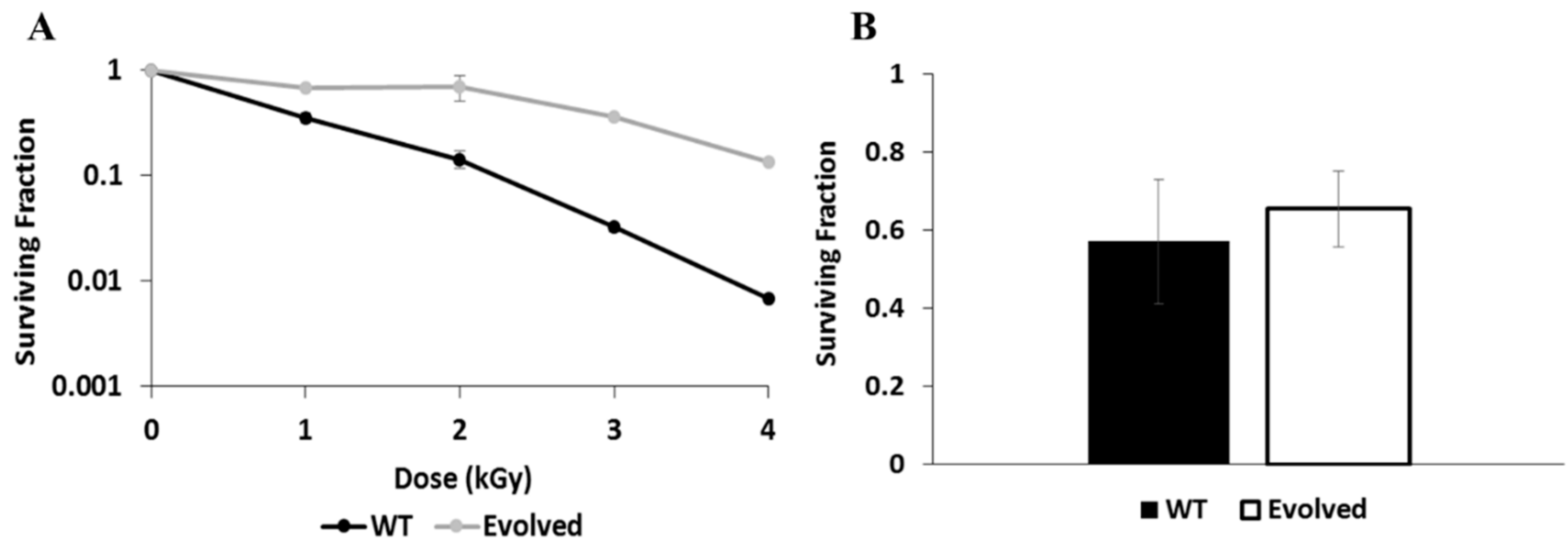
3.1. Radiation Resistance of Wild Type and Evolved E. dermatitidis Strains
3.2. Pressure-Assisted Lysis of E. dermatitidis Cells for Protein Extraction and Shotgun Proteomics
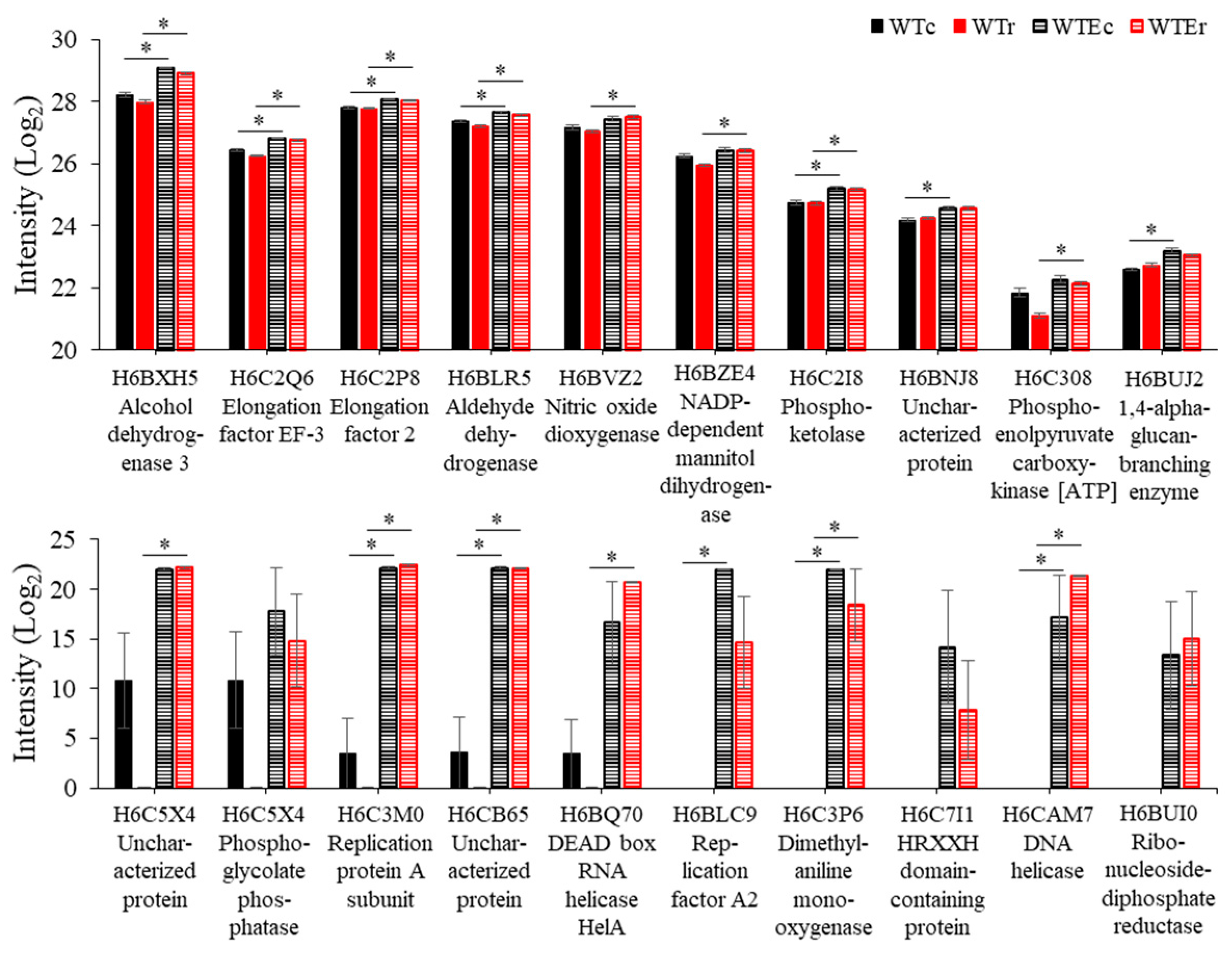
3.3. The Proteomic Responses of Two E. dermatitidis Strains to γ-Radiation Exposure
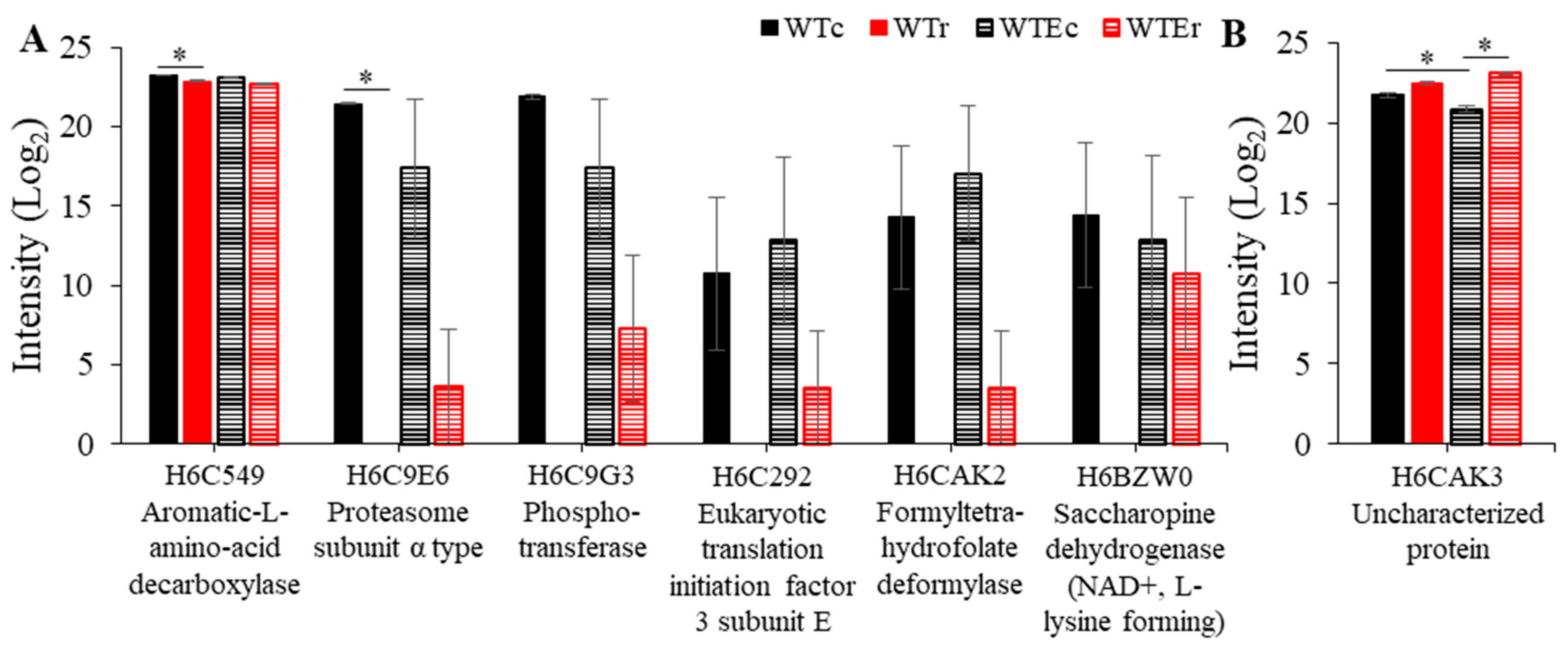
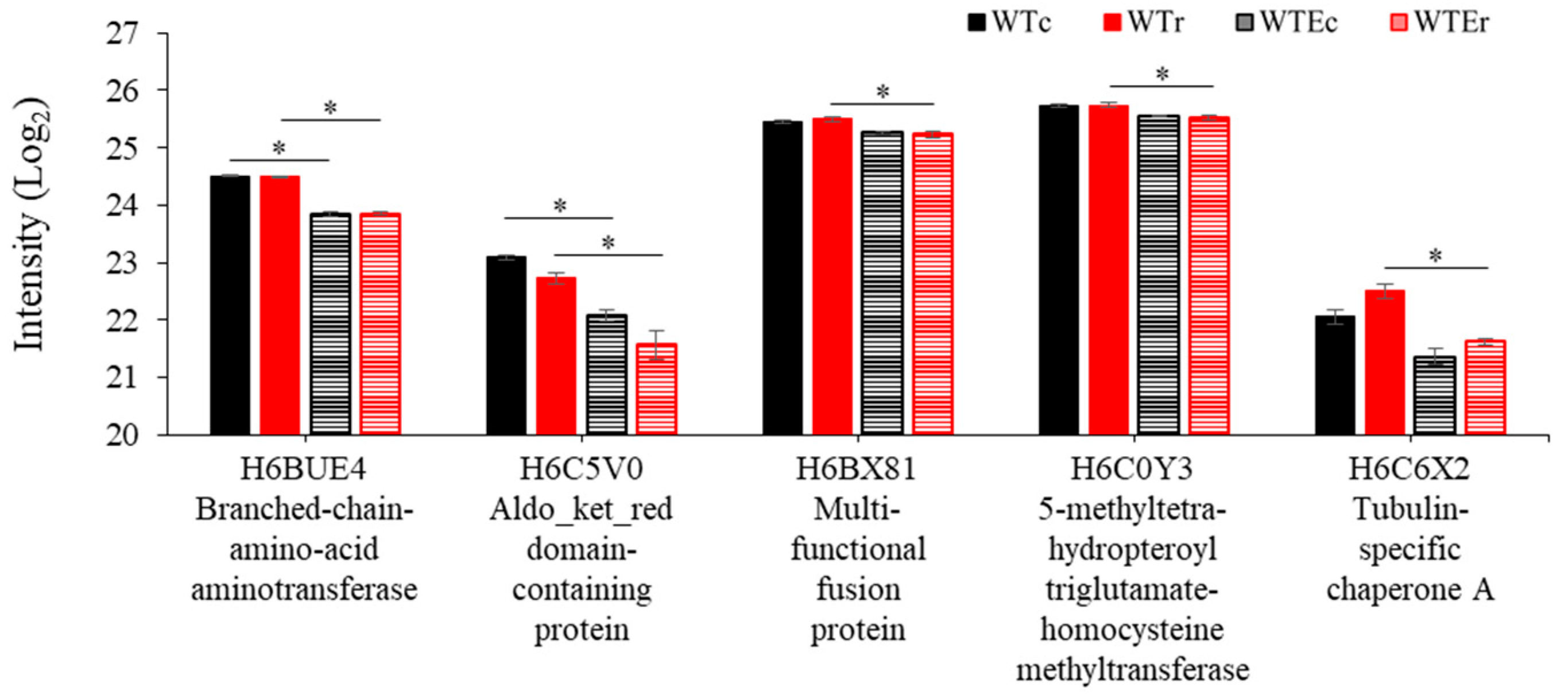
3.4. Proteins Associated with Evolution of Higher Radioresistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- LaVerne, J.A. OH radicals and oxidizing products in the gamma radiolysis of water. Radiat. Res. 2000, 153, 196–200. [Google Scholar] [PubMed]
- Lliakis, G. The role of DNA double strand breaks in lonizing radiation—Induced killing of eukaryotic cells. Bioessays 1991, 13, 641–648. [Google Scholar]
- Resnick, M.A. The repair of double-strand breaks in DNA: A model involving recombination. J. Theor. Biol. 1976, 59, 97–106. [Google Scholar]
- Critchlow, S.E.; Jackson, S.P. DNA end-joining: From yeast to man. Trends Biochem. Sci. 1998, 23, 394–398. [Google Scholar] [PubMed]
- Daly, M.J. Death by protein damage in irradiated cells. DNA Repair 2012, 11, 12–21. [Google Scholar] [PubMed]
- Aravind, L.; Walker, D.R.; Koonin, E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999, 27, 1223–1242. [Google Scholar] [PubMed]
- Paithankar, J.G.; Raghu, S.V.; Patil, R.K. Concomitant changes in radiation resistance and trehalose levels during life stages of Drosophila melanogaster suggest radio-protective function of trehalose. Int. J. Radiat. Biol. 2018, 94, 576–589. [Google Scholar]
- Vaisnav, M.; Xing, C.; Ku, H.-C.; Hwang, D.; Stojadinovic, S.; Pertsemlidis, A.; Abrams, J.M. Genome-wide association analysis of radiation resistance in Drosophila melanogaster. PLoS ONE 2014, 9, e104858. [Google Scholar]
- Schultzhaus, Z.; Romsdahl, J.; Chen, A.; Tschirhart, T.; Kim, S.; Leary, D.H.; Wang, Z. The response of the melanized yeast Exophiala dermatitidis to gamma radiation exposure. Environ. Microbiol. 2020, 22, 1310–1326. [Google Scholar]
- Sharma, A.; Gaidamakova, E.K.; Grichenko, O.; Matrosova, V.Y.; Hoeke, V.; Klimenkova, P.; Conze, I.H.; Volpe, R.P.; Gostinčar, C.; Gunde-Cimerman, N.; et al. Across the tree of life, radiation resistance is governed by antioxidant Mn2+, gauged by paramagnetic resonance. Proc. Natl. Acad. Sci. USA 2017, 114, E9253–E9260. [Google Scholar]
- Mitchel, R.; Morrison, D. Heat-shock induction of ionizing radiation resistance in Saccharomyces cerevisiae, and correlation with stationary growth phase. Radiat. Res. 1982, 90, 284–291. [Google Scholar] [PubMed]
- Schultzhaus, Z.; Cuomo, C.A.; Wang, Z. Genome Sequence of the Black Yeast Exophiala lecanii-corni. Microbiol. Resour. Announc. 2019, 8, e01709–e01718. [Google Scholar] [PubMed]
- Babič, M.N.; Zupančič, J.; Gunde-Cimerman, N.; De Hoog, S.; Zalar, P. Ecology of the human opportunistic black yeast Exophiala dermatitidis indicates preference for human-made habitats. Mycopathologia 2018, 183, 201–212. [Google Scholar] [PubMed]
- Blasi, B.; Tafer, H.; Tesei, D.; Sterflinger, K. From glacier to sauna: RNA-Seq of the human pathogen black fungus Exophiala dermatitidis under varying temperature conditions exhibits common and novel fungal response. PLoS ONE 2015, 10, e0127103. [Google Scholar]
- Robertson, K.L.; Mostaghim, A.; Cuomo, C.A.; Soto, C.M.; Lebedev, N.; Bailey, R.F.; Wang, Z. Adaptation of the black yeast Wangiella dermatitidis to ionizing radiation: Molecular and cellular mechanisms. PLoS ONE 2012, 7, e48674. [Google Scholar] [CrossRef]
- Zeng, J.; Sutton, D.; Fothergill, A.; Rinaldi, M.; Harrak, M.; De Hoog, G. Spectrum of clinically relevant Exophiala species in the United States. J. Clin. Microbiol. 2007, 45, 3713–3720. [Google Scholar]
- Cheng, Q.; Kinney, K.A.; Whitman, C.P.; Szaniszlo, P.J. Characterization of two polyketide synthase genes in Exophiala lecanii-corni, a melanized fungus with bioremediation potential. Bioorg. Chem. 2004, 32, 92–108. [Google Scholar]
- Teixeira, M.M.; Moreno, L.F.; Stielow, B.; Muszewska, A.; Hainaut, M.; Gonzaga, L.; Abouelleil, A.; Patané, J.; Priest, M.; Souza, R. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud. Mycol. 2017, 86, 1–28. [Google Scholar]
- Feng, B.; Wang, X.; Hauser, M.; Kaufmann, S.; Jentsch, S.; Haase, G.; Becker, J.M.; Szaniszlo, P.J. Molecular cloning and characterization of WdPKS1, a gene involved in dihydroxynaphthalene melanin biosynthesis and virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 2001, 69, 1781–1794. [Google Scholar]
- Paolo, W.F.; Dadachova, E.; Mandal, P.; Casadevall, A.; Szaniszlo, P.J.; Nosanchuk, J.D. Effects of disrupting the polyketide synthase gene WdPKS1 in Wangiella [Exophiala] dermatitidis on melanin production and resistance to killing by antifungal compounds, enzymatic degradation, and extremes in temperature. BMC Microbiol. 2006, 6, 55. [Google Scholar]
- Poyntner, C.; Mirastschijski, U.; Sterflinger, K.; Tafer, H. Transcriptome study of an Exophiala dermatitidis PKS1 mutant on an ex vivo skin model: Is melanin important for infection? Front. Microbiol. 2018, 9, 1457. [Google Scholar] [CrossRef]
- De Hoog, G.; Takeo, K.; Yoshida, S.; Göttlich, E.; Nishimura, K.; Miyaji, M. Pleoanamorphic life cycle of Exophiala (Wangiella) dermatitidis. Antonie Van Leeuwenhoek 1994, 65, 143–153. [Google Scholar] [PubMed]
- Dixon, D.M.; Polak, A.; Szaniszlo, P.J. Pathogenicity and virulence of wild-type and melanin-deficient Wangiella dermatitidis. J. Med. Vet. Mycol. 1987, 25, 97–106. [Google Scholar] [PubMed]
- Chen, Z.; Martinez, D.A.; Gujja, S.; Sykes, S.M.; Zeng, Q.; Szaniszlo, P.J.; Wang, Z.; Cuomo, C.A. Comparative genomic and transcriptomic analysis of Wangiella dermatitidis, a major cause of phaeohyphomycosis and a model black yeast human pathogen. G3 Genes Genomes Genet. 2014, 4, 561–578. [Google Scholar]
- Birrell, G.W.; Brown, J.A.; Wu, H.I.; Giaever, G.; Chu, A.M.; Davis, R.W.; Brown, J.M. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 2002, 99, 8778–8783. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Schultzhaus, J.N.; Dean, S.N.; Leary, D.H.; Hervey, W.J.; Fears, K.P.; Wahl, K.J.; Spillmann, C.M. Pressure cycling technology for challenging proteomic sample processing: Application to barnacle adhesive. Integr. Biol. 2019, 11, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Li, C.; Smejkal, G.; Lazarev, A.; Lawrence, N.; Schumacher, R.T. Pressure cycling technology (PCT) applications in extraction of biomolecules from challenging biological samples. High Press. Biosci. Biotechnol. 2007, 1, 166–173. [Google Scholar]
- López-Ferrer, D.; Petritis, K.; Hixson, K.K.; Heibeck, T.H.; Moore, R.J.; Belov, M.E.; Camp, D.G.; Smith, R.D. Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: Proteomics on the fly. J. Proteome Res. 2008, 7, 3276–3281. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Guo, T.; Gross, V.; Lazarev, A.; Koh, C.C.; Gillessen, S.; Joerger, M.; Jochum, W.; Aebersold, R. Reproducible tissue homogenization and protein extraction for quantitative proteomics using micropestle-assisted pressure-cycling technology. J. Proteome Res. 2016, 15, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Kouvonen, P.; Koh, C.C.; Gillet, L.C.; Wolski, W.E.; Röst, H.L.; Rosenberger, G.; Collins, B.C.; Blum, L.C.; Gillessen, S. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 2015, 21, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Schultzhaus, J.N.; Taitt, C.R.; Leary, D.H.; Shriver-Lake, L.C.; Snellings, D.; Sturiale, S.; North, S.H.; Orihuela, B.; Rittschof, D. Characterization of longitudinal canal tissue in the acorn barnacle Amphibalanus amphitrite. PLoS ONE 2018, 13, e0208352. [Google Scholar] [CrossRef] [PubMed]
- Spangler, J.R.; Dean, S.N.; Leary, D.H.; Walper, S.A. Response of Lactobacillus plantarum WCFS1 to the Gram-negative pathogen-associated quorum sensing molecule N-3-oxododecanoyl homoserine lactone. Front. Microbiol. 2019, 10, 715. [Google Scholar] [CrossRef]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Ovaa, H.; Huber, W.; Vermeulen, M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018, 13, 530. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 1–7. [Google Scholar] [CrossRef]
- Schultzhaus, Z.; Chen, A.; Shuryak, I.; Wang, Z. The transcriptomic and phenotypic response of the melanized yeast Exophiala dermatitidis to ionizing particle exposure. BMC Genom. 2020. [Google Scholar] [CrossRef]
- Priebe, S.; Linde, J.; Albrecht, D.; Guthke, R.; Brakhage, A.A. FungiFun: A web-based application for functional categorization of fungal genes and proteins. Fungal Genet. Biol. 2011, 48, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Priebe, S.; Kreisel, C.; Horn, F.; Guthke, R.; Linde, J. FungiFun2: A comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 2014, 31, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Harder, A.; Wildgruber, R.; Nawrocki, A.; Fey, S.J.; Mose Larsen, P.; Görg, A. Comparison of yeast cell protein solubilization procedures for two—Dimensional electrophoresis. Electrophor. Int. J. 1999, 20, 826–829. [Google Scholar] [CrossRef]
- Shieh, I.-F.; Lee, C.-Y.; Shiea, J. Eliminating the interferences from TRIS buffer and SDS in protein analysis by fused-droplet electrospray ionization mass spectrometry. J. Proteome Res. 2005, 4, 606–612. [Google Scholar] [CrossRef]
- Schultzhaus, Z.; Chen, A.; Kim, S.; Shuryak, I.; Chang, M.; Wang, Z. Transcriptomic analysis reveals the relationship of melanization to growth and resistance to gamma radiation in Cryptococcus neoformans. Environ. Microbiol. 2019, 21, 2613–2628. [Google Scholar] [CrossRef]
- Riley, P. Free radicals in biology: Oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 1994, 65, 27–33. [Google Scholar] [CrossRef]
- Basu, B.; Apte, S.K. Gamma radiation-induced proteome of Deinococcus radiodurans primarily targets DNA repair and oxidative stress alleviation. Mol. Cell. Proteom. 2012, 11, 011734. [Google Scholar] [CrossRef]
- Cabiscol, E.; Piulats, E.; Echave, P.; Herrero, E.; Ros, J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 27393–27398. [Google Scholar] [CrossRef]
- Feng, C.; Chen, Y.; Pan, J.; Yang, A.; Niu, L.; Min, J.; Meng, X.; Liao, L.; Zhang, K.; Shen, L. Redox proteomic identification of carbonylated proteins in autism plasma: Insight into oxidative stress and its related biomarkers in autism. Clin. Proteom. 2017, 14, 2. [Google Scholar] [CrossRef]
- Ghezzi, P.; Chan, P. Redox proteomics applied to the thiol secretome. Antioxid. Redox Signal. 2017, 26, 299–312. [Google Scholar] [CrossRef]
- Lennicke, C.; Rahn, J.; Heimer, N.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Redox proteomics: Methods for the identification and enrichment of redox-modified proteins and their applications. Proteomics 2016, 16, 197–213. [Google Scholar] [CrossRef]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Alterzon-Baumel, S.; Hertz, R.; Methling, K.; Lalk, M.; Ankri, S. Proteomic identification of oxidized proteins in Entamoeba histolytica by resin-assisted capture: Insights into the role of arginase in resistance to oxidative stress. PLoS Negl. Trop. Dis. 2016, 10, e0004340. [Google Scholar] [CrossRef] [PubMed]
- Dornfeld, K.J.; Livingston, D.M. Effects of controlled RAD52 expression on repair and recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 2013–2017. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eden, A.; Simchen, G.; Benvenisty, N. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J. Biol. Chem. 1996, 271, 20242–20245. [Google Scholar] [CrossRef] [PubMed]
- Deuschle, K.; Funck, D.; Forlani, G.; Stransky, H.; Biehl, A.; Leister, D.; van der Graaff, E.; Kunze, R.; Frommer, W.B. The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 2004, 16, 3413–3425. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, D.; Stephan, D. 5-Methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase. In Enzyme Handbook 11; Springer: Berlin/Heidelberg, Germany, 1996; pp. 63–66. [Google Scholar]
- Han, T.-L.; Cannon, R.D.; Gallo, S.M.; Villas-Bôas, S.G. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ Biofilms Microb. 2019, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Longhese, M.P.; Plevani, P.; Lucchini, G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol. Cell. Biol. 1994, 14, 7884–7890. [Google Scholar] [CrossRef]
- Holloman, W.K.; Schirawski, J.; Holliday, R. Towards understanding the extreme radiation resistance of Ustilago maydis. Trends Microbiol. 2007, 15, 525–529. [Google Scholar] [CrossRef]
- Jung, K.-W.; Yang, D.-H.; Kim, M.-K.; Seo, H.S.; Lim, S.; Bahn, Y.-S. Unraveling fungal radiation resistance regulatory networks through the genome-wide transcriptome and genetic analyses of Cryptococcus neoformans. MBio 2016, 7. [Google Scholar] [CrossRef]
- Frankenberg, D.; Frankenberg-Schwager, M.; Harbich, R. Split-dose recovery is due to the repair of DNA double-strand breaks. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1984, 46, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Resnick, M.A.; Martin, P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. MGG 1976, 143, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, J.; Omelchenko, M.V.; Beliaev, A.S.; Venkateswaran, A.; Stair, J.; Wu, L.; Thompson, D.K.; Xu, D.; Rogozin, I.B.; et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 2003, 100, 4191–4196. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Daly, M.J.; Vasilenko, A.; Omelchenko, M.V.; Gaidamakova, E.K.; Wu, L.; Zhou, J.; Sundin, G.W.; Tiedje, J.M. Transcriptome analysis applied to survival of Shewanella oneidensis MR-1 exposed to ionizing radiation. J. Bacteriol. 2006, 188, 1199–1204. [Google Scholar] [CrossRef]
- Park, M.S. Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem. 1995, 270, 15467–15470. [Google Scholar] [CrossRef]
- Campa, A.; Ballarini, F.; Belli, M.; Cherubini, R.; Dini, V.; Esposito, G.; Friedland, W.; Gerardi, S.; Molinelli, S.; Ottolenghi, A. DNA DSB induced in human cells by charged particles and gamma rays: Experimental results and theoretical approaches. Int. J. Radiat. Biol. 2005, 81, 841–854. [Google Scholar] [CrossRef]
- McIlwrath, A.J.; Vasey, P.A.; Ross, G.M.; Brown, R. Cell cycle arrests and radiosensitivity of human tumor cell lines: Dependence on wild-type p53 for radiosensitivity. Cancer Res. 1994, 54, 3718–3722. [Google Scholar]
- Goldman, G.H.; Kafer, E. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 2004, 41, 428–442. [Google Scholar] [CrossRef]
- Lisby, M.; Rothstein, R.; Mortensen, U.H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 2001, 98, 8276–8282. [Google Scholar] [CrossRef]
- Shinohara, A.; Ogawa, H.; Ogawa, T. Rad51 protein involved in repair and recombination in Saccharomyces cerevisiae is a RecA-like protein. Cell 1992, 69, 457–470. [Google Scholar] [CrossRef]
- Meyer, V.; Arentshorst, M.; El-Ghezal, A.; Drews, A.-C.; Kooistra, R.; van den Hondel, C.A.; Ram, A.F. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 2007, 128, 770–775. [Google Scholar] [CrossRef]
- Cortesão, M.; de Haas, A.; Unterbusch, R.; Fujimori, A.; Schütze, T.; Meyer, V.; Moeller, R. Aspergillus niger Spores Are Highly Resistant to Space Radiation. Front. Microbiol. 2020, 11, 560. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, U.H.; Erdeniz, N.; Feng, Q.; Rothstein, R. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 2002, 161, 549–562. [Google Scholar] [PubMed]
- Benson, F.E.; Stasiak, A.; West, S.C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO 1994, 13, 5764–5771. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, M.; Gao, Y.; Lu, D.; Li, W.; Zhou, L. Repair characteristics and time-dependent effects in response to heavy-ion beam irradiation in Saccharomyces cerevisiae: A comparison with X-ray irradiation. Appl. Microbiol. Biotechnol. 2020, 104, 4043–4057. [Google Scholar] [CrossRef]
- Cole, G.; Mortimer, R. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol. Cell. Biol. 1989, 9, 3314–3322. [Google Scholar] [CrossRef]
- Watson, A.; Mata, J.; Bahler, J.; Carr, A.; Humphrey, T. Global gene expression responses of fission yeast to ionizing radiation. Mol. Biol. Cell 2004, 15, 851–860. [Google Scholar] [CrossRef]
- Kimura, S.; Ishidou, E.; Kurita, S.; Suzuki, Y.; Shibato, J.; Rakwal, R.; Iwahashi, H. DNA microarray analyses reveal a post-irradiation differential time-dependent gene expression profile in yeast cells exposed to X-rays and gamma-rays. Biochem. Biophys. Res. Commun. 2006, 346, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Matuo, Y.; Izumi, Y.; N Sakamoto, A.; Hase, Y.; Satoh, K.; Shimizu, K. Molecular Analysis of Carbon Ion-Induced Mutations in DNA Repair-Deficient Strains of Saccharomyces cerevisiae. Quantum Beam Sci. 2019, 3, 14. [Google Scholar] [CrossRef]
- Milisavljevic, M.; Petkovic, J.; Samardzic, J.; Kojic, M. Bioavailability of nutritional resources from cells killed by oxidation supports expansion of survivors in Ustilago maydis populations. Front. Microbiol. 2018, 9, 990. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E.; Meselson, M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. USA 2008, 105, 5139–5144. [Google Scholar] [CrossRef] [PubMed]
- Krisko, A.; Leroy, M.; Radman, M.; Meselson, M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 2012, 109, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Radman, M. Protein damage, radiation sensitivity and aging. DNA Repair 2016, 44, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, S.; Badura, M.L.; Xi, Q.; Formenti, S.C.; Schneider, R.J. Regulation of protein synthesis by ionizing radiation. Mol. Cell. Biol. 2009, 29, 5645–5656. [Google Scholar] [CrossRef]
- Haynes, R.H. The interpretation of microbial inactivation and recovery phenomena. Radiat. Res. Suppl. 1966, 6, 1–29. [Google Scholar] [CrossRef]
- Kiefer, J.; Wienhard, I. Delayed plating recovery and protein synthesis in X-irradiated yeast. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1980, 37, 343–346. [Google Scholar] [CrossRef]
- Koval, T.M. Inducible repair of ionizing radiation damage in higher eukaryotic cells. Mutat. Res. Lett. 1986, 173, 291–293. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Ducret, A.; Khoueiry, P.; Lignon, S.; Longhi, S.; Talla, E.; Dukan, S. Rules governing selective protein carbonylation. PLoS ONE 2009, 4, e7269. [Google Scholar] [CrossRef]
- Blomberg, A. Global changes in protein synthesis during adaptation of the yeast Saccharomyces cerevisiae to 0.7 M NaCl. J. Bacteriol. 1995, 177, 3563–3572. [Google Scholar] [CrossRef][Green Version]
- Brill, S.J.; Stillman, B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature 1989, 342, 92–95. [Google Scholar] [CrossRef]
- Shcherbik, N.; Pestov, D.G. The impact of oxidative stress on ribosomes: From injury to regulation. Cells 2019, 8, 1379. [Google Scholar]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [PubMed]
- Bernstein, K.A.; Bleichert, F.; Bean, J.M.; Cross, F.R.; Baserga, S.J. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 2007, 18, 953–964. [Google Scholar] [PubMed]
- Jorgensen, P.; Rupeš, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [PubMed]
- Ide, S.; Miyazaki, T.; Maki, H.; Kobayashi, T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 2010, 327, 693–696. [Google Scholar] [PubMed]
- Zhao, Y.; Tan, M.; Liu, X.; Xiong, X.; Sun, Y. Inactivation of ribosomal protein S27-like confers radiosensitivity via the Mdm2-p53 and Mdm2–MRN–ATM axes. Cell Death Dis. 2018, 9, 1–11. [Google Scholar]
- Trotter, E.W.; Rand, J.D.; Vickerstaff, J.; Grant, C.M. The yeast Tsa1 peroxiredoxin is a ribosome-associated antioxidant. Biochem. J. 2008, 412, 73–80. [Google Scholar]
- Hanway, D.; Chin, J.K.; Xia, G.; Oshiro, G.; Winzeler, E.A.; Romesberg, F.E. Previously uncharacterized genes in the UV-and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA 2002, 99, 10605–10610. [Google Scholar]
- Pestov, D.G.; Shcherbik, N. Rapid cytoplasmic turnover of yeast ribosomes in response to rapamycin inhibition of TOR. Mol. Cell. Biol. 2012, 32, 2135–2144. [Google Scholar]
- Nagata, Y.; Takahashi, A.; Ohnishi, K.; Ota, I.; Ohnishi, T.; Tojo, T.; Taniguchi, S. Effect of rapamycin, an mTOR inhibitor, on radiation sensitivity of lung cancer cells having different p53 gene status. Int. J. Oncol. 2010, 37, 1001–1010. [Google Scholar]
- Lü, X.; de la Peña, L.; Barker, C.; Camphausen, K.; Tofilon, P.J. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006, 66, 1052–1061. [Google Scholar] [PubMed]

| Condition | Gene Name | Annotation | Proteomics FC | Transcriptomics FC |
|---|---|---|---|---|
| WTc vs. WTr | HMPREF1120_02715 | 40S ribosomal protein S9 | −0.47 | −1.05 |
| HMPREF1120_01741 | Mannose-1-phosphate guanyltransferase | −0.74 | −1.29 | |
| HMPREF1120_08664 | Proteasome subunit alpha type | Not observed in WTr | NS | |
| WTEc vs. WTEr | HMPREF1120_08138 | Uncharacterized Protein | 2.28 | 3.91 |
| HMPREF1120_06350 | NADH-ubiquinone oxidoreductase subunit, mitochondrial | 1.22 | NS | |
| HMPREF1120_07744 | Aromatic-L-amino-acid decarboxylase | −0.36 | NS | |
| HMPREF1120_00283 | (R,R)-butanediol dehydrogenase | Not observed in WTEr | NS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schultzhaus, Z.S.; Schultzhaus, J.N.; Romsdahl, J.; Chen, A.; Hervey IV, W.J.; Leary, D.H.; Wang, Z. Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala dermatitidis. Genes 2020, 11, 1128. https://doi.org/10.3390/genes11101128
Schultzhaus ZS, Schultzhaus JN, Romsdahl J, Chen A, Hervey IV WJ, Leary DH, Wang Z. Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala dermatitidis. Genes. 2020; 11(10):1128. https://doi.org/10.3390/genes11101128
Chicago/Turabian StyleSchultzhaus, Zachary S., Janna N. Schultzhaus, Jillian Romsdahl, Amy Chen, W. Judson Hervey IV, Dagmar H. Leary, and Zheng Wang. 2020. "Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala dermatitidis" Genes 11, no. 10: 1128. https://doi.org/10.3390/genes11101128
APA StyleSchultzhaus, Z. S., Schultzhaus, J. N., Romsdahl, J., Chen, A., Hervey IV, W. J., Leary, D. H., & Wang, Z. (2020). Proteomics Reveals Distinct Changes Associated with Increased Gamma Radiation Resistance in the Black Yeast Exophiala dermatitidis. Genes, 11(10), 1128. https://doi.org/10.3390/genes11101128





