Ring 1 and YY1 Binding Protein is Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Germ-Cell-Specific Rybp-Knockout Mice
2.2. Quantitative RT-PCR
2.3. Histological Analysis
2.4. Immunohistochemical Staining
2.5. Fertility Test
2.6. Chromosome Spreads of Mouse Spermatocytes and Staining
2.7. Statistical Analysis
3. Results
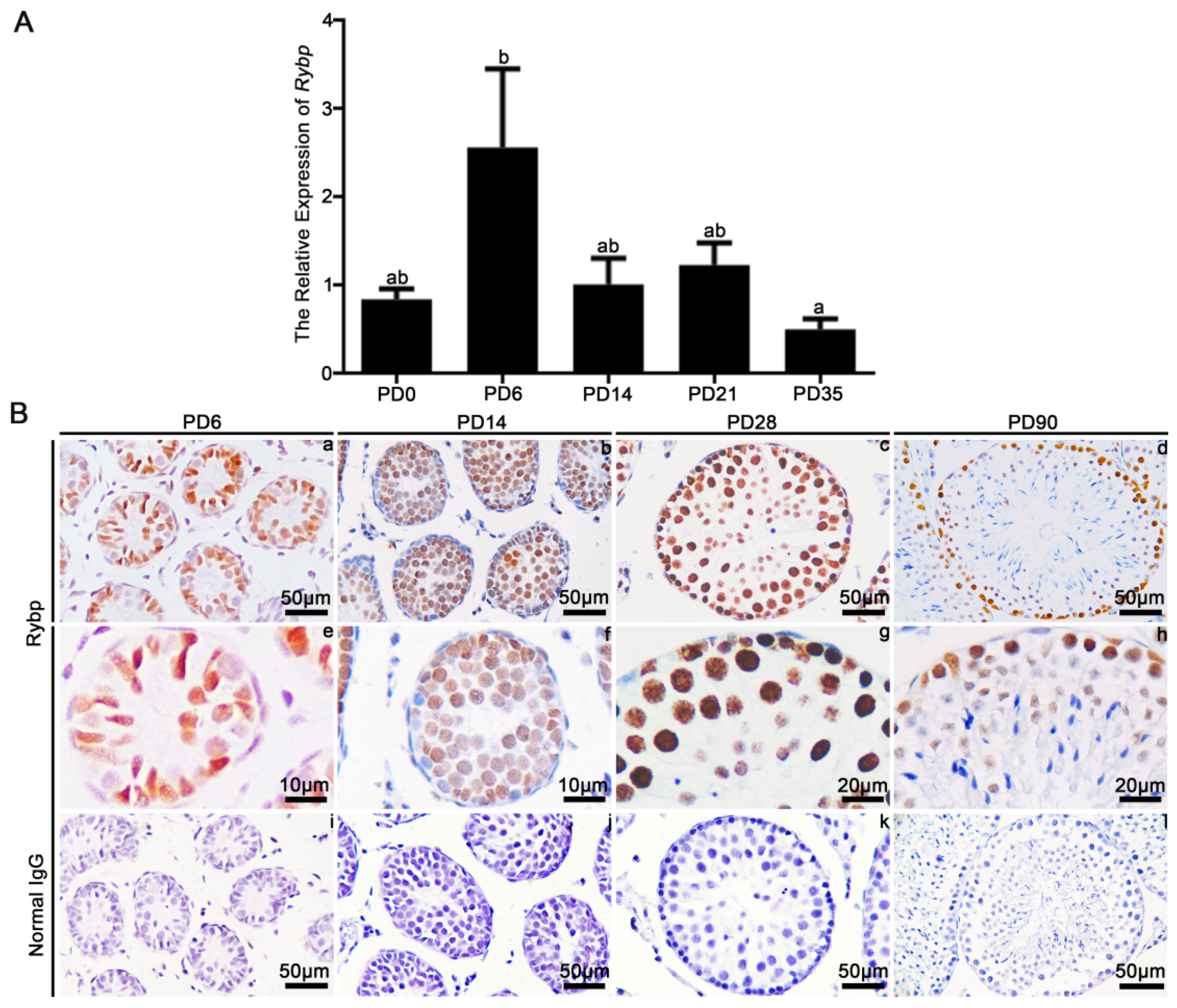
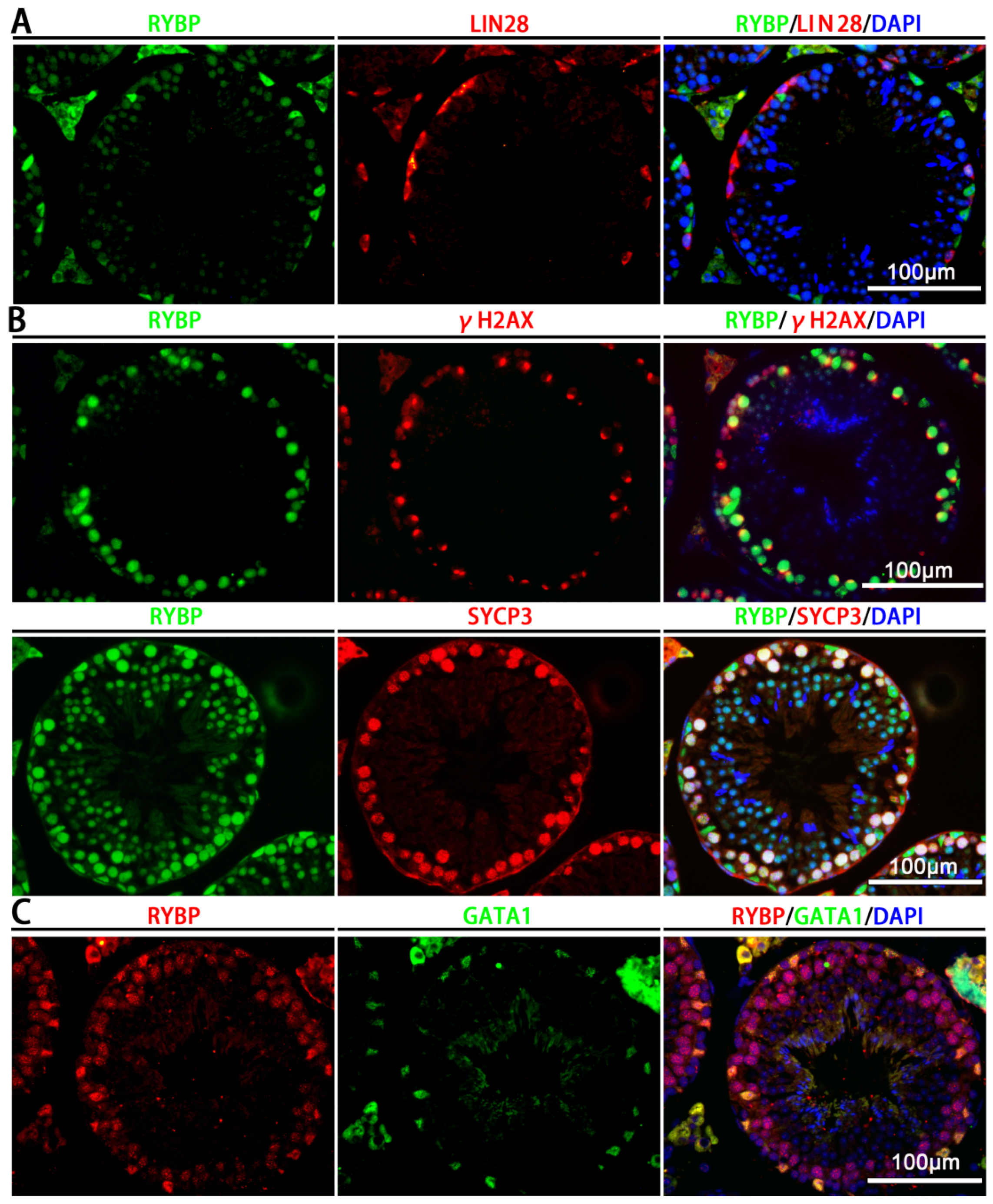
3.1. Relative mRNA Expression and Protein Localization of Rybp in Postnatal Mouse Testes
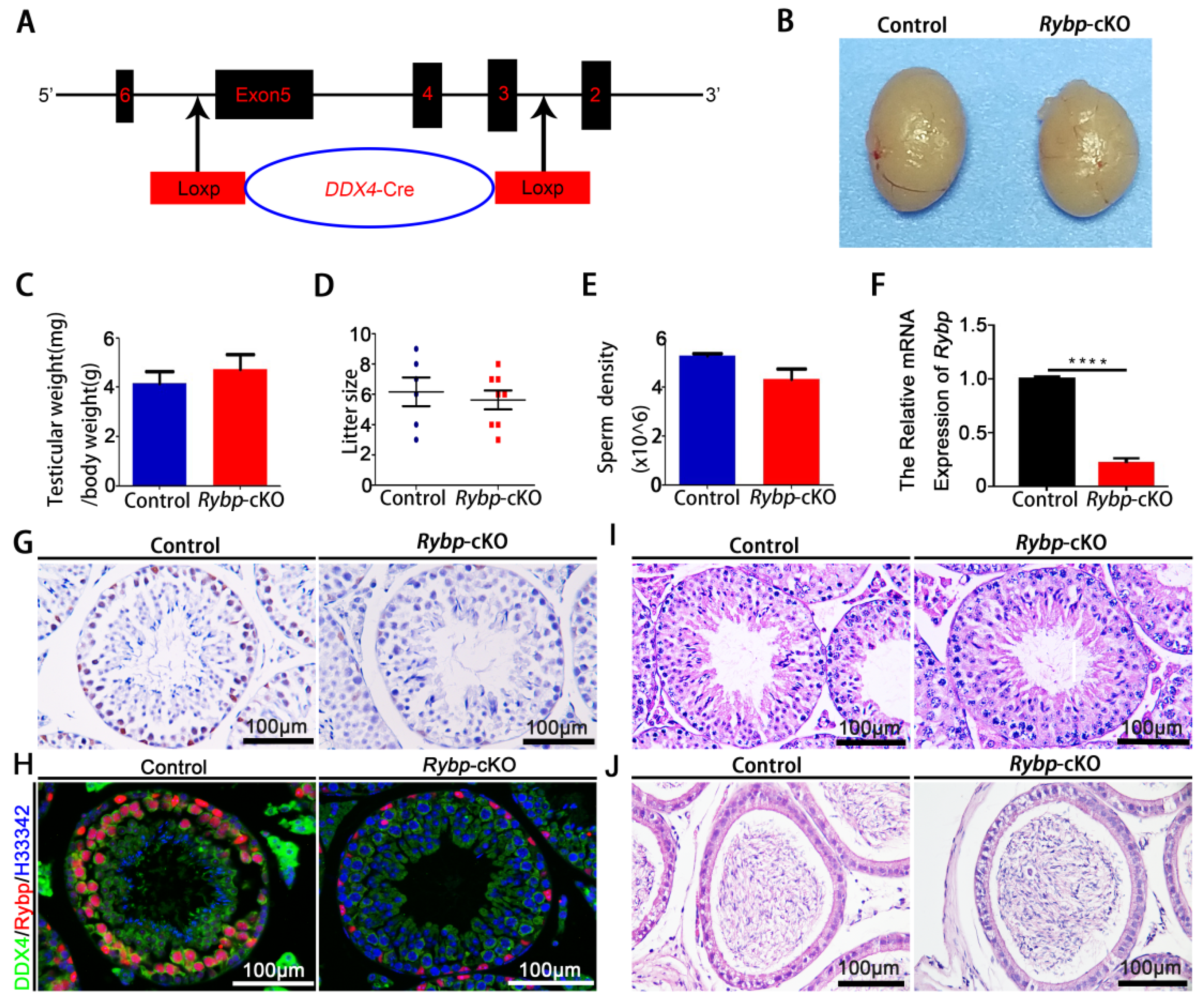
3.2. Phenotypic Analyses of Germ-Cell-Specific Rybp-Knockout Mice
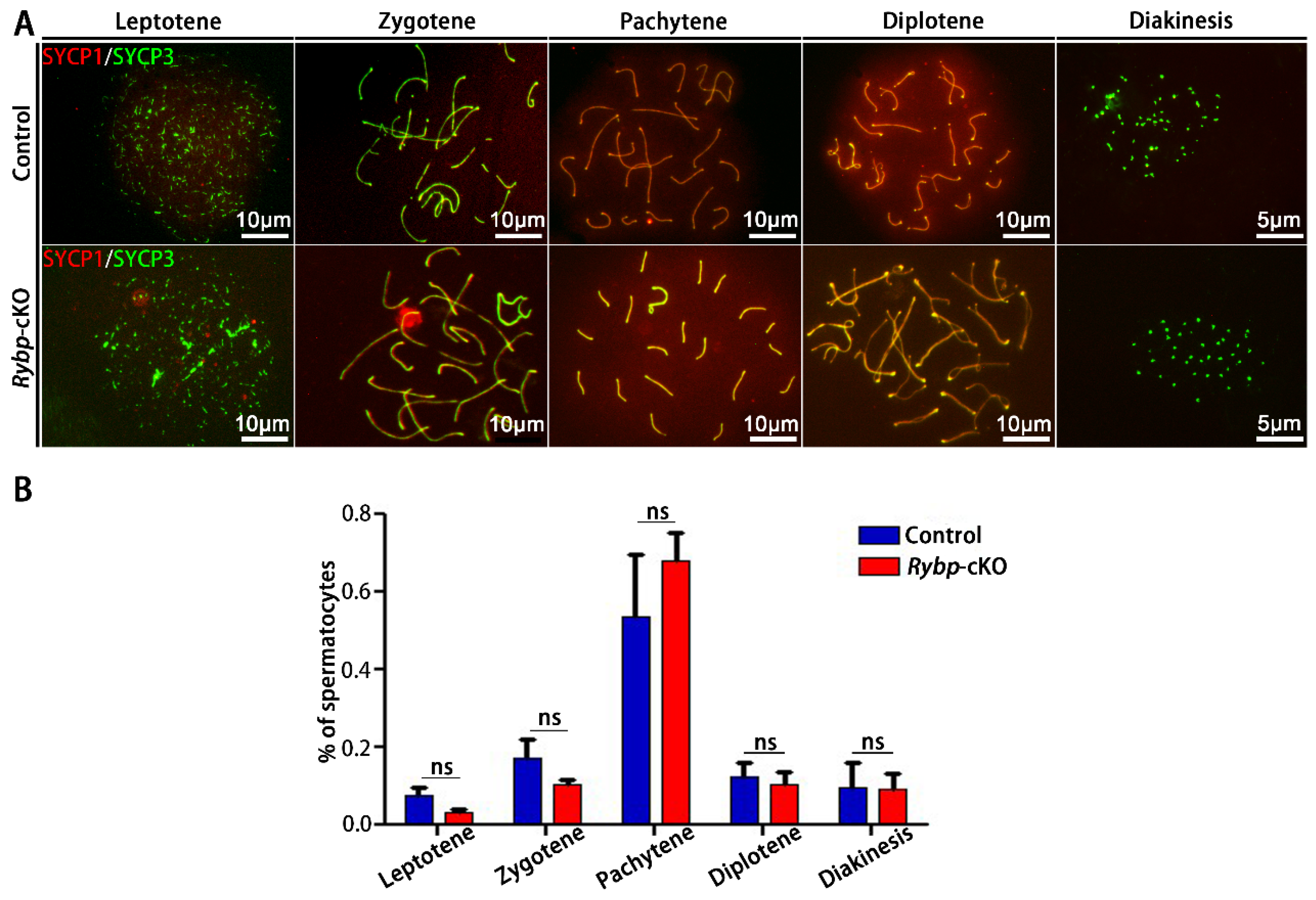
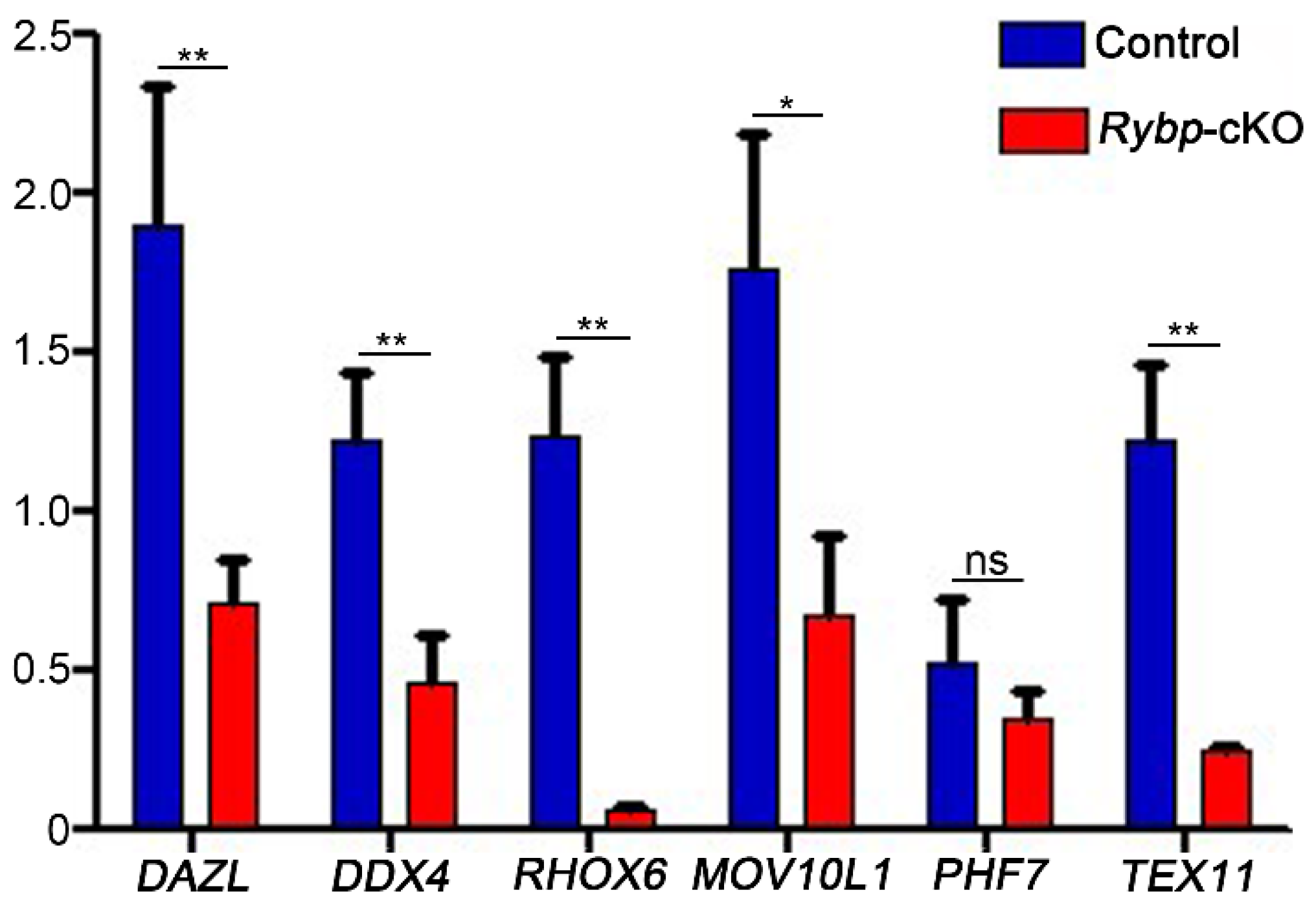
3.3. Analysis of Meiosis Progression and Meiosis-Related Gene Expression in Germ-Cell-Specific Rybp-Knockout Testes
4. Discussion
5. Conclusions
Data Availability
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Rybp | Ring 1 and YY1 binding protein |
| PcG | Polycomb group |
| PRC1 | Polycomb repressive complex 1 |
| PRC2 | Polycomb repressive complex 2 |
| qRT-PCR | Quantitative real-time PCR |
| H&E | Hematoxylin and eosin |
| PBS | Phosphate-buffered saline |
| DAB | 3,3-diaminobenzidine |
| RT | Room temperature |
| H33342 | Hoechst33342 |
| PFA | Paraformaldehyde |
| BSA | Bovine serum albumin |
| bp | Base pair(s) |
| PD | Postnatal day |
References
- De Rooij, D.G.; Russell, L.D. All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 2000, 21, 776–798. [Google Scholar] [PubMed]
- Hunt, P.A. Meiosis in mammals: Recombination, non-disjunction and the environment. Biochem. Soc. Trans. 2006, 34, 574–577. [Google Scholar] [CrossRef] [PubMed]
- de Rooij, D.G.; Griswold, M.D. Questions about spermatogonia posed and answered since 2000. J Androl. 2012, 33, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Lígia, T.; Emilia, D.; David, O.; Judith, W.; Raymond, P.; Jeroen, D.; Karel, B.; Stephen, T.; Hiroki, U.; Hiroshi, K. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar]
- Huang, Y.; Jiang, L.; Liu, B.Y.; Tan, C.F.; Chen, D.H.; Shen, W.H.; Ruan, Y. Evolution and conservation of polycomb repressive complex 1 core components and putative associated factors in the green lineage. BMC Genom. 2019, 20, 533. [Google Scholar] [CrossRef]
- Morey, L.; Aloia, L.; Cozzuto, L.; Benitah, S.A.; Dicroce, L. RYBP and Cbx7 Define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2012, 3, 60–69. [Google Scholar] [CrossRef]
- Wei, W.; Jiang-Jiang, Q.; Sukesh, V.; Subhasree, N.; Jianwei, Z.; Ruiwen, Z. Polycomb group (pcg) proteins and human cancers: Multifaceted functions and therapeutic implications. Med. Res. Rev. 2015, 35, 1220–1267. [Google Scholar]
- Zhao, W.; Huang, Y.; Zhang, J.; Liu, M.; Ji, H.; Wang, C.; Cao, N.; Li, C.; Xia, Y.; Jiang, Q.; et al. Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 2017, 292, 21527–21537. [Google Scholar] [CrossRef]
- Hasegawa, K.; Sin, H.S.; Maezawa, S.; Broering, T.; Kartashov, A.; Alavattam, K.; Ichijima, Y.; Zhang, F.; Bacon, W.C.; Greis, K. SCML2 establishes the male germline epigenome through regulation of histone h2a ubiquitination. Dev. Cell 2015, 32, 574–588. [Google Scholar] [CrossRef]
- Maezawa, S.; Hasegawa, K.; Yukawa, M.; Sakashita, A.; Alavattam, K.G.; Andreassen, P.R.; Vidal, M.; Koseki, H.; Barski, A.; Namekawa, S.H. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017, 31, 117. [Google Scholar] [CrossRef]
- Suzuki, A.; Hirasaki, M.; Hishida, T.; Wu, J.; Okamura, D.; Ueda, A.; Nishimoto, M.; Nakachi, Y.; Mizuno, Y.; Okazaki, Y. Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat. Commun. 2016, 7, 11056. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Jian, W.; Lin, H.; Yi, L.; Ji, W. Knockdown of polycomb-group RING finger 6 modulates mouse male germ cell differentiation in vitro. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 35, 339. [Google Scholar]
- Baumann, C.; De La Fuente, R. Role of polycomb group protein cbx2/m33 in meiosis onset and maintenance of chromosome stability in the Mammalian germline. Genes 2011, 2, 59–80. [Google Scholar] [CrossRef] [PubMed]
- Pirity, M.K.; Joseph, L.; Nicole, S.A. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol. Cell. Biol. 2005, 25, 7193–7202. [Google Scholar] [CrossRef]
- Pirity, M.K.; Wang, W.L.; Wolf, L.V.; Tamm, E.R.; Schreiber-Agus, N.; Cvekl, A. Rybp, a polycomb complex-associated protein, is required for mouse eye development. BMC Dev. Biol. 2007, 7, 39. [Google Scholar] [CrossRef]
- Kovacs, G.; Szabo, V.; Pirity, M.K. Absence of Rybp compromises neural differentiation of embryonic stem cells. Stem Cells Internat. 2016, 2016, 4034620. [Google Scholar] [CrossRef]
- Ujhelly, O.; Szabo, V.; Kovacs, G.; Vajda, F.; Mallok, S.; Prorok, J.; Acsai, K.; Hegedus, Z.; Krebs, S.; Dinnyes, A. Lack of rybp in mouse embryonic stem cells impairs cardiac differentiation. Stem Cells Dev. 2015, 24, 2193. [Google Scholar] [CrossRef]
- Hisada, K.; Sanchez, C.; Endo, T.A.; Endoh, M.; Roman-Trufero, M.; Sharif, J.; Koseki, H.; Vidal, M. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 2012, 32, 1139–1149. [Google Scholar] [CrossRef]
- Bajusz, I.; Henry, S.; Sutus, E.; Kovacs, G.; Pirity, M.K. Evolving role of RING1 and YY1 binding protein in the regulation of germ-cell-specific transcription. Genes 2019, 10. [Google Scholar] [CrossRef]
- Wang, Y.J.; Jia, G.X.; Yan, R.G.; Guo, S.C.; Tian, F.; Ma, J.B.; Zhang, R.N.; Li, C.; Zhang, L.Z.; Du, Y.R. Testosterone-retinoic acid signaling directs spermatogonial differentiation and seasonal spermatogenesis in the Plateau pika (Ochotona curzoniae). Theriogenology 2019, 123, 74–82. [Google Scholar] [CrossRef]
- Wang, G.; Li, Y.; Yang, Q.; Xu, S.; Ma, S.; Yan, R.; Zhang, R.; Jia, G.; Ai, D.; Yang, Q. Gene expression dynamics during the gonocyte to spermatogonia transition and spermatogenesis in the domestic yak. J. Anim. Sci. Biotechnol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Choi, E.H.; Kim, J.W.; Kim, K.P. Structured illumination microscopy imaging reveals localization of replication protein A between chromosome lateral elements during mammalian meiosis. Exp. Mol. Med. 2018, 50, 112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wu, X.; Kaestner, K.H.; Wang, P.J. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 2009, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Hunter, N.; Borner, G.V.; Lichten, M.; Kleckner, N. Gamma-H2AX illuminates meiosis. Nat. Genet. 2001, 27, 236–238. [Google Scholar] [CrossRef]
- Kim, J.S.; Griswold, M.D. E2F and GATA-1 are required for the Sertoli cell-specific promoter activity of the follicle-stimulating hormone receptor gene. J. Androl. 2001, 22, 629–639. [Google Scholar]
- Guiraldelli, M.F.; Eyster, C.; Wilkerson, J.L.; Dresser, M.E.; Pezza, R.J. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 2013, 9, e1003383. [Google Scholar] [CrossRef]
- Gallardo, T.; Shirley, L.; John, G.B.; Castrillon, D.H. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007, 45, 413–417. [Google Scholar] [CrossRef]
- Saitou, M.; Yamaji, M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012, 4. [Google Scholar] [CrossRef]
- Yang, Q.E.; Oatley, J.M. Spermatogonial stem cell functions in physiological and pathological conditions. Curr. Top. Dev. Biol. 2014, 107, 235–267. [Google Scholar]
- Ali, M.A.M.; Strickfaden, H.; Lee, B.L.; Spyracopoulos, L.; Hendzel, M.J. RYBP is a k63-ubiquitin-chain-binding protein that inhibits homologous recombination repair. Cell Rep. 2018, 22, 383–395. [Google Scholar] [CrossRef]
- Basu, A.; Wilkinson, F.H.; Colavita, K.; Fennelly, C.; Atchison, M.L. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014, 42, 2208–2223. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Schrans-Stassen, B.H.; Saunders, P.T.; Cooke, H.J.; de Rooij, D.G. Nature of the spermatogenic arrest in Dazl -/- mice. Biol. Reprod. 2001, 65, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.S.; Toyooka, Y.; Akasu, R.; Katoh-Fukui, Y.; Nakahara, Y.; Suzuki, R.; Yokoyama, M.; Noce, T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000, 14, 841–853. [Google Scholar] [PubMed]
- Schlisio, S.; Halperin, T.; Vidal, M.; Nevins, J.R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002, 21, 5775–5786. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Yan, R.-G.; Zhang, X.-N.; Yang, Q.-E. Ring 1 and YY1 Binding Protein is Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis. Genes 2020, 11, 84. https://doi.org/10.3390/genes11010084
He Z, Yan R-G, Zhang X-N, Yang Q-E. Ring 1 and YY1 Binding Protein is Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis. Genes. 2020; 11(1):84. https://doi.org/10.3390/genes11010084
Chicago/Turabian StyleHe, Zhen, Rong-Ge Yan, Xiao-Na Zhang, and Qi-En Yang. 2020. "Ring 1 and YY1 Binding Protein is Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis" Genes 11, no. 1: 84. https://doi.org/10.3390/genes11010084
APA StyleHe, Z., Yan, R.-G., Zhang, X.-N., & Yang, Q.-E. (2020). Ring 1 and YY1 Binding Protein is Expressed in Murine Spermatocytes but Dispensable for Spermatogenesis. Genes, 11(1), 84. https://doi.org/10.3390/genes11010084




