Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Wide Association Study
2.2. Candidate Gene Identification
2.3. Putative Causal Variant Genotyping
2.4. ATCAY Expression Quantification
2.5. Rederived Atcayji-hes Mice
2.6. Atcayji-hes Genotyping
2.7. Atcayji-hes Phenotyping
2.8. Statistical Analysis
2.9. Central Nervous System Histopathology
3. Results
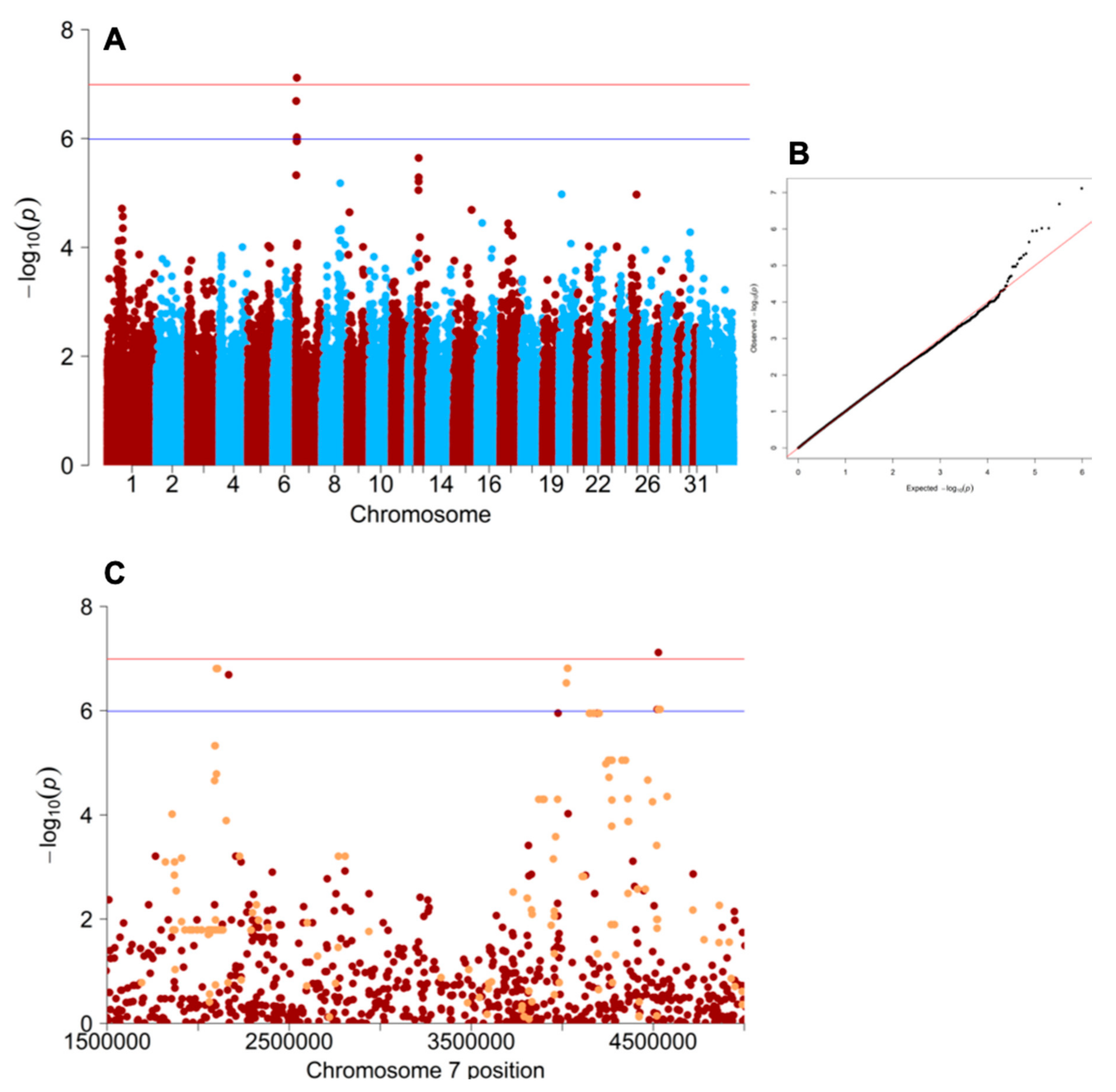
3.1. Genome-Wide Association Study
3.2. Equine ATCAY Genotyping
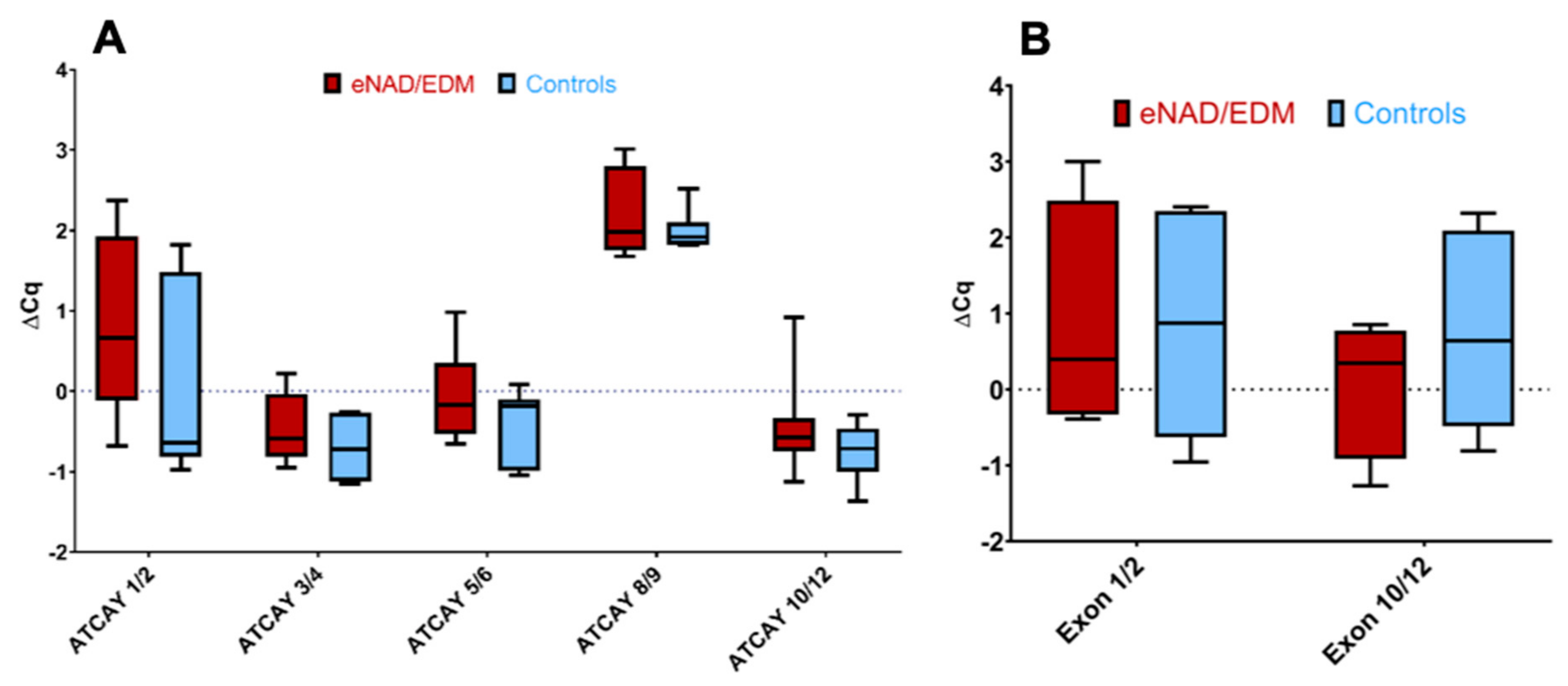
3.3. Equine ATCAY Expression Quantification
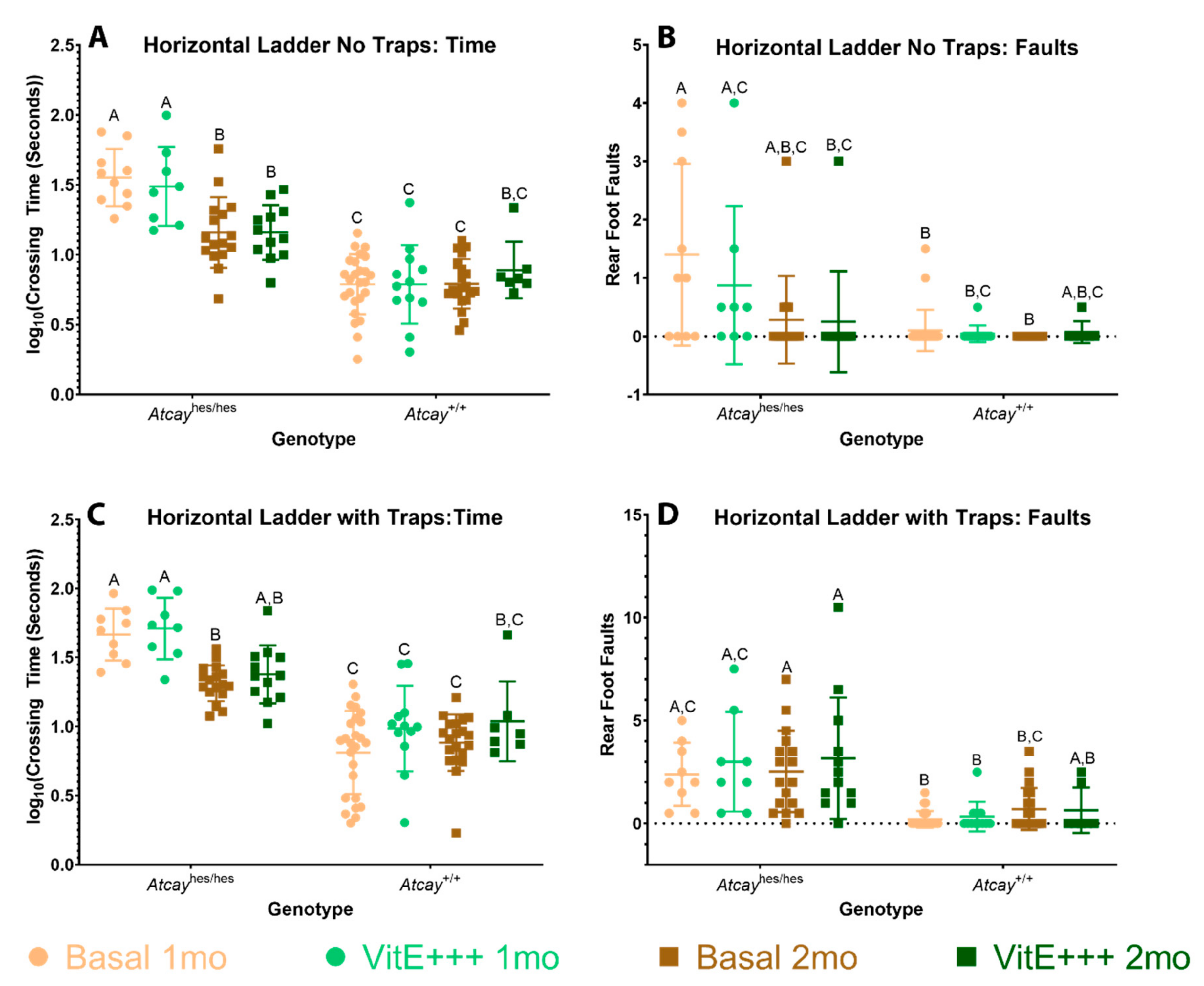
3.4. Phenotype of Atcayji-hes Mice
3.5. Atcayji-hes Histology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- BVSc, I.G.M.; Brown, C.M.; Stowe, H.D.; Trapp, A.L.; Derksen, F.J.; Clement, S.F. Equine degenerative myeloencephalopathy: A vitamin E deficiency that may be familial. J. Vet. Intern. Med. 1987, 1, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Finno, C.J.; Famula, T.; Aleman, M.; Higgins, R.J.; Madigan, J.E.; Bannasch, D.L. Pedigree analysis and exclusion of alpha-tocopherol transfer protein (TTPA) as a candidate gene for neuroaxonal dystrophy in the American quarter horse. J. Vet. Intern. Med. 2013, 27, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Aleman, M.; Finno, C.J.; Higgins, R.J.; Puschner, B.; Gericota, B.; Gohil, K.; LeCouteur, R.A.; Madigan, J.E. Evaluation of epidemiological, clinical, and pathological features of neuroaxonal dystrophy in Quarter Horses. J. Am. Vet. Med. Assoc. 2011, 239, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Finno, C.J.; Higgins, R.J.; Aleman, M.; Ofri, R.; Hollingsworth, S.R.; Bannasch, D.L.; Reilly, C.M.; Madigan, J.E. Equine degenerative myeloencephalopathy in Lusitano horses. J. Vet. Intern. Med. 2011, 25, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, I.G.; de Lahunta, A.; Whitlock, R.; Geary, J.C. Equine degenerative myeloencephalopathy. J. Am. Vet. Med. Assoc. 1977, 170, 195–201. [Google Scholar]
- Finno, C.J.; Valberg, S.J.; Shivers, J.; D’Almeida, E.; Armién, A.G. Evidence of the primary afferent tracts undergoing neurodegeneration in horses with equine degenerative myeloencephalopathy based on calretinin immunohistochemical localization. Vet. Pathol. 2016, 53, 77–86. [Google Scholar] [CrossRef]
- Burns, E.N.; Finno, C.J. Equine degenerative myeloencephalopathy: Prevalence, impact, and management. Vet. Med. Res. Reports 2018, 9, 63–67. [Google Scholar] [CrossRef]
- Ouahchi, K.; Arita, M.; Kayden, H.; Hentati, F.; Hamida, M.B.; Sokol, R.; Arai, H.; Inoue, K.; Mandel, J.-L.; Koenig, M. Ataxia with isolated vitamin E deficiency is caused by mutations in the α–tocopherol transfer protein. Nat. Genet. 1995, 9, 141–145. [Google Scholar] [CrossRef]
- Finno, C.J.; Aleman, M.; Higgins, R.J.; Madigan, J.E.; Bannasch, D.L. Risk of false positive genetic associations in complex traits with underlying population structure: A case study. Vet. J. 2014, 202, 543–549. [Google Scholar] [CrossRef]
- Schaefer, R.J.; Schubert, M.; Bailey, E.; Bannasch, D.L.; Barrey, E.; Bar-Gal, G.K.; Brem, G.; Brooks, S.A.; Distl, O.; Fries, R.; et al. Developing a 670k genotyping array to tag ~2M SNPs across 24 horse breeds. BMC Genomics 2017, 18, 1–18. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Bomar, J.M.; Benke, P.J.; Slattery, E.L.; Puttagunta, R.; Taylor, L.P.; Seong, E.; Nystuen, A.; Chen, W.; Albin, R.L.; Patel, P.D.; et al. Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nat. Genet. 2003, 35, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Kapfhamer, D.; Sweet, H.O.; Sufalko, D.; Warren, S.; Johnson, K.R.; Burmeister, M. The neurological mouse mutations jittery and hesitant are allelic and map to the region of mouse Chromosome 10 homologous to 19p13.3. Genomics 1996, 35, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Hata, S.; Nakao, T.; Tanigawa, Y.; Oka, C.; Kawaichi, M. Cayman ataxia protein caytaxin is transported by kinesin along neurites through binding to kinesin light chains. J. Cell Sci. 2009, 122, 4177–4185. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Q.; Low, B.C. Functional plasticity of the BNIP-2 and Cdc42GAP Homology (BCH) domain in cell signaling and cell dynamics. FEBS Lett. 2012, 586, 2674–2691. [Google Scholar] [CrossRef][Green Version]
- Sikora, K.M.; Nosavanh, L.M.; Kantheti, P.; Burmeister, M.; Hortsch, M. Expression of caytaxin protein in cayman ataxia mouse models correlates with phenotype severity. PLoS One 2012, 7, 1–12. [Google Scholar] [CrossRef]
- Luna-Cancalon, K.; Sikora, K.M.; Pappas, S.S.; Singh, V.; Wulff, H.; Paulson, H.L.; Burmeister, M.; Shakkottai, V.G. Alterations in cerebellar physiology are associated with a stiff-legged gait in Atcayji-hes mice. Neurobiol. Dis. 2014, 67, 140–148. [Google Scholar] [CrossRef]
- Lakes, E.H.; Allen, K.D. Gait analysis methods for rodent models of arthritic disorders: Reviews and recommendations. Osteoarthr. Cartil. 2016, 24, 1837–1849. [Google Scholar] [CrossRef]
- Cummings, B.J.; Engesser-cesar, C.; Anderson, A.J. Adaptation of a ladder beam walking task to assess locomotor recovery in mice following spinal cord injury Adaptation of a ladder beam walking task to assess locomotor recovery in mice. Brain 2007, 177, 232–241. [Google Scholar]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Beeson, S.K.; Schaefer, R.J.; Mason, V.C.; McCue, M.E. Robust remapping of equine SNP array coordinates to EquCab3. Anim. Genet. 2018, 50, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. Am. J. Hum. Genet. 2014. [Google Scholar] [CrossRef]
- Vuckovic, D.; Gasparini, P.; Soranzo, N.; Iotchkova, V. MultiMeta: An R package for meta-analyzing multi-phenotype genome-wide association studies. Bioinformatics 2015, 31, 2754–2756. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Manley, G. Animal models of dystonia: Lessons from a mutant Rat. Neurobiol. Dis. 2013, 71, 233–236. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Marth, G.; Garrison, E. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012, 3, 1–9. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Ruden, D.M.; Lu, X. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin). 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef]
- Finno, C.J.; Bordbari, M.H.; Valberg, S.J.; Lee, D.; Herron, J.; Hines, K.; Monsour, T.; Scott, E.; Bannasch, D.L.; Mickelson, J.; et al. Transcriptome profiling of equine vitamin E deficient neuroaxonal dystrophy identifies upregulation of liver X receptor target genes. Free Radic. Biol. Med. 2016, 101, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Finno, C.J.; Bordbari, M.H.; Gianino, G.; Ming-Whitfield, B.; Burns, E.; Merkel, J.; Britton, M.; Durbin-Johnson, B.; Sloma, E.A.; McMackin, M.; et al. An innate immune response and altered nuclear receptor activation defines the spinal cord transcriptome during alpha-tocopherol deficiency in Ttpa-null mice. Free Radic. Biol. Med. 2018, 120, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Itoh, M.; Yamada, A.; Mitsuda, T.; Nakagawa, T. Expression and localization of Cayman ataxia-related protein, Caytaxin, is regulated in a developmental- and spatial-dependent manner. Brain Res. 2007. [Google Scholar] [CrossRef] [PubMed]
- Beare, J.E.; Morehouse, J.R.; DeVries, W.H.; Enzmann, G.U.; Burke, D.A.; Magnuson, D.S.K.; Whittemore, S.R. Gait analysis in normal and spinal contused mice using the TreadScan system. J. Neurotrauma 2009, 26, 2045–2056. [Google Scholar] [CrossRef]
- Finno, C.J.; Estell, K.E.; Katzman, S.; Winfield, L.; Rendahl, A.; Textor, J.; Bannasch, D.L.; Puschner, B. Blood and cerebrospinal fluid α-tocopherol and selenium concentrations in neonatal foals with neuroaxonal dystrophy. J. Vet. Intern. Med. 2015, 29, 1667–1675. [Google Scholar] [CrossRef]
- Sun, J.; Pan, C.Q.; Chew, T.W.; Liang, F.; Burmeister, M.; Low, B.C. BNIP-H recruits the cholinergic machinery to neurite terminals to promote acetylcholine signaling and neuritogenesis. Dev. Cell 2015, 34, 555–568. [Google Scholar] [CrossRef]
- Ito-Smith, K.M. Characterization of Caytaxin Protein in Animal Models of cerebellar dysfunction. PhD Thesis, University of Michigan, Ann Arbor, MI, USA, 2012. [Google Scholar]
- Gago, S.; Overton, N.L.D.; Ben-Ghazzi, N.; Novak-Frazer, L.; Read, N.D.; Denning, D.W.; Bowyer, P. Lung colonization by Aspergillus fumigatus is controlled by ZNF77. Nat. Commun. 2018, 9, 3835. [Google Scholar] [CrossRef]
- Pei, Z.; Jia, Z.; Watkins, P.A. The second member of the human and murine “Bubblegum” family is a testis- and brainstem-specific Acyl-CoA synthetase. J. Biol. Chem. 2006, 281, 6632–6641. [Google Scholar] [CrossRef]
- Sisó, S.; Ferrer, I.; Pumarola, M. Abnormal synaptic protein expression in two Arabian horses with equine degenerative myeloencephalopathy. Vet. J. 2003, 166, 238–243. [Google Scholar] [CrossRef]
- Finno, C.J.; Kaese, H.J.; Miller, A.D.; Gianino, G.; Divers, T.; Valberg, S.J. Pigment retinopathy in warmblood horses with equine degenerative myeloencephalopathy and equine motor neuron disease. Vet. Ophthalmol. 2017, 20, 304–309. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hales, E.N.; Esparza, C.; Peng, S.; Dahlgren, A.R.; Peterson, J.M.; Miller, A.D.; Finno, C.J. Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models. Genes 2020, 11, 82. https://doi.org/10.3390/genes11010082
Hales EN, Esparza C, Peng S, Dahlgren AR, Peterson JM, Miller AD, Finno CJ. Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models. Genes. 2020; 11(1):82. https://doi.org/10.3390/genes11010082
Chicago/Turabian StyleHales, Erin N, Christina Esparza, Sichong Peng, Anna R Dahlgren, Janel M Peterson, Andrew D Miller, and Carrie J Finno. 2020. "Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models" Genes 11, no. 1: 82. https://doi.org/10.3390/genes11010082
APA StyleHales, E. N., Esparza, C., Peng, S., Dahlgren, A. R., Peterson, J. M., Miller, A. D., & Finno, C. J. (2020). Genome-Wide Association Study and Subsequent Exclusion of ATCAY as a Candidate Gene Involved in Equine Neuroaxonal Dystrophy Using Two Animal Models. Genes, 11(1), 82. https://doi.org/10.3390/genes11010082





