Genetic Mapping Identifies Consistent Quantitative Trait Loci for Yield Traits of Rice under Greenhouse Drought Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Mapping Population
2.2. Drought Screening and Phenotypic Data Analysis
2.3. Molecular Markers and Genotyping
2.4. Genotyping, Linkage and QTL Mapping
2.5. The Identification of Candidate Genes in QTL Region
3. Results
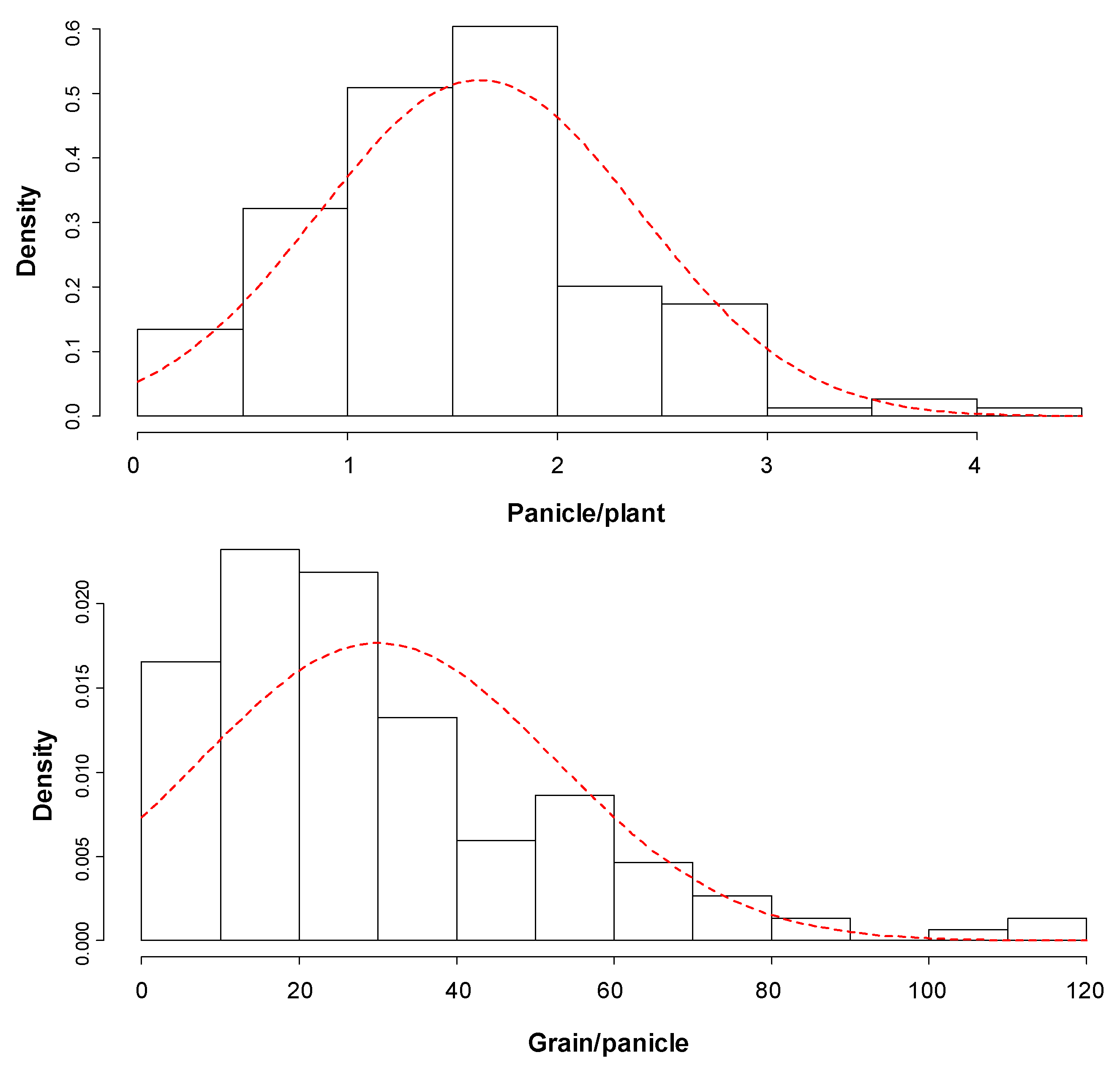
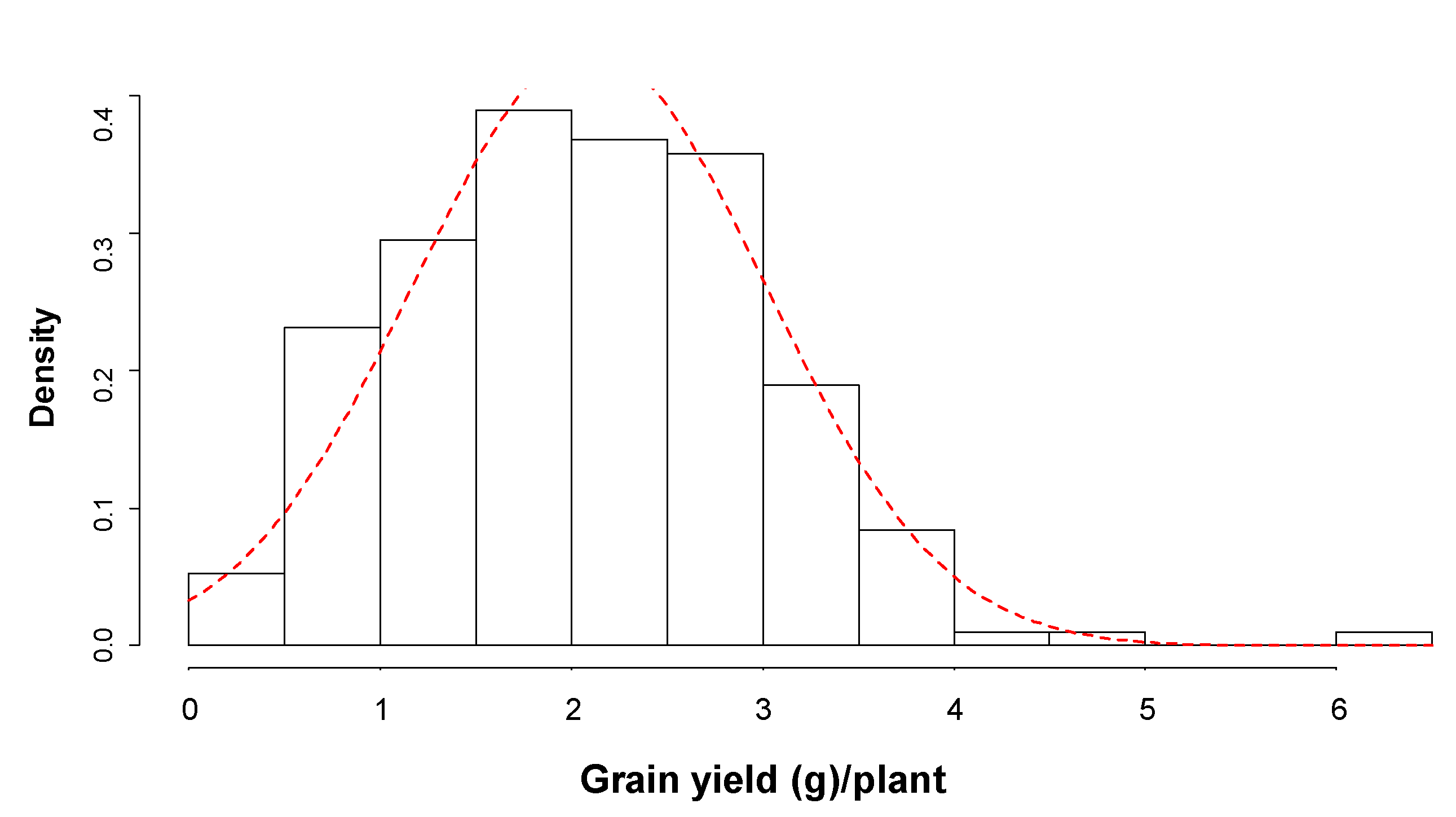
3.1. Genetic Variation for Grain Yield and Yield Traits under Drought
3.2. QTLs Controlling the Yield Traits
3.3. Allelic Contribution at the Drought Yield Traits QTLs
3.4. Genes Underlying QTL Regions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sandhu, K.S.; You, F.M.; Conner, R.L.; Balasubramanian, P.M.; Hou, A. Genetic Analysis and QTL Mapping of the Seed Hardness Trait in a Black Common Bean (Phaseolus vulgaris) Recombinant Inbred Line (RIL) Population. Mol. Breed. 2018, 38, 34. [Google Scholar] [CrossRef]
- Sandhu, N.; Kumar, A. Bridging the Rice Yield Gaps under Drought: QTLs, Genes, and Their Use in Breeding Programs. Agron. J. 2017, 7, 27. [Google Scholar] [CrossRef]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef]
- Kumar, A.; Bernier, J.; Verulkar, S.; Lafitte, H.R.; Atlin, G.N. Breeding for Drought Tolerance: Direct Selection for Yield, Response to Selection and Use of Drought-Tolerant Donors in Upland and Lowland-Adapted Populations. Field Crops Res. 2008, 107, 221–231. [Google Scholar] [CrossRef]
- Baisakh, N.; Famoso, A.; Yabes, J.; Tabien, R.; Harrell, D. Developing rice varieties suitable for alternative irrigation regime in Louisiana. La. Agric. 2019, 62, 18–19. [Google Scholar]
- Swain, P.; Anumalla, M.; Prusty, S.; Marndi, B.C.; Rao, G.J.N. Characterization of some Indian native land race rice accessions for drought tolerance at seedling stage. Aust. J. Crop Sci. 2014, 8, 324–331. [Google Scholar]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef]
- Kumar, S.; Sachdeva, S.; Bhat, K.V.; Vats, S. Plant Responses to Drought Stress: Physiological, Biochemical and Molecular Basis. In Biotic and Abiotic Stress Tolerance in Plants; Vats, S., Ed.; Springer Nature: Singapore, 2018. [Google Scholar]
- Kumar, A.; Dixit, S.; Ram, T.; Yadaw, R.B.; Mishra, K.K.; Mandal, N.P. Breeding High-Yielding Drought-Tolerant Rice: Genetic Variations and Conventional and Molecular Approaches. J. Exp. Bot. 2014, 65, 6265–6278. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-Analysis of Grain Yield QTL Identified during Agricultural Drought in Grasses Showed Consensus. BMC Genom. 2010, 12, 319. [Google Scholar] [CrossRef]
- Bernier, J.; Kumar, A.; Ramaiah, V.; Spaner, D.; Atlin, G.N. A Large-Effect QTL for Grain Yield under Reproductive-Stage Drought Stress in Upland Rice. Crop Sci. 2007, 47, 507–516. [Google Scholar] [CrossRef]
- Bernier, J.; Kumar, A.; Venuprasad, R.; Spaner, D.; Verulkar, S.; Mandal, N.P.; Sinha, P.K.; Peeraju, P.; Dongre, P.R.; Mahto, R.N.; et al. Characterization of the Effect of a QTL for Drought Resistance in Rice, qtl12.1, over a Range of Environments in the Philippines and Eastern INDIA. Euphytica 2009, 166, 207–217. [Google Scholar] [CrossRef]
- Mishra, K.K.; Vikram, P.; Yadaw, R.B.; Swamy, B.P.M.; Dixit, S.; Cruz, M.T.S.; Maturan, P.; Marker, S.; Kumar, A. qDTY12.1: A Locus with a Consistent Effect on Grain Yield under Drought in Rice. BMC Genet. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.; Cruz, M.T.; Singh, A.K.; Kumar, A. qDTY1.1, a Major QTL for Rice Grain Yield under Reproductive-Stage Drought Stress with a Consistent Effect in Multiple elite Genetic Backgrounds. BMC Genet. 2011, 12. [Google Scholar] [CrossRef]
- Bhattarai, U.; Subudhi, P.K. Genetic Analysis of Yield and Agronomic Traits under Reproductive-Stage Drought Stress in Rice Using a High-Resolution Linkage Map. Gene 2018, 669, 69–76. [Google Scholar] [CrossRef]
- Ghimire, K.H.; Quiatchon, L.A.; Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.; Hernandez, J.E.; Borromeo, T.H.; Kumar, A. Identification and Mapping of a QTL (qDTY1.1) with a Consistent Effect on Grain Yield under Drought. Field Crops Res. 2012, 131, 88–96. [Google Scholar] [CrossRef]
- Yadaw, R.B.; Dixit, S.; Raman, A.; Mishra, K.K.; Vikram, P.; Swamy, B.P.M.; Cruz, M.T.S.; Maturan, P.T.; Pandey, M.; Kumar, A. A QTL for High Grain Yield under Lowland Drought in the Background of Popular Rice Variety SABITRI from Nepal. Field Crops Res. 2013, 144, 281–287. [Google Scholar] [CrossRef]
- Saikumar, S.; Gouda, K.P.; Saiharini, A.; Varma, C.M.K.; Vineesha, O.; Padmavathi, G.; Shenoy, V.V. Major QTL for Enhancing Rice Grain Yield under Lowland Reproductive Drought Stress Identified Using an O. sativa/O. glaberrima Introgression Line. Field Crops Res. 2014, 163, 119–131. [Google Scholar] [CrossRef]
- Tang, S.Q.; Shao, G.N.; Wei, X.J.; Chen, M.L.; Shen, Z.H.; Luo, J.; Jiao, G.A.; Xie, L.H.; Hu, P.S. QTL Mapping of Grain Weight in Rice and the Validation of the QTL qTGW3.2. Gene. 2013, 527, 201–206. [Google Scholar] [CrossRef]
- Dixit, S.; Swamy, B.P.M.; Vikram, P.; Bernier, J.; Cruz, M.T.; Amante, M.; Atri, D.; Kumar, A. Increased drought tolerance and wider adaptability of qDTY 12.1 conferred by its interaction with qDTY 2.3 and qDTY 3.2. Mol. Breed. 2012, 30, 1767–1779. [Google Scholar] [CrossRef]
- Lang, N.T.; Nha, C.T.; Ha, P.T.T.; Buu, B.C. Quantitative Trait Loci (QTLs) Associated with Drought Tolerance in Rice (Oryza sativa L.). SABRAO J. Breed. Genet. 2013, 45, 409–421. [Google Scholar]
- Zhang, Y.D.; Zhang, Y.H.; Dong, S.L.; Chen, T.; Zhao, Q.Y.; Zhu, Z.; Zhou, L.H.; Yao, S.; Zhao, L.; Yu, X.; et al. QTL Mapping for Grain Size Traits Based on Extra-Large Grain Rice Line TD70. Rice Sci. 2014, 20, 400–406. [Google Scholar] [CrossRef]
- Dilday, R.H. Contribution of Ancestral Lines in the Development of New Cultivars of Rice. Crop Sci. 1990, 30, 905–911. [Google Scholar] [CrossRef]
- Solis, J.; Gutierrez, A.; Mangu, V.; Sanchez, E.; Bedre, R.; Linscombe, S.; Baisakh, N. Genetic Mapping of Quantitative Trait Loci for Grain Yield under Drought in Rice under Controlled Greenhouse Conditions. Front. Chem. 2018, 5, 129. [Google Scholar] [CrossRef]
- Linscombe, S.D.; Jodari, F.; Bollich, P.K.; Groth, D.E.; White, L.M.; Chu, Q.R.; Dunand, R.T.; Sanders, D.E. Registration of “Cocodrie” Rice. Crop Sci. 2000, 40, 294. [Google Scholar] [CrossRef]
- Jagadish, S.V.; Muthurajan, R.; Oane, R.; Wheeler, T.R.; Heuer, S.; Bennett, J.; Craufurd, P.Q. Physiological and Proteomic Approaches to Address Heat Tolerance during Anthesis in Rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 143–156. [Google Scholar] [CrossRef]
- Batlang, U.; Baisakh, N.; Ambavaram, M.M.R.; Pereira, A. Phenotypic and Physiological Evaluation for Drought and Salinity Stress Responses in Rice. In Rice Protocols. Methods in Molecular Biology (Methods and Protocols); Yang, Y., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 956. [Google Scholar] [CrossRef]
- SAS Institute. SAS 9.3; SAS Institute Inc.: Cary, NC, USA, 2011. [Google Scholar]
- Gutierrez, A.F. Development of Functional Markers for Resistance to Leaf Scald in Sugarcane; [Dissertation/doctoral dissertation etd-11102016-152405]; Louisiana State University: Baton Rouge, LA, USA, 2016; pp. 57–58. [Google Scholar]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Wu, D.-H.; Wu, H.-P.; Wang, C.-S.; Tseng, H.-Y.; Hwu, K.-K. Genome-Wide InDel Marker System for Application in Rice Breeding and Mapping Studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Zhang, L.; Meng, L. Users’ Manual of QTL IciMapping; The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS): Beijing, China; Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 2014. [Google Scholar]
- Mansueto, L.; Fuentes, R.R.; Borja, F.N.; Detras, J.; Abriol-Santos, J.M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A. Rice SNP-Seek Database Update: New SNPs, Indels, and Queries. Nucleic Acids Res. 2017, 45, D1075–D1081. [Google Scholar] [CrossRef]
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome Analysis in Different Rice Cultivars Provides Novel Insights into Desiccation and Salinity Stress Responses. Sci. Rep. 2016, 6, 23719. [Google Scholar] [CrossRef]
- Dixit, S.; Singh, A.; Kumar, A. Rice Breeding for High Grain Yield under Drought: A Strategic Solution to a Complex Problem. Int. J. Agron. 2014, 2014. [Google Scholar] [CrossRef]
- Lanceras, J.C.; Pantuwan, G.; Jongdee, B.; Toojinda, T. Quantitative Trait Loci Associated with Drought Tolerance at Reproductive Stage in Rice. Plant Physiol. 2004, 135, 384–399. [Google Scholar] [CrossRef]
- Venuprasad, R.; Dalid, C.O.; Del Valle, M.; Zhao, D.; Espiritu, M.; Cruz, M.T.; Amante, M.; Kumar, A.; Atlin, G.N. Identification and Characterization of Large-Effect Quantitative Trait Loci for Grain Yield under Lowland Drought Stress in Rice Using Bulk-Segregant Analysis. Theor. Appl. Genet. 2009, 120, 177–190. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Ahmed, H.U.; Henry, A.; Mauleon, R.; Dixit, S.; Vikram, P.; Tilatto, R.; Verulkar, S.B.; Perraju, P.; Mandal, N.P.; et al. Genetic, Physiological, and Gene Expression Analyses Reveal that Multiple QTL Enhance Yield of Rice Mega-Variety IR64 under Drought. PLoS ONE 2013, 8, e62795. [Google Scholar] [CrossRef]
- Sandhu, N.; Singh, A.; Dixit, S.; Sta Cruz, M.T.; Maturan, P.C.; Kumar, R.; Kumar, A. Identification and Mapping of Stable QTL with Main and Epistasis Effect on Rice Grain Yield under Upland Drought Stress. BMC Genet. 2014, 15, 63. [Google Scholar] [CrossRef]
- Manickavelu, A.; Nadarajan, N.; Ganesh, S.K.; Gnanamalar, R.P.; Babu, R.C. Drought Tolerance in Rice: Morphological and Molecular Genetic Consideration. Plant Growth Regul. 2006, 50, 121–138. [Google Scholar] [CrossRef]
- Venuprasad, R.; Bool, M.E.; Quiatchon, L.; Cruz, M.T.; Amante, M.; Atlin, G.N. A Large-Effect QTL for Rice Grain Yield under Upland Drought Stress on Chromosome 1. Mol. Breed. 2012, 30, 535–547. [Google Scholar] [CrossRef]
- Trijatmiko, K.R.; Prasetiyono, S.K.; Thomson, M.J.; Vera Cruz, C.M.; Moeljopawiro, S.; Pereira, A. Meta-Analysis of Quantitative Trait Loci for Grain Yield and Component Traits under Reproductive-Stage Drought Stress in an Upland Rice Population. Mol. Breed. 2014, 34, 283–295. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Singh, R.; Singh, B.P.; Miro, B.; Kohli, A.; Henry, A.; Singh, N.K.; Kumar, A. Drought Susceptibility of Modern Rice Varieties: An Effect of Linkage of Drought Tolerance with Undesirable Traits. Sci. Rep. 2015, 5, 14799. [Google Scholar] [CrossRef]
- Xing, W.; Zhao, H.; Zou, D. Detection of Main-Effect and Epistatic QTL for Yield-Related Traits in Rice under Drought Stress and Normal Conditions. Can. J. Plant Sci. 2014, 94, 633–641. [Google Scholar] [CrossRef]
- Sandhu, N.; Jain, S.; Kumar, A.; Mehla, B.S.; Jain, R. Genetic Variation, Linkage Mapping of QTL and Correlation Studies for Yield, Root, and Agronomic Traits for Aerobic Adaptation. BMC Genet. 2013, 14. [Google Scholar] [CrossRef]
- Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.; Cruz, M.T.; Singh, A.K.; Ye, G.; Kumara, A. Bulk Segregant Analysis: “An Effective Approach for Mapping Consistent-Effect Drought Grain Yield QTLs in Rice.”. Field Crops Res. 2012, 134, 185–192. [Google Scholar] [CrossRef]
- Zhou, J.; You, A.; Ma, Z.; Zhu, L.; He, G. Association Analysis of Important Agronomic Traits in Japonica Rice Germplasm. Afr. J. Biotechnol. 2012, 11, 2957–2970. [Google Scholar] [CrossRef]
- Kaladhar, K.; Swamy, B.P.M.; Babu, A.P.; Reddy, C.; Sarala, N. Mapping Quantitative Trait Loci for Yield Traits in BC2F2 Population Derived from “Swarna” x O. Nivara Cross. Rice Genet Newslett. 2008, 24, 34–36. [Google Scholar]
- Prince, S.J.; Beena, R.; Gomez, S.M.; Senthivel, S.; Babu, R.C. Mapping Consistent Rice (Oryza sativa L.) Yield QTLs under Drought Stress in Target Rainfed Environments. Rice 2015, 8, 25. [Google Scholar] [CrossRef]
- Sabar, M.; Shabir, G.; Shah, S.M.; Aslam, K.; Naveed, S.A.; Arif, M. Identification and Mapping of QTLs Associated with Drought Tolerance Traits in Rice by a Cross between Super Basmati and IR55419-04. Breed. Sci. 2019, 69, 169–178. [Google Scholar] [CrossRef]
- Subashri, M.; Robin, S.; Vinod, K.K.; Rajeswari, S.; Mohanasundaram, K.; Raveendran, T.S. Trait Identification and QTL Validation for Reproductive Stage Drought Resistance in Rice Using Selective Genotyping of Near Flowering RILs. Euphytica 2009, 166, 291. [Google Scholar] [CrossRef]
- Wang, X.; Pang, Y.; Zhang, J.; Tao, Y.; Feng, B.; Zheng, T.; Xu, J.; Li, Z. Genetic Background Effects on QTL and QTL × Environment Interaction for Yield and Its Component Traits as Revealed by Reciprocal Introgression Lines in Rice. Crop, J. 2014, 2, 345–357. [Google Scholar] [CrossRef]
- Yue, B.; Xue, W.; Xiong, L.; Yu, X.; Luo, L.; Cui, K.; Jin, D.; Xing, Y.; Zhang, Q. Genetic Basis of Drought Resistance at Reproductive Stage in Rice: Separation of Drought Tolerance from Drought Avoidance. Genetics 2006, 172, 1213–1228. [Google Scholar] [CrossRef]
- Weber, V.S.; Melchinger, A.E.; Magorokosho, C.; Makumbi, D.; Banziger, M.; Atlin, G.N. Efficiency of Managed-Stress Screening of Elite Maize Hybrids under Drought and Low Nitrogen for Yield under rainfed Conditions in South Africa. Crop Sci. 2012, 52, 1011–1020. [Google Scholar] [CrossRef]
- Pandey, S.; Bhandari, H.; Ding, S.; Prapertchob, P.; Sharan, R.; Naik, D.; Taunk, S.K.; Sastri, A. Coping with Drought in Rice Farming in Asia: Insights from a Cross-Country Comparative Study. Agric. Econ. 2007, 37, 213–224. [Google Scholar] [CrossRef]
- Kamoshita, A.; Babu, R.C.; Boopathi, N.; Fukai, S. Phenotypic and Genotypic Analysis of Drought-Resistance Traits for Development of Rice Cultivars Adapted to Rainfed Environments. Field Crops Res. 2008, 109, 1–23. [Google Scholar] [CrossRef]
- Mackill, D.J.; Coffman, W.R.; Garrity, D.P. Rainfed lowland Rice Improvement; International Rice Research Institute: Manila, Philippines, 1996; p. 242. [Google Scholar]
- Wade, L.J.; McLaren, C.G.; Quintana, L.; Harnpichitvitaya, D.; Rajatasereekul, S.; Sarawgi, A.K.; Kumar, A.; Ahmed, H.U.; Singh, A.K.; Rodriguez, R.; et al. Genotype by Environment Interactions in Diverse Rainfed Lowland Rice Environment. Field Crops Res. 1999, 64, 35–50. [Google Scholar] [CrossRef]
- Arai-Sanoh, Y.; Takai, T.; Yoshinaga, S.; Nakano, H.; Kojima, M.; Sakakibara, H.; Kondo, M.; Uga, Y. Deep Rooting Conferred by DEEPER ROOTING 1 Enhances Rice Yield in Paddy Fields. Sci. Rep. 2015, 4, 5563. [Google Scholar] [CrossRef]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a Major QTL Involved in Deep Rooting of Rice under Upland Field Conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef]
- Serraj, R.; McNally, K.; Slamet-Loedin, I.; Kohli, A.; Haefele, S.; Atlin, G.; Kumar, A. Drought Resistance Improvement in Rice: An Integrated Genetic and Resource Management Strategy. Plant Prod. Sci. 2011, 14, 1–14. [Google Scholar] [CrossRef]

| Trait | QTL | Chr | Position (cM) | Left Marker | Right Marker | LOD | PVE | Add | Dom | Left CI | Right CI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Panicle/plant | |||||||||||
| qPN1.1 | 1 | 279.6078 | SNPID280 | RD0105_2 | 15.104 | 37.732 | 2.566 | −2.6217 | 276.6577 | 282.558 | |
| Grain/panicle | |||||||||||
| qGN3.1 | 3 | 31.4001 | RM1278 | RM1867 | 5.956 | 3.824 | 21.694 | −21.3334 | 20.95 | 43.0499 | |
| qGN3.2 | 3 | 130.6988 | RM514 | RM5755 | 4.339 | 3.753 | −21.515 | −21.0113 | 119.8487 | 140.4494 | |
| qGN5.1 | 5 | 107.2989 | RM440 | CVSSR21 | 3.354 | 3.770 | −21.117 | −22.9362 | 104.549 | 114.1488 | |
| Grain yield/plant | |||||||||||
| qGY1.1 | 1 | 287.7083 | SNPID280 | RD0105_2 | 2.787 | 11.147 | 0.983 | −1.395 | 282.7581 | 294.8588 | |
| qGY7.1 | 7 | 113.8988 | RM10 | RM47 | 2.661 | 12.652 | 0.370 | −1.0751 | 98.5491 | 124.2487 | |
| qGY8.1 | 8 | 30.6001 | RM22926 | SNPID457 | 5.777 | 13.264 | 0.424 | 0.2553 | 22.8501 | 37.65 | |
| qGY11.1 | 11 | 111.7989 | SNPID452 | SNPID202 | 3.767 | 7.859 | 0.396 | 0.0759 | 105.349 | 118.3488 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baisakh, N.; Yabes, J.; Gutierrez, A.; Mangu, V.; Ma, P.; Famoso, A.; Pereira, A. Genetic Mapping Identifies Consistent Quantitative Trait Loci for Yield Traits of Rice under Greenhouse Drought Conditions. Genes 2020, 11, 62. https://doi.org/10.3390/genes11010062
Baisakh N, Yabes J, Gutierrez A, Mangu V, Ma P, Famoso A, Pereira A. Genetic Mapping Identifies Consistent Quantitative Trait Loci for Yield Traits of Rice under Greenhouse Drought Conditions. Genes. 2020; 11(1):62. https://doi.org/10.3390/genes11010062
Chicago/Turabian StyleBaisakh, Niranjan, Jonalyn Yabes, Andres Gutierrez, Venkata Mangu, Peiyong Ma, Adam Famoso, and Andy Pereira. 2020. "Genetic Mapping Identifies Consistent Quantitative Trait Loci for Yield Traits of Rice under Greenhouse Drought Conditions" Genes 11, no. 1: 62. https://doi.org/10.3390/genes11010062
APA StyleBaisakh, N., Yabes, J., Gutierrez, A., Mangu, V., Ma, P., Famoso, A., & Pereira, A. (2020). Genetic Mapping Identifies Consistent Quantitative Trait Loci for Yield Traits of Rice under Greenhouse Drought Conditions. Genes, 11(1), 62. https://doi.org/10.3390/genes11010062





