The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Populations
2.2.1. Roma Living in Segregated Colonies
2.2.2. Hungarian General Population
2.3. DNA Isolation, Selection of SNPs and Genotyping
2.4. Statistical Analyses
3. Results
3.1. Characteristics of the Study Populations
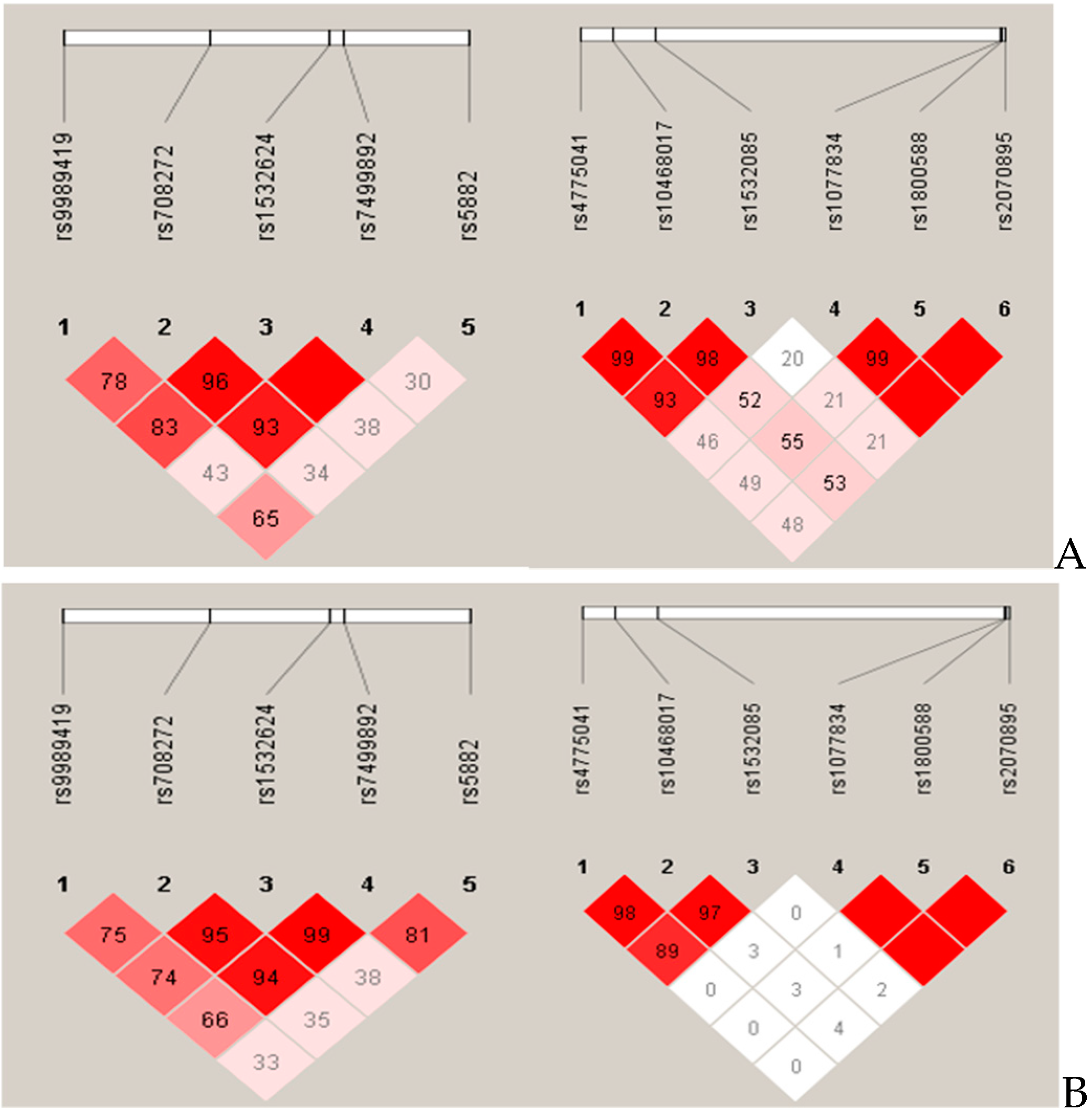
3.2. Haplotypes in the CETP and LIPC Genes and Their Linkage Disequilibrium Structure and Frequencies in the Roma and Hungarian General Populations
3.3. Association of Haplotypes in CETP and LIPC Genes with HDL-C and TG Levels in Combined Population
3.4. Association of Haplotypes in CETP and LIPC Genes with TG/HDL-C Ratio
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Torkamani, A.; Wineinger, N.E.; Topol, E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018, 19, 581–590. [Google Scholar] [CrossRef]
- European Commission. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee, and the Committee of the Regions: An EU Framework for National Roma Integration Strategies up to 2020; European Commission: Brussels, Belgium, 2011.
- Hungarian Central Statistical Office. Population Census of Hungary in 2011; Hungarian Central Statistical Office: Budapest, Hungary, 2011.
- Council of Europe. Estimates of Roma population in European Countries; Council of Europe: Strasbourg, France, 2014. [Google Scholar]
- Kosa, K.; Darago, L.; Adany, R. Environmental survey of segregated habitats of Roma in Hungary: A way to be empowering and reliable in minority research. Eur. J. Public Health 2011, 21, 463–468. [Google Scholar] [CrossRef]
- Foldes, M.E.; Covaci, A. Research on Roma health and access to healthcare: State of the art and future challenges. Int. J. Public Health 2012, 57, 37–39. [Google Scholar] [CrossRef]
- Kosa, K.; Adany, R. Studying vulnerable populations: Lessons from the Roma minority. Epidemiology 2007, 18, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Voko, Z.; Csepe, P.; Nemeth, R.; Kosa, K.; Kosa, Z.; Szeles, G.; Adany, R. Does socioeconomic status fully mediate the effect of ethnicity on the health of Roma people in Hungary? J. Epidemiol. Community Health 2009, 63, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Zeljko, H.; Skaric-Juric, T.; Narancic, N.S.; Salihovic, M.P.; Klaric, I.M.; Barbalic, M.; Starcevic, B.; Lauc, L.B.; Janicijevic, B. Traditional CVD risk factors and socio-economic deprivation in Roma minority population of Croatia. Coll Antropol. 2008, 32, 667–676. [Google Scholar]
- Dobranici, M.; Buzea, A.; Popescu, R. The cardiovascular risk factors of the Roma (gypsies) people in Central-Eastern Europe: A review of the published literature. J. Med. Life 2012, 5, 382–389. [Google Scholar] [PubMed]
- Zeljko, H.M.; Skaric-Juric, T.; Narancic, N.S.; Baresic, A.; Tomas, Z.; Petranovic, M.Z.; Milicic, J.; Salihovic, M.P.; Janicijevic, B. Age trends in prevalence of cardiovascular risk factors in Roma minority population of Croatia. Econ. Hum. Biol. 2013, 11, 326–336. [Google Scholar] [CrossRef]
- Weiss, E.; Japie, C.; Balahura, A.M.; Bartos, D.; Badila, E. Cardiovascular risk factors in a Roma sample population from Romania. Rom. J. Intern. Med. 2018, 56, 193–202. [Google Scholar] [CrossRef]
- Hernandez-PerezdelaBlanca, M.; Rebora-Mariano, T.; Ramirez-Robles, R.; Berna-Guisado, C.; Vides-Batanero, M.C.; Castro-Gomez, J.A. Predictors of cardiovascular risk in a population of diabetic adults of Gypsy origin, in Granada. Bratislavske Lekarske Listy 2014, 115, 579–584. [Google Scholar] [CrossRef]
- Fedacko, J.; Pella, D.; Jarcuska, P.; Siegfried, L.; Janicko, M.; Veseliny, E.; Pella, J.; Sabol, F.; Jarcuska, P.; Marekova, M.; et al. Prevalence of cardiovascular risk factors in relation to metabolic syndrome in the Roma population compared with the non-Roma population in the eastern part of Slovakia. Cent. Eur. J. Public Health 2014, 22, S69–S74. [Google Scholar] [CrossRef] [PubMed]
- Kosa, Z.; Moravcsik-Kornyicki, A.; Dioszegi, J.; Roberts, B.; Szabo, Z.; Sandor, J.; Adany, R. Prevalence of metabolic syndrome among Roma: A comparative health examination survey in Hungary. Eur. J. Public Health 2015, 25, 299–304. [Google Scholar] [CrossRef] [PubMed]
- NIH Consensus Conference. Triglyceride, high-density lipoprotein, and coronary heart disease. NIH Consensus Development Panel on Triglyceride, High-Density Lipoprotein, and Coronary Heart Disease. JAMA 1993, 269, 505–510. [Google Scholar] [CrossRef]
- Tosheska Trajkovska, K.; Topuzovska, S. High-density lipoprotein metabolism and reverse cholesterol transport: Strategies for raising HDL cholesterol. Anatol. J. Cardiol. 2017, 18, 149–154. [Google Scholar] [CrossRef]
- Rader, D.J.; Hovingh, G.K. HDL and cardiovascular disease. Lancet 2014, 384, 618–625. [Google Scholar] [CrossRef]
- Hokanson, J.E.; Austin, M.A. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: A meta-analysis of population-based prospective studies. J. Cardiovasc. Risk 1996, 3, 213–219. [Google Scholar] [CrossRef]
- Da Luz, P.L.; Favarato, D.; Faria-Neto, J.R., Jr.; Lemos, P.; Chagas, A.C. High ratio of triglycerides to HDL-cholesterol predicts extensive coronary disease. Clinics (Sao Paulo) 2008, 63, 427–432. [Google Scholar] [CrossRef]
- Ul Ain, Q.; Asif, N.; Gilani, M.; Waqas Sheikh, N.; Akram, A. To Determine Cutoff Value of Triglycerides to HDL Ratio in Cardio Vascular Risk Factors. Biochem. Anal. Biochem. 2017, 7, 2. [Google Scholar] [CrossRef]
- Piko, P.; Fiatal, S.; Kosa, Z.; Sandor, J.; Adany, R. Genetic factors exist behind the high prevalence of reduced high-density lipoprotein cholesterol levels in the Roma population. Atherosclerosis 2017, 263, 119–126. [Google Scholar] [CrossRef]
- Piko, P.; Fiatal, S.; Kosa, Z.; Sandor, J.; Adany, R. Generalizability and applicability of results obtained from populations of European descent regarding the effect direction and size of HDL-C level-associated genetic variants to the Hungarian general and Roma populations. Gene 2019, 686, 187–193. [Google Scholar] [CrossRef]
- Zarkesh, M.; Daneshpour, M.S.; Faam, B.; Fallah, M.S.; Hosseinzadeh, N.; Guity, K.; Hosseinpanah, F.; Momenan, A.A.; Azizi, F. Heritability of the metabolic syndrome and its components in the Tehran Lipid and Glucose Study (TLGS). Genet. Res. (Camb.) 2012, 94, 331–337. [Google Scholar] [CrossRef]
- Todur, S.P.; Ashavaid, T.F. Association of CETP and LIPC Gene Polymorphisms with HDL and LDL Sub-fraction Levels in a Group of Indian Subjects: A Cross-Sectional Study. Indian J. Clin. Biochem. IJCB 2013, 28, 116–123. [Google Scholar] [CrossRef][Green Version]
- Wang, J.; Wang, L.J.; Zhong, Y.; Gu, P.; Shao, J.Q.; Jiang, S.S.; Gong, J.B. CETP gene polymorphisms and risk of coronary atherosclerosis in a Chinese population. Lipids Health Dis. 2013, 12, 176. [Google Scholar] [CrossRef] [PubMed]
- Clifford, A.J.; Rincon, G.; Owens, J.E.; Medrano, J.F.; Moshfegh, A.J.; Baer, D.J.; Novotny, J.A. Single nucleotide polymorphisms in CETP, SLC46A1, SLC19A1, CD36, BCMO1, APOA5, and ABCA1 are significant predictors of plasma HDL in healthy adults. Lipids Health Dis. 2013, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.F.; Lira, M.E.; Durham, L.K.; Clark, R.W.; Bamberger, M.J.; Milos, P.M. Polymorphisms in the CETP gene and association with CETP mass and HDL levels. Atherosclerosis 2003, 167, 195–204. [Google Scholar] [CrossRef]
- Barter, P. CETP and atherosclerosis. Arter. Thromb. Vasc. Biol. 2000, 20, 2029–2031. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.C.; Brunzell, J.D.; Deeb, S.S. Ethnic differences in hepatic lipase and HDL in Japanese, black, and white Americans: Role of central obesity and LIPC polymorphisms. J. Lipid Res. 2004, 45, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Hodoglugil, U.; Williamson, D.W.; Mahley, R.W. Polymorphisms in the hepatic lipase gene affect plasma HDL-cholesterol levels in a Turkish population. J. Lipid Res. 2010, 51, 422–430. [Google Scholar] [CrossRef]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef]
- Daniels, T.F.; Killinger, K.M.; Michal, J.J.; Wright, R.W., Jr.; Jiang, Z. Lipoproteins, cholesterol homeostasis and cardiac health. Int. J. Biol. Sci. 2009, 5, 474–488. [Google Scholar] [CrossRef]
- Szeles, G.; Voko, Z.; Jenei, T.; Kardos, L.; Pocsai, Z.; Bajtay, A.; Papp, E.; Pasti, G.; Kosa, Z.; Molnar, I.; et al. A preliminary evaluation of a health monitoring programme in Hungary. Eur. J. Public Health 2005, 15, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Szigethy, E.; Szeles, G.; Horvath, A.; Hidvegi, T.; Jermendy, G.; Paragh, G.; Blasko, G.; Adany, R.; Voko, Z. Epidemiology of the metabolic syndrome in Hungary. Public Health 2012, 126, 143–149. [Google Scholar] [CrossRef]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Pers, T.H.; Timshel, P.; Hirschhorn, J.N. SNPsnap: A Web-based tool for identification and annotation of matched SNPs. Bioinformatics 2015, 31, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Wan, K.; Zhao, J.; Huang, H.; Zhang, Q.; Chen, X.; Zeng, Z.; Zhang, L.; Chen, Y. The association between triglyceride/high-density lipoprotein cholesterol ratio and all-cause mortality in acute coronary syndrome after coronary revascularization. PLoS ONE 2015, 10, e0123521. [Google Scholar] [CrossRef] [PubMed]
- Radovica, I.; Fridmanis, D.; Vaivade, I.; Nikitina-Zake, L.; Klovins, J. The association of common SNPs and haplotypes in CETP gene with HDL cholesterol levels in Latvian population. PLoS ONE 2013, 8, e64191. [Google Scholar] [CrossRef]
- McCaskie, P.A.; Beilby, J.P.; Chapman, C.M.; Hung, J.; McQuillan, B.M.; Thompson, P.L.; Palmer, L.J. Cholesteryl ester transfer protein gene haplotypes, plasma high-density lipoprotein levels and the risk of coronary heart disease. Hum. Genet. 2007, 121, 401–411. [Google Scholar] [CrossRef]
- Feitosa, M.F.; Myers, R.H.; Pankow, J.S.; Province, M.A.; Borecki, I.B. LIPC variants in the promoter and intron 1 modify HDL-C levels in a sex-specific fashion. Atherosclerosis 2009, 204, 171–177. [Google Scholar] [CrossRef]
- Guerra, R.; Wang, J.; Grundy, S.M.; Cohen, J.C. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc. Natl. Acad. Sci. USA 1997, 94, 4532–4537. [Google Scholar] [CrossRef]
- Lahoz, C.; Pena, R.; Mostaza, J.M.; Laguna, F.; Garcia-Iglesias, M.F.; Taboada, M.; Pinto, X.; Respuesta Ambulatoria a Pravastatina Study Group. The -514C/T polymorphism of the hepatic lipase gene significantly modulates the HDL-cholesterol response to statin treatment. Atherosclerosis 2005, 182, 129–134. [Google Scholar] [CrossRef]
- Tall, A.R. Plasma cholesteryl ester transfer protein. J. Lipid Res. 1993, 34, 1255–1274. [Google Scholar] [PubMed]
- Shrestha, S.; Wu, B.J.; Guiney, L.; Barter, P.J.; Rye, K.A. Cholesteryl ester transfer protein and its inhibitors. J. Lipid Res. 2018, 59, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, B.R.; Hoffmann, M.M.; Nauck, M.; Kumar, A.M.; Nandabalan, K.; Judson, R.S.; Boehm, B.O.; Tall, A.R.; Ruano, G.; Marz, W. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharm. J. 2003, 3, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Bercovich, D.; Friedlander, Y.; Korem, S.; Houminer, A.; Hoffman, A.; Kleinberg, L.; Shochat, C.; Leitersdorf, E.; Meiner, V. The association of common SNPs and haplotypes in the CETP and MDR1 genes with lipids response to fluvastatin in familial hypercholesterolemia. Atherosclerosis 2006, 185, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.R. Personalized medicine: Striding from genes to medicines. Perspect. Clin. Res. 2010, 1, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.T.; Yoon, J.H.; Kim, J.Y.; Ahn, S.K.; Linton, J.A.; Koh, S.B.; Kim, J.K. The association between the ratio of triglyceride to HDL-C and insulin resistance according to waist circumference in a rural Korean population. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 1054–1060. [Google Scholar] [CrossRef]
- Zhou, M.; Zhu, L.; Cui, X.; Feng, L.; Zhao, X.; He, S.; Ping, F.; Li, W.; Li, Y. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance but not of beta cell function in a Chinese population with different glucose tolerance status. Lipids Health Dis. 2016, 15, 104. [Google Scholar] [CrossRef]
- Iwani, N.A.; Jalaludin, M.Y.; Zin, R.M.; Fuziah, M.Z.; Hong, J.Y.; Abqariyah, Y.; Mokhtar, A.H.; Wan Nazaimoon, W.M. Triglyceride to HDL-C Ratio is Associated with Insulin Resistance in Overweight and Obese Children. Sci. Rep. 2017, 7, 40055. [Google Scholar] [CrossRef]
- Young, K.A.; Maturu, A.; Lorenzo, C.; Langefeld, C.D.; Wagenknecht, L.E.; Chen, Y.I.; Taylor, K.D.; Rotter, J.I.; Norris, J.M.; Rasouli, N. The triglyceride to high-density lipoprotein cholesterol (TG/HDL-C) ratio as a predictor of insulin resistance, beta-cell function, and diabetes in Hispanics and African Americans. J. Diabetes Complicat. 2019, 33, 118–122. [Google Scholar] [CrossRef]
- De Courten, B.V.; de Courten, M.; Hanson, R.L.; Zahorakova, A.; Egyenes, H.P.; Tataranni, P.A.; Bennett, P.H.; Vozar, J. Higher prevalence of type 2 diabetes, metabolic syndrome and cardiovascular diseases in gypsies than in non-gypsies in Slovakia. Diabetes Res. Clin. Pract. 2003, 62, 95–103. [Google Scholar] [CrossRef]
| Hungarian General (n = 1494) | Roma (n = 613) | ||
|---|---|---|---|
| Mean (95% CI) | p-Value | ||
| Age (year) | 44.16 (43.53–44.78) | 40.3 (39.39–41.21) | <0.001 |
| Body mass index (kg/m2) | 27.43 (27.15–27.70) | 27.47 (26.64–28.30) | 0.898 |
| Systolic blood pressure (mmHg) | 126.81 (125.95–127.66) | 125.21 (123.67–126.76) | 0.059 |
| Diastolic blood pressure (mmHg) | 80.26 (79.79–80.72) | 78.43 (77.63–79.23) | <0.001 |
| Fasting glucose level (mmol/L) | 4.83 (4.74–4.92) | 5.44 (5.29–5.59) | <0.001 |
| High-density lipoprotein-cholesterol (HDL-C) level (mmol/L) | 1.43 (1.40–1.45) | 1.21 (1.18–1.24) | <0.001 |
| Triglyceride (TG) level (mmol/L) | 1.63 (1.55–1.72) | 1.61 (1.50–1.71) | 0.760 |
| Triglycerides to HDL-C ratio (TG/HDL-C) ratio | 1.48 (1.35–1.61) | 1.67 (1.47–1.86) | 0.115 |
| Prevalence (%) | p-Value | ||
| Sex (female/male) | 52.7/47.3 | 61.7/38.3 | <0.001 |
| Antihypertensive treatment | 29.90 | 25.40 | 0.042 |
| Lipid-lowering treatment | 13.90 | 10.80 | 0.050 |
| Antidiabetic treatment | 5.40 | 5.20 | 0.901 |
| Reduced HDL-C level 1 | 28.20 | 53.00 | <0.001 |
| Elevated TG level 2 | 30.32 | 28.71 | 0.463 |
| Elevated TG/HDL-C ratio (≥1) 3 | 42.44 | 53.83 | <0.001 |
| Highly elevated TG/HDL-C ratio (>4) 4 | 5.09 | 5.87 | 0.465 |
| Haplotypes | Rs1532624 | Rs5882 | Rs708272 | Rs7499892 | Rs9989419 | Frequency in Combined Population (n = 2107) | Frequency in Hungarian General Population (n = 1494) | Frequency in Roma Population (n = 613) | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| H1 | A | G | A | C | G | 20.57% | 17.32% | 28.29% | <0.001 |
| H2 | A | A | A | C | G | 19.55% | 21.78% | 14.24% | <0.001 |
| H3 | C | A | G | C | A | 13.98% | 15.05% | 11.45% | 0.015 |
| H4 | C | A | G | C | G | 13.69% | 14.79% | 11.09% | 0.015 |
| H5 | C | A | G | T | A | 12.63% | 11.60% | 15.07% | 0.019 |
| H6 | C | G | G | C | A | 5.55% | 5.95% | 4.60% | 0.170 |
| H7 | A | A | A | C | A | 2.77% | 2.92% | 2.43% | 0.467 |
| H8 | C | G | G | T | G | 2.26% | 0.14% | 7.28% | <0.001 |
| H9 | C | G | G | C | G | 2.60% | 3.04% | 1.56% | 0.039 |
| H10 | C | A | G | T | G | 2.43% | 2.86% | 1.41% | 0.038 |
| Haplotypes | Rs10468017 | Rs1077834 | Rs1532085 | Rs1800588 | Rs2070895 | Rs4775041 | Frequency in Combined Population (n = 2107) | Frequency in Hungarian General Population (n = 1494) | Frequency in Roma Population (n = 613) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| H1 | C | T | G | C | G | G | 44.41% | 48.38% | 34.97% | <0.001 |
| H2 | T | T | A | C | G | C | 21.08% | 20.40% | 22.70% | 0.073 |
| H3 | C | C | G | T | A | G | 13.81% | 13.63% | 14.25% | 0.470 |
| H4 | C | T | A | C | G | G | 10.20% | 7.23% | 17.24% | <0.001 |
| H5 | T | C | A | C | G | G | 4.69% | 5.48% | 2.81% | 0.006 |
| H6 | C | C | A | T | A | G | 3.43% | 2.08% | 6.63% | <0.001 |
| Haplotypes | HDL-C | TG | ||||||
|---|---|---|---|---|---|---|---|---|
| β (95% CI) | p-Value | OR (95% CI) | p-Value | β (95% CI) | p-Value | OR (95% CI) | p-Value | |
| H1 | reference | --- | reference | --- | reference | --- | reference | --- |
| H2 | 0.02 (−0.01–0.06) | 0.220 | 0.88 (0.67–1.16) | 0.370 | −0.01 (−0.15–0.13) | 0.890 | 0.94 (0.71–1.24) | 0.650 |
| H3 | −0.05 (−0.09)–(−0.01) | 0.016 * | 1.34 (1.01–1.76) | 0.040 * | −0.16 (−0.30)–(−0.01) | 0.033 * | 0.85 (0.63–1.14) | 0.270 |
| H4 | −0.01 (−0.06–0.03) | 0.470 | 0.91 (0.68–1.20) | 0.490 | −0.03 (−0.18–0.11) | 0.650 | 1.02 (0.76–1.36) | 0.900 |
| H5 | −0.11 (−0.16)–(−0.07) | <0.001 ** | 1.74 (1.32–2.30) | <0.001 ** | −0.01 (−0.16–0.13) | 0.840 | 0.92 (0.68–1.23) | 0.560 |
| H6 | 0.01 (−0.06–0.07) | 0.880 | 0.86 (0.55–1.34) | 0.500 | 0.16 (−0.08–0.39) | 0.180 | 0.99 (0.63–1.57) | 0.980 |
| H7 | 0.04 (−0.05–0.13) | 0.370 | 0.93 (0.50–1.72) | 0.810 | −0.03 (−0.36–0.29) | 0.850 | 0.75 (0.37–1.53) | 0.430 |
| H8 | −0.14 (−0.22)–(−0.06) | 0.001 ** | 2.60 (1.43–4.72) | 0.002 ** | −0.33 (−0.62)–(−0.03) | 0.032 * | 0.50 (0.27–0.92) | 0.026 * |
| H9 | −0.07 (−0.17–0.03) | 0.140 | 1.04 (0.50–2.13) | 0.920 | −0.31 (−0.68–0.05) | 0.095 | 0.47 (0.19–1.12) | 0.087 |
| H10 | −0.07 (−0.16–0.02) | 0.130 | 1.80 (0.98–3.30) | 0.058 | −0.09 (−0.41–0.23) | 0.570 | 1.11 (0.60–2.04) | 0.740 |
| Haplotypes | HDL-C | TG | ||||||
|---|---|---|---|---|---|---|---|---|
| β (95% CI) | p-Value | OR (95% CI) | p-Value | β (95% CI) | p-Value | OR (95% CI) | p-Value | |
| H1 | reference | --- | reference | --- | reference | --- | reference | --- |
| H2 | 0.05 (0.01–0.08) | 0.004 ** | 0.75 (0.60–0.92) | 0.007 ** | 0.16 (0.05–0.27) | 0.005 ** | 1.30 (1.03–1.63) | 0.025 * |
| H3 | 0.07 (0.03–0.11) | 0.001 ** | 0.78 (0.59–1.03) | 0.079 | 0.21 (0.07–0.35) | 0.004 ** | 1.57 (1.18–2.10) | 0.002 ** |
| H4 | 0.03 (−0.01–0.07) | 0.160 | 0.97 (0.73–1.29) | 0.840 | 0.04 (−0.11–0.19) | 0.590 | 0.98 (0.71–1.35) | 0.890 |
| H5 | 0.09 (0.03–0.15) | 0.005 ** | 0.50 (0.31–0.81) | 0.005 ** | 0.16 (−0.06–0.39) | 0.150 | 1.23 (0.78–1.95) | 0.380 |
| H6 | 0.05 (−0.02–0.13) | 0.170 | 0.91 (0.56–1.46) | 0.680 | 0.45 (0.20–0.69) | <0.001 ** | 2.39 (1.46–3.92) | 0.001 ** |
| Haplotypes | β (95% CI) | p-Value | ORCMR a (95% CI) | p-Value | ORCD b (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| H1 | reference | --- | reference | --- | reference | --- |
| H2 | −0.12 (−0.36–0.13) | 0.350 | 0.82 (0.64–1.06) | 0.130 | 0.58 (0.37–0.91) | 0.019 * |
| H3 | −0.02 (−0.27–0.22) | 0.870 | 0.92 (0.71–1.18) | 0.510 | 0.96 (0.62–1.49) | 0.860 |
| H4 | 0.01 (−0.24–0.25) | 0.970 | 0.98 (0.76–1.26) | 0.850 | 1.05 (0.69–1.58) | 0.820 |
| H5 | 0.56 (0.31–0.82) | <0.001 ** | 1.51 (1.16–1.95) | 0.002 ** | 1.59 (1.09–2.33) | 0.017 * |
| H6 | 0.24 (−0.17–0.65) | 0.250 | 1.06 (0.70–1.59) | 0.790 | 1.25 (0.66–2.34) | 0.500 |
| H7 | −0.24 (−0.79–0.31) | 0.400 | 0.80 (0.45–1.40) | 0.430 | 0.98 (0.38–2.51) | 0.960 |
| H8 | 0.14 (−0.36–0.64) | 0.580 | 1.27 (0.74–2.17) | 0.390 | 0.88 (0.38–2.02) | 0.760 |
| H9 | −0.05 (−0.67–0.56) | 0.870 | 0.69 (0.36–1.32) | 0.260 | 0.61 (0.15–2.54) | 0.500 |
| H10 | 0.14 (−0.40–0.68) | 0.610 | 1.00 (0.58–1.75) | 0.990 | 1.37 (0.60–3.10) | 0.450 |
| Haplotypes | β (95% CI) | p-Value | ORCMR a (95% CI) (Cut-Off: 1) | p-Value | ORCD b (95% CI) (Cut-Off: 4) | p-Value |
|---|---|---|---|---|---|---|
| H1 | reference | --- | reference | --- | reference | --- |
| H2 | 0.07 (−0.12–0.26) | 0.480 | 1.00 (0.82–1.21) | 0.970 | 1.23 (0.89–1.70) | 0.220 |
| H3 | 0.05 (−0.19–0.29) | 0.690 | 0.99 (0.77–1.27) | 0.940 | 1.57 (1.06–2.32) | 0.024 ** |
| H4 | −0.07 (−0.33–0.18) | 0.570 | 0.81 (0.62–1.06) | 0.130 | 1.15 (0.72–1.83) | 0.560 |
| H5 | −0.15 (−0.55–0.24) | 0.440 | 0.95 (0.64–1.42) | 0.820 | 0.36 (0.11–1.19) | 0.093 |
| H6 | 0.52 (0.10–0.95) | 0.015 ** | 1.87 (1.17–2.98) | 0.009 ** | 1.44 (0.68–3.03) | 0.340 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piko, P.; Fiatal, S.; Werissa, N.A.; Bekele, B.B.; Racz, G.; Kosa, Z.; Sandor, J.; Adany, R. The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations. Genes 2020, 11, 56. https://doi.org/10.3390/genes11010056
Piko P, Fiatal S, Werissa NA, Bekele BB, Racz G, Kosa Z, Sandor J, Adany R. The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations. Genes. 2020; 11(1):56. https://doi.org/10.3390/genes11010056
Chicago/Turabian StylePiko, Peter, Szilvia Fiatal, Nardos Abebe Werissa, Bayu Begashaw Bekele, Gabor Racz, Zsigmond Kosa, Janos Sandor, and Roza Adany. 2020. "The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations" Genes 11, no. 1: 56. https://doi.org/10.3390/genes11010056
APA StylePiko, P., Fiatal, S., Werissa, N. A., Bekele, B. B., Racz, G., Kosa, Z., Sandor, J., & Adany, R. (2020). The Effect of Haplotypes in the CETP and LIPC Genes on the Triglycerides to HDL-C Ratio and Its Components in the Roma and Hungarian General Populations. Genes, 11(1), 56. https://doi.org/10.3390/genes11010056









