Transcriptional Differences of Coding and Non-Coding Genes Related to the Absence of Melanocyte in Skins of Bama Pig
Abstract
1. Introduction
2. Materials and Methods
2.1. Pig Skin Sampling
2.2. Total RNA Extraction, Sequencing, and Read Mapping
2.3. LncRNA Identification
2.4. Expression Analysis of mRNA and lncRNA
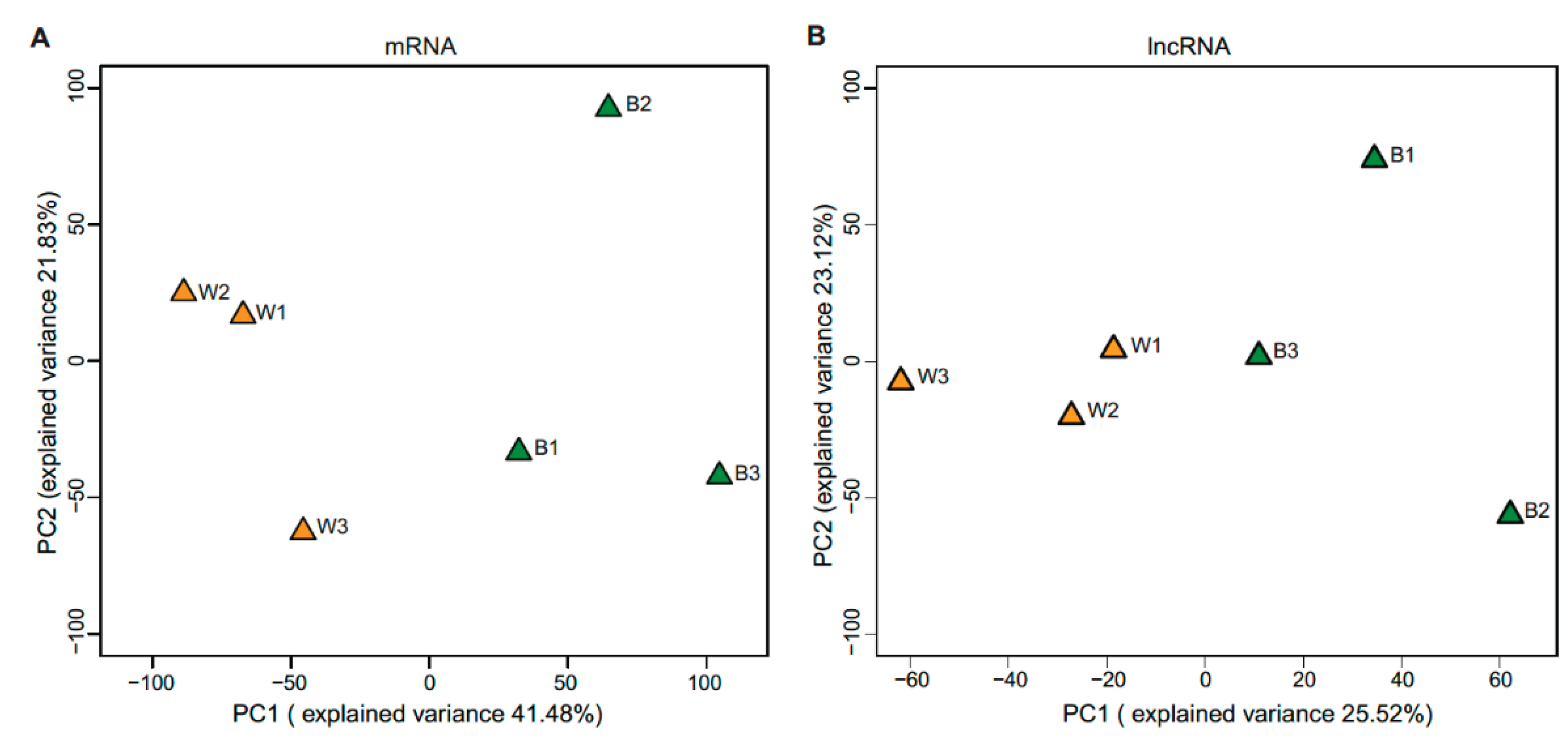
2.5. Clustering and Principal Component Analysis
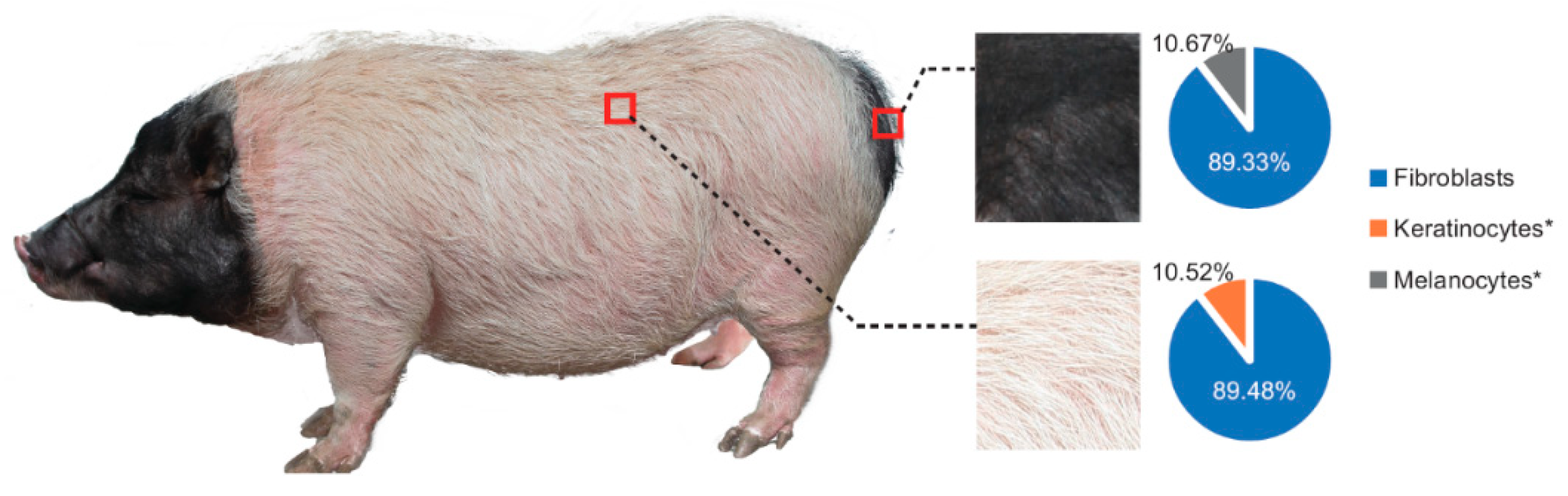
2.6. Profiling Melanocyte Proportion with CIBERSORT
2.7. Construction of lncRNA-mRNA Interaction Network
2.8. Functional Enrichment Analysis
2.9. Validation of Genes and lncRNAs by Real-Time PCR
3. Results
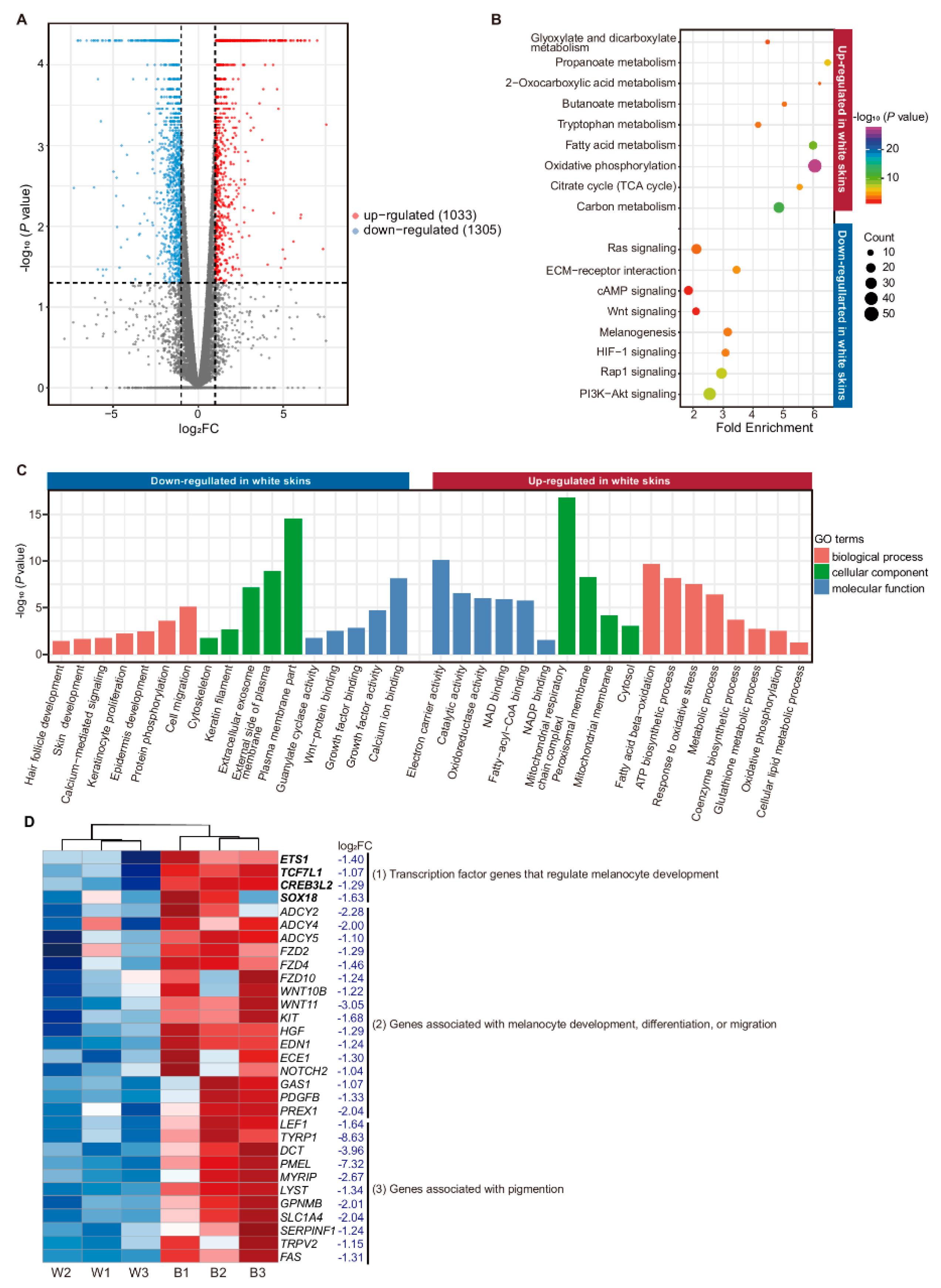
3.1. Expression Profiles of mRNAs and lncRNAs
3.2. Functional Enrichment Analysis of DE mRNA
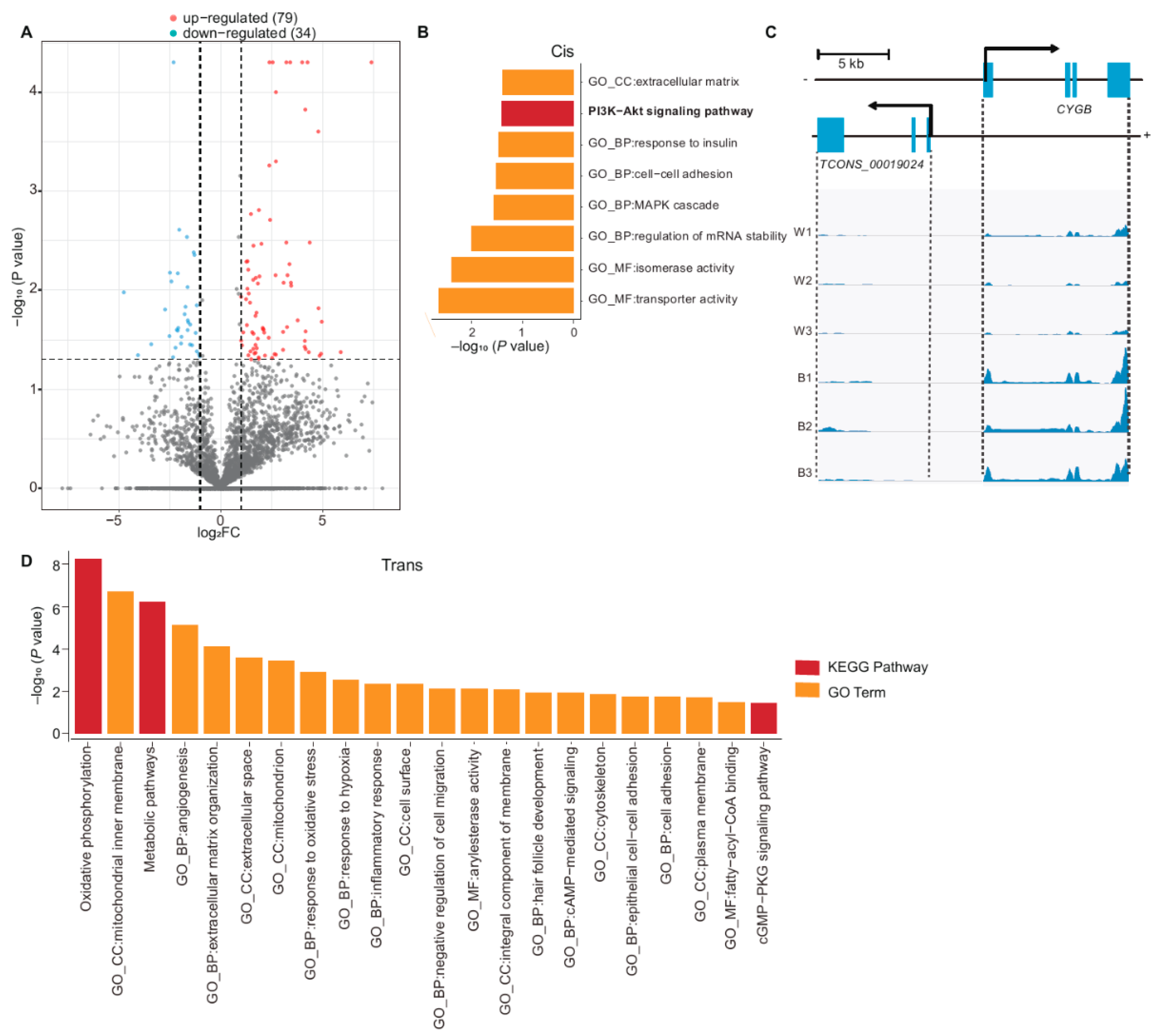
3.3. Functional Enrichment Analysis of DE lncRNAs
3.4. Quantitative Real-Time PCR Validation
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Founding
Conflicts of Interest
References
- Kanitakis, J. Anatomy, histology and immunohistochemistry of normal human skin. Eur. J. Dermatol. 2002, 12, 390–399. [Google Scholar] [PubMed]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tyminska, A. Skin melanocytes: Biology and development. Adv. Dermatol. Allergol. 2013, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.G.; Nair, N.; Begum, G.; Joshi, N.B.; Sinkar, V.P.; Vora, S. Melanocyte-keratinocyte interaction induces calcium signalling and melanin transfer to keratinocytes. Pigment Cell Res. 2007, 20, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.D.; Simon, J.D. Quantification of Ca2+ binding to melanin supports the hypothesis that melanosomes serve a functional role in regulating calcium homeostasis. Pigment Cell Res. 2007, 20, 134–139. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, J.Y.; Shang, H.T.; Liu, C.E.; Wang, Y.; Niu, R.; Wu, J.; Wei, H. Light microscopic, electron microscopic, and immunohistochemical comparison of bama minipig (sus scrofa domestica) and human skin. Comp. Med. 2010, 60, 142–148. [Google Scholar]
- Liu, B.; Liu, Y.; Wang, L.; Hou, C.; An, M. Rna-seq-based analysis of the hypertrophic scarring with and without pressure therapy in a bama minipig model. Sci. Rep. 2018, 8, 11831. [Google Scholar] [CrossRef]
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef]
- Tachibana, M.; Kobayashi, Y.; Matsushima, Y. Mouse models for four types of waardenburg syndrome. Pigment Cell Res. 2003, 16, 448–454. [Google Scholar] [CrossRef]
- Lerner, A.B.; Shiohara, T.; Boissy, R.E.; Jacobson, K.A.; Lamoreux, M.L.; Moellmann, G.E. A mouse model for vitiligo. J. Investig. Dermatol. 1986, 87, 299–304. [Google Scholar] [CrossRef]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding rnas with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The landscape of long noncoding rna classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Xie, J.; Bai, J.; Wang, H.; Tian, X.; Bai, R.; Jia, X.; Yang, L.; Song, Y.; Herrid, M.; et al. Skin transcriptome profiles associated with coat color in sheep. BMC Genom. 2013, 14, 389. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling tumor infiltrating immune cells with cibersort. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [PubMed]
- Ghosh, S.; Chan, C.K. Analysis of rna-seq data using tophat and cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. Hmmer web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. Cpc: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hedan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. Feelnc: A tool for long non-coding rna annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from rna-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Reemann, P.; Reimann, E.; Ilmjärv, S.; Porosaar, O.; Silm, H.; Jaks, V.; Vasar, E.; Kingo, K.; Kõks, S. Melanocytes in the skin--comparative whole transcriptome analysis of main skin cell types. PLoS ONE 2014, 9, e115717. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Wgcna: An r package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Hu, S.; Tu, T.; Huang, Z.; Tang, Q.; Ma, J.; Wang, X.; Li, X.; Zhou, X.; Shuai, S.; et al. Global long noncoding rna and mrna expression changes between prenatal and neonatal lung tissue in pigs. Genes 2018, 9, 443. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncrnas regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tsoi, L.C.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Johnston, A.; Ding, J.; Stuart, P.E.; Xing, X.; Kochkodan, J.J.; et al. Transcriptome analysis of psoriasis in a large case-control sample: Rna-seq provides insights into disease mechanisms. J. Investig. Dermatol. 2014, 134, 1828–1838. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Iyer, M.K.; Stuart, P.E.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Sarkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding rnas highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 16, 24. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.J.; Erickson, C.A.; Takada, S.; Burrus, L.W. Wnt and bmp signaling govern lineage segregation of melanocytes in the avian embryo. Dev. Biol. 2001, 233, 22–37. [Google Scholar] [CrossRef]
- Rabbani, P.; Takeo, M.; Chou, W.; Myung, P.; Bosenberg, M.; Chin, L.; Taketo, M.M.; Ito, M. Coordinated activation of wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell 2011, 145, 941–955. [Google Scholar] [CrossRef]
- Larribere, L.; Khaled, M.; Tartare-Deckert, S.; Busca, R.; Luciano, F.; Bille, K.; Valony, G.; Eychene, A.; Auberger, P.; Ortonne, J.P.; et al. Pi3k mediates protection against trail-induced apoptosis in primary human melanocytes. Cell Death Differ. 2004, 11, 1084–1091. [Google Scholar] [CrossRef]
- Rodriguez, C.I.; Setaluri, V. Cyclic amp (camp) signaling in melanocytes and melanoma. Arch. Biochem. Biophys. 2014, 563, 22–27. [Google Scholar] [CrossRef]
- D’Orazio, J.; Fisher, D.E. Central role for camp signaling in pigmentation and uv resistance. Cell Cycle 2011, 10, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.; Singh, A.; Iqbal, Z.; Kaushik, J.K.; Rao, A.R.; Ahmad, S.M.; Bhat, H.; Ayaz, A.; Sheikh, F.D.; Kalra, S.; et al. Comparative transcriptome analysis reveals the genetic basis of coat color variation in pashmina goat. Sci. Rep. 2019, 9, 6361. [Google Scholar] [CrossRef] [PubMed]
- Sheinboim, D.; Maza, I.; Dror, I.; Parikh, S.; Krupalnik, V.; Bell, R.E.; Zviran, A.; Suita, Y.; Hakim, O.; Mandel-Gutfreund, Y.; et al. Oct4 impedes cell fate redirection by the melanocyte lineage master regulator mitf in mouse escs. Nat. Commun. 2017, 8, 1022. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.; Kedlaya, R.; Maddodi, N.; Bhat, K.M.; Weber, C.S.; Valdivia, H.; Setaluri, V. Calcium homeostasis in human melanocytes: Role of transient receptor potential melastatin 1 (trpm1) and its regulation by ultraviolet light. Am. J. Physiol.-Cell Physiol. 2009, 297, C679–C687. [Google Scholar] [CrossRef] [PubMed]
- Toshihiko, H.; Hidenori, W.; Jacqueline, M.; Yuji, Y.; Vieira, W.D.; Hearing, V.J. Mart-1 is required for the function of the melanosomal matrix protein pmel17/gp100 and the maturation of melanosomes. J. Biol. Chem. 2005, 280, 14006–14016. [Google Scholar]
- Hellstrom, A.R.; Watt, B.; Fard, S.S.; Tenza, D.; Mannstrom, P.; Narfstrom, K.; Ekesten, B.; Ito, S.; Wakamatsu, K.; Larsson, J.; et al. Inactivation of pmel alters melanosome shape but has only a subtle effect on visible pigmentation. PLoS Genet. 2011, 7, e1002285. [Google Scholar] [CrossRef] [PubMed]
- Seberg, H.E.; Van Otterloo, E.; Loftus, S.K.; Liu, H.; Bonde, G.; Sompallae, R.; Gildea, D.E.; Santana, J.F.; Manak, J.R.; Pavan, W.J.; et al. Tfap2 paralogs regulate melanocyte differentiation in parallel with mitf. PLoS Genet. 2017, 13, e1006636. [Google Scholar] [CrossRef]
- Saldana-Caboverde, A.; Perera, E.M.; Watkins-Chow, D.E.; Hansen, N.F.; Vemulapalli, M.; Mullikin, J.C.; Program, N.C.S.; Pavan, W.J.; Kos, L. The transcription factors ets1 and sox10 interact during murine melanocyte development. Dev. Biol. 2015, 407, 300–312. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. A J. Tech. Methods Pathol. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Schouwey, K.; Delmas, V.; Larue, L.; Zimber-Strobl, U.; Strobl, L.J.; Radtke, F.; Beermann, F. Notch1 and notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev. Dyn. 2007, 236, 282–289. [Google Scholar] [CrossRef]
- Lindsay, C.R.; Lawn, S.; Campbell, A.D.; Faller, W.J.; Rambow, F.; Mort, R.L.; Timpson, P.; Li, A.; Cammareri, P.; Ridgway, R.A.; et al. P-rex1 is required for efficient melanoblast migration and melanoma metastasis. Nat. Commun. 2011, 2, 555. [Google Scholar] [CrossRef] [PubMed]
- Park, P.J.; Lee, T.R.; Cho, E.G. Substance p stimulates endothelin 1 secretion via endothelin-converting enzyme 1 and promotes melanogenesis in human melanocytes. J. Investig. Dermatol. 2015, 135, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, T.S.; Mitsunori, F. Functional analysis of slac2-c/myrip as a linker protein between melanosomes and myosin viia. J. Biol. Chem. 2005, 280, 28015–28022. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Jung, S.H. Recent development of signaling pathways inhibitors of melanogenesis. Cell. Signal. 2017, 40, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Takeo, M.; Lee, W.; Rabbani, P.; Sun, Q.; Hu, H.; Lim, C.H.; Manga, P.; Ito, M. Ednrb governs regenerative response of melanocyte stem cells by crosstalk with wnt signaling. Cell Rep. 2016, 15, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Mao, H.; Zhang, Z.; Xiao, S.; Ding, N.; Huang, L. A 6-bp deletion in the tyrp1 gene causes the brown colouration phenotype in chinese indigenous pigs. Heredity 2011, 106, 862–868. [Google Scholar] [CrossRef]
- Pielberg, G.; Olsson, C.; Syvanen, A.C.; Andersson, L. Unexpectedly high allelic diversity at the kit locus causing dominant white color in the domestic pig. Genetics 2002, 160, 305–311. [Google Scholar]
- Southard-Smith, E.M.; Kos, L.; Pavan, W.J. Sox10 mutation disrupts neural crest development in dom hirschsprung mouse model. Nat. Genet. 1998, 18, 60–64. [Google Scholar] [CrossRef]
- Liao, C.P.; Booker, R.C.; Morrison, S.J.; Le, L.Q. Identification of hair shaft progenitors that create a niche for hair pigmentation. Genes Dev. 2017, 31, 744–756. [Google Scholar] [CrossRef]
- Andersson, L.S.; Wilbe, M.; Viluma, A.; Cothran, G.; Ekesten, B.; Ewart, S.; Lindgren, G. Equine multiple congenital ocular anomalies and silver coat colour result from the pleiotropic effects of mutant pmel. PLoS ONE 2013, 8, e75639. [Google Scholar] [CrossRef]
- Irvine, A.D.; McKenna, K.E.; Jenkinson, H.; Hughes, A.E. A mutation in the v1 domain of keratin 5 causes epidermolysis bullosa simplex with mottled pigmentation. J. Investig. Dermatol. 1997, 108, 809–810. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geller, L.; Kristal, L.; Morel, K.D. Epidermolysis bullosa simplex with mottled pigmentation due to a rare keratin 5 mutation: Cutaneous findings in infancy. Pediatr. Dermatol. 2013, 30, 631–632. [Google Scholar] [CrossRef] [PubMed]
- Lo Cicero, A.; Delevoye, C.; Gilles-Marsens, F.; Loew, D.; Dingli, F.; Guere, C.; Andre, N.; Vie, K.; van Niel, G.; Raposo, G. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat. Commun. 2015, 6, 7506. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Galan, A.K.; Xin, X.; Dong, F.; Abdul-Ghani, M.A.; Zhou, L.; Wang, C.; Li, C.; Holmes, B.M.; Sloane, L.B.; et al. Appl1 potentiates insulin sensitivity by facilitating the binding of irs1/2 to the insulin receptor. Cell Rep. 2014, 7, 1227–1238. [Google Scholar] [CrossRef]
- De Launoit, Y.; Adamski, J. Unique multifunctional hsd17b4 gene product: 17beta-hydroxysteroid dehydrogenase 4 and d-3-hydroxyacyl-coenzyme a dehydrogenase/hydratase involved in zellweger syndrome. J. Mol. Endocrinol. 1999, 22, 227–240. [Google Scholar] [CrossRef]
- Fujita, Y.; Koinuma, S.; De Velasco, M.A.; Bolz, J.; Togashi, Y.; Terashima, M.; Hayashi, H.; Matsuo, T.; Nishio, K. Melanoma transition is frequently accompanied by a loss of cytoglobin expression in melanocytes: A novel expression site of cytoglobin. PLoS ONE 2014, 9, e94772. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Lee, Y.; Hwang, K. Skin thickness of korean adults. Surg. Radiol. Anat. 2002, 24, 183–189. [Google Scholar]
- Wilhelm, K.P.; Cua, A.B.; Maibach, H.I. Skin aging. Effect on transepidermal water loss, stratum corneum hydration, skin surface ph, and casual sebum content. Arch. Dermatol. 1991, 127, 1806–1809. [Google Scholar] [CrossRef]
- Ghahary, A.; Ghaffari, A. Role of keratinocyte-fibroblast cross-talk in development of hypertrophic scar. Wound Repair Regen. 2007, 15 (Suppl. 1), S46–S53. [Google Scholar] [CrossRef]
- Rennekampff, H.O.; Busche, M.N.; Knobloch, K.; Tenenhaus, M. Is uv radiation beneficial in postburn wound healing? Med. Hypotheses 2010, 75, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.L.; Jin, R.; Zhang, L.; Zhang, Y.G. The contribution of melanocytes to pathological scar formation during wound healing. Int. J. Clin. Exp. Med. 2013, 6, 609–613. [Google Scholar] [PubMed]
- Gallant-Behm, C.L.; Tsao, H.; Reno, C.; Olson, M.E.; Hart, D.A. Skin wound healing in the first generation (f1) offspring of yorkshire and red duroc pigs: Evidence for genetic inheritance of wound phenotype. Burn. J. Int. Soc. Burn Inj. 2006, 32, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Johansson Moller, M.; Chaudhary, R.; Hellmen, E.; Hoyheim, B.; Chowdhary, B.; Andersson, L. Pigs with the dominant white coat color phenotype carry a duplication of the kit gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 1996, 7, 822–830. [Google Scholar] [CrossRef]
- Gallant-Behm, C.L.; Hart, D.A. Genetic analysis of skin wound healing and scarring in a porcine model. Wound Repair Regen. 2006, 14, 46–54. [Google Scholar] [CrossRef]
- Ren, H.; Wang, G.; Chen, L.; Jiang, J.; Liu, L.; Li, N.; Zhao, J.; Sun, X.; Zhou, P. Genome-wide analysis of long non-coding rnas at early stage of skin pigmentation in goats (capra hircus). BMC Genom. 2016, 17, 67. [Google Scholar] [CrossRef]
- Ji, K.; Fan, R.; Zhang, J.; Yang, S.; Dong, C. Long non-coding rna expression profile in cdk5-knockdown mouse skin. Gene 2018, 672, 195–201. [Google Scholar] [CrossRef]
- Li, C.X.; Li, H.G.; Huang, L.T.; Kong, Y.W.; Chen, F.Y.; Liang, J.Y.; Yu, H.; Yao, Z.R. H19 lncrna regulates keratinocyte differentiation by targeting mir-130b-3p. Cell Death Dis. 2017, 8, e3174. [Google Scholar] [CrossRef]

| Gene Name | Up/Down-Regulated in White | FC | p-Value | Functions |
|---|---|---|---|---|
| TYR | Down | Un-expressed in white | 5.0 × 10−5 | Melanin synthesis [2] |
| TYRP1 | Down | B/W = 388:1 | 3.3 × 10−2 | Melanosomal protein [2] |
| TRPM1 | Down | B/W = 235:1 | 2.4 × 10−1 | Regulate tyrosinase activity [34] |
| MLANA | Down | B/W = 164:1 | 1.5 × 10−1 | Regulating PMEL processing [35] |
| PMEL (sliver) | Down | B/W = 157:1 | 3.5 × 10−3 | Melanosome complex [36] |
| DCT | Down | B/W = 15:1 | 5.0 × 10−5 | Melanosomal protein [2] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Zhao, L.; Hu, S.; Long, K.; Liu, P.; Liu, R.; Zhou, X.; Wang, Y.; Huang, Z.; Lin, X.; et al. Transcriptional Differences of Coding and Non-Coding Genes Related to the Absence of Melanocyte in Skins of Bama Pig. Genes 2020, 11, 47. https://doi.org/10.3390/genes11010047
Jin L, Zhao L, Hu S, Long K, Liu P, Liu R, Zhou X, Wang Y, Huang Z, Lin X, et al. Transcriptional Differences of Coding and Non-Coding Genes Related to the Absence of Melanocyte in Skins of Bama Pig. Genes. 2020; 11(1):47. https://doi.org/10.3390/genes11010047
Chicago/Turabian StyleJin, Long, Lirui Zhao, Silu Hu, Keren Long, Pengliang Liu, Rui Liu, Xuan Zhou, Yixin Wang, Zhiqing Huang, Xuxu Lin, and et al. 2020. "Transcriptional Differences of Coding and Non-Coding Genes Related to the Absence of Melanocyte in Skins of Bama Pig" Genes 11, no. 1: 47. https://doi.org/10.3390/genes11010047
APA StyleJin, L., Zhao, L., Hu, S., Long, K., Liu, P., Liu, R., Zhou, X., Wang, Y., Huang, Z., Lin, X., Tang, Q., & Li, M. (2020). Transcriptional Differences of Coding and Non-Coding Genes Related to the Absence of Melanocyte in Skins of Bama Pig. Genes, 11(1), 47. https://doi.org/10.3390/genes11010047





