Transcriptomic Analysis Suggests Genes Expressed Stage-Independently and Stage–Dependently Modulating the Wing Dimorphism of the Brown Planthopper
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
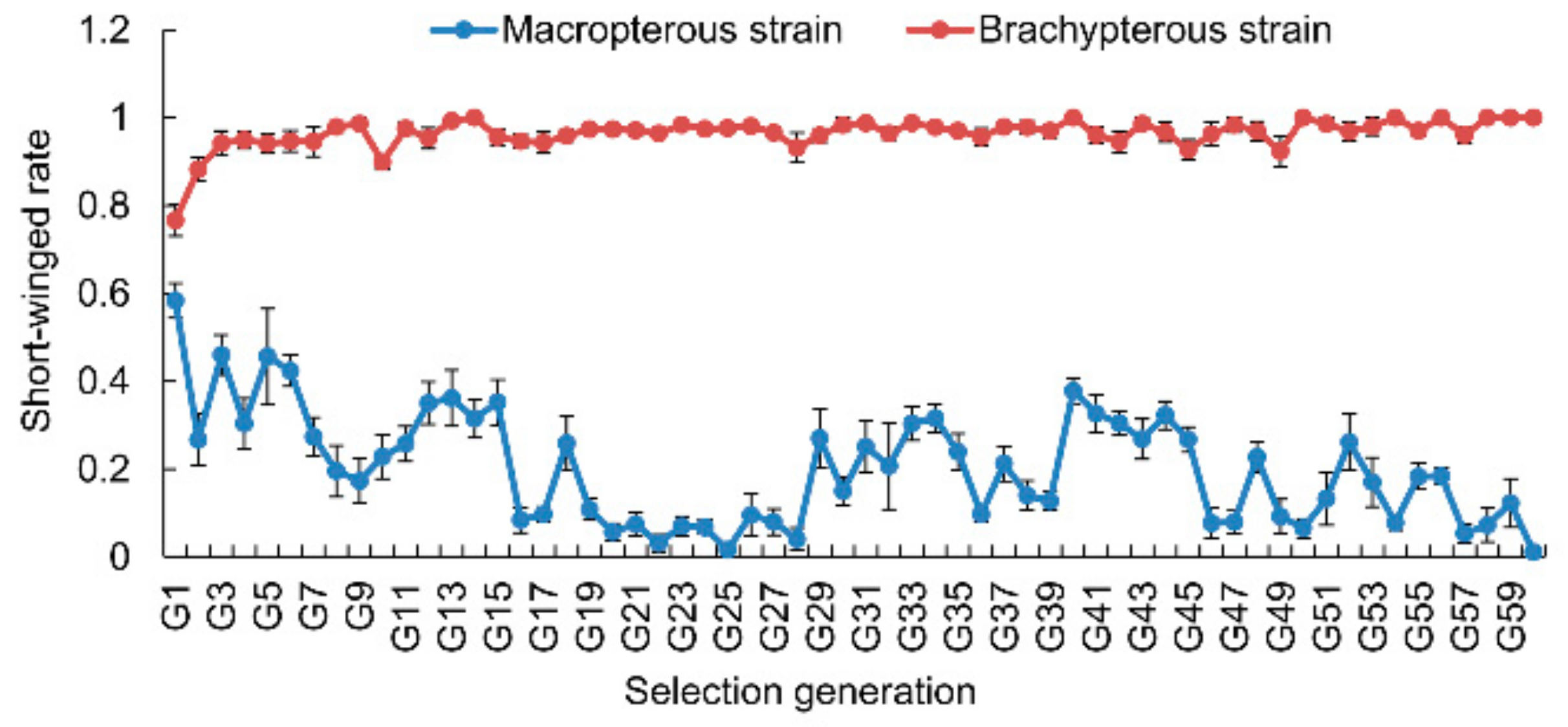
2.2. Selection of Macropterous and Brachypterous Pure-Bred Strains
2.3. RNA Extraction and cDNA Library Construction for Transcriptome Sequencing
2.4. cDNA Library Construction and Sequencing
2.5. De novo Assembly and Annotation
2.6. Analysis of Differential Gene Expression Profiles
2.7. Expression Levels of Genes Related to Wing Development in Macropterous and Brachypterous Strains
3. Results
3.1. Selection Response of Wing Morphs
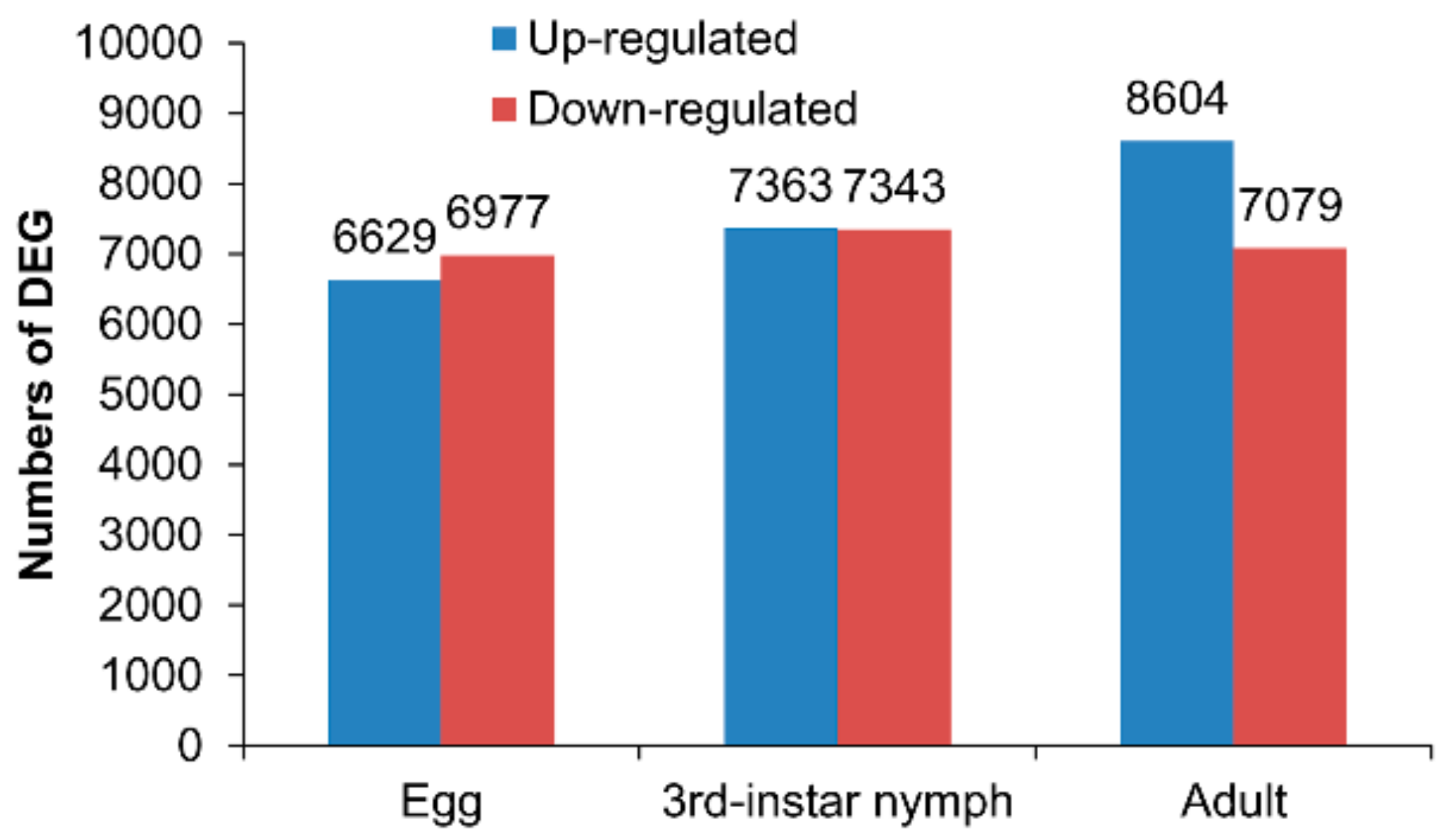
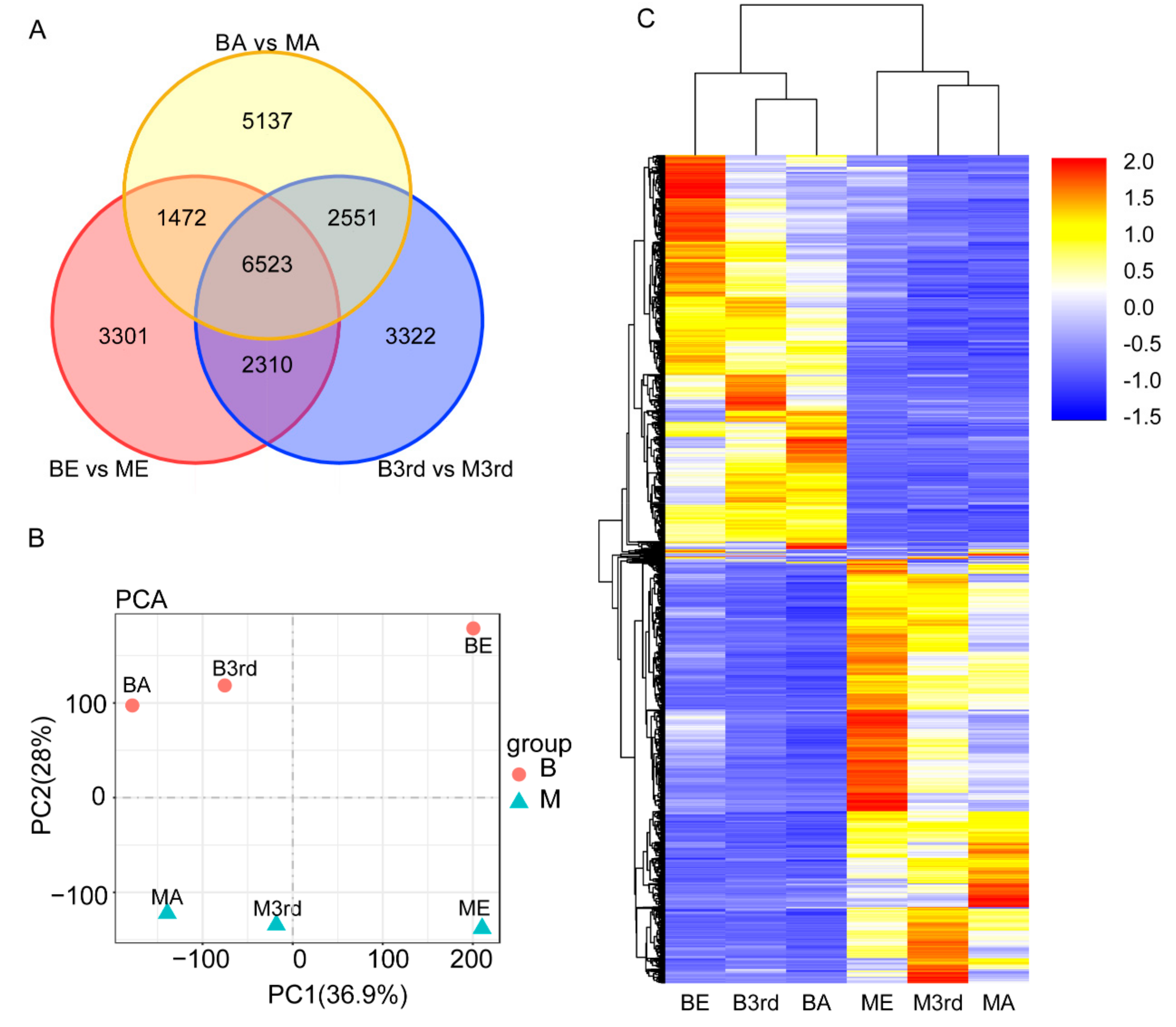
3.2. Differentially Expressed Genes between the Macropterous and Brachypterous Strains
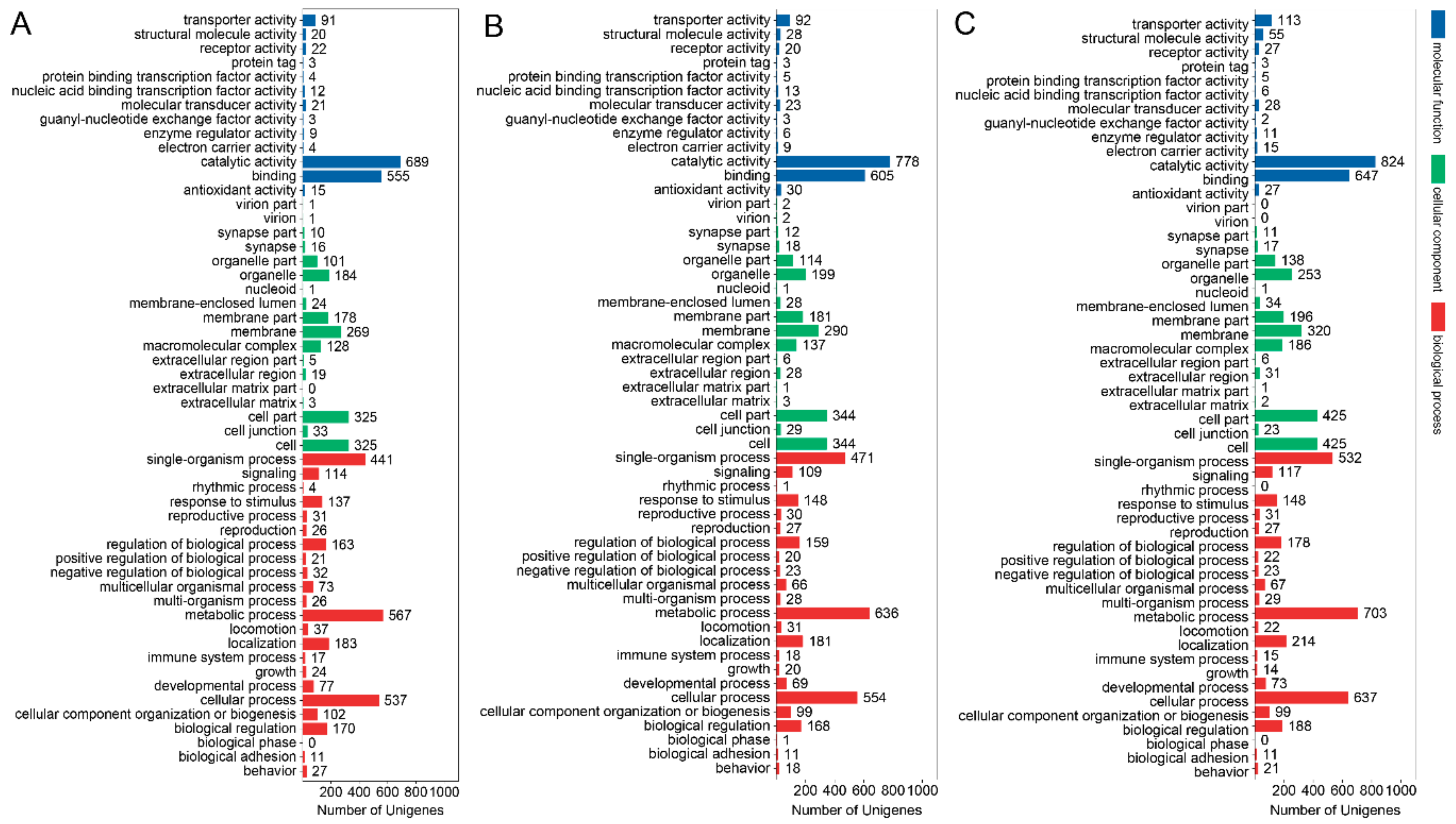
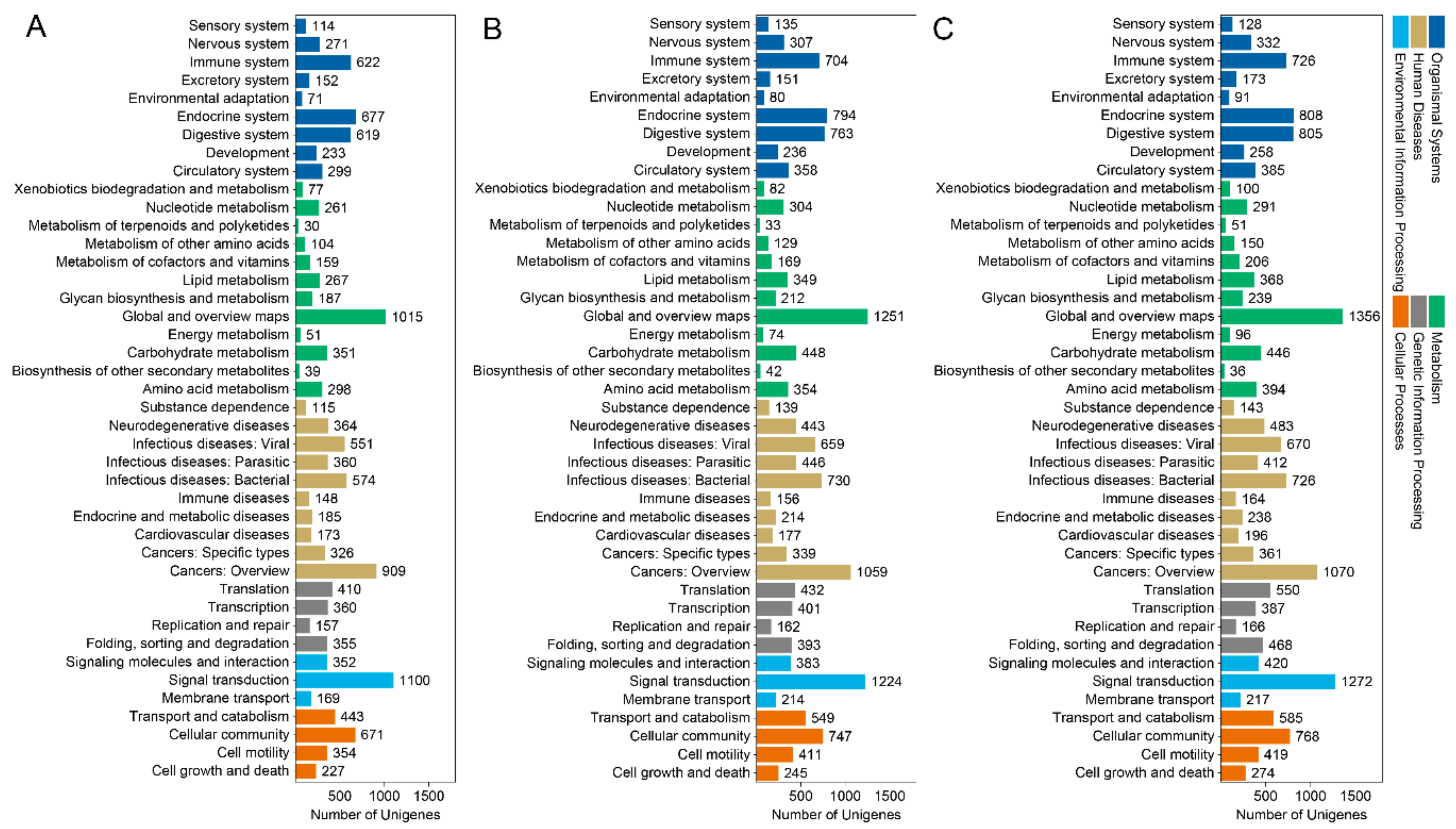
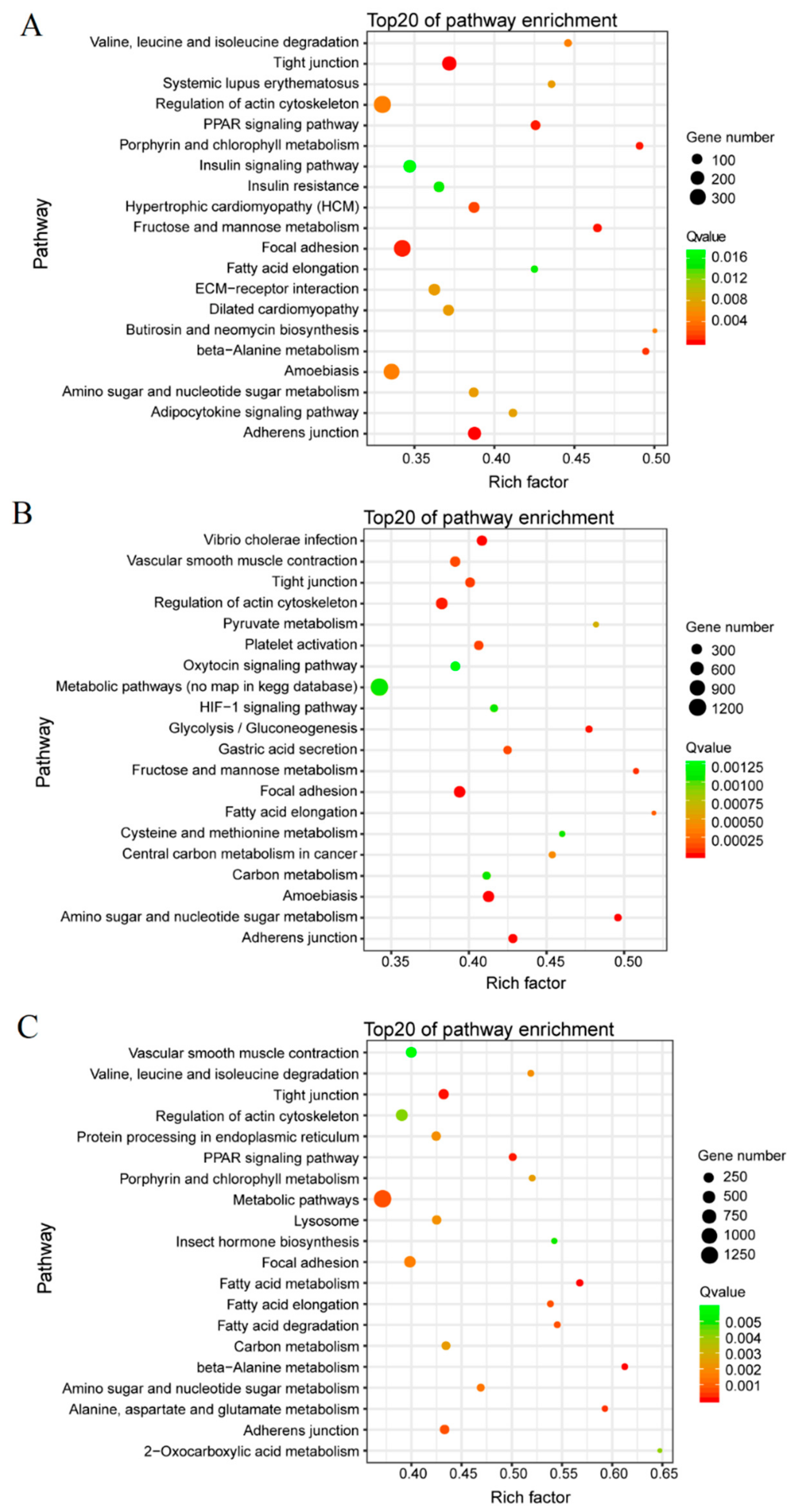
3.3. GO Term and KEGG Classification for DEGs between the Macropterous and Brachypterous Strains
3.4. Molecular Differentiation of Macropterous and Brachypterous Strains
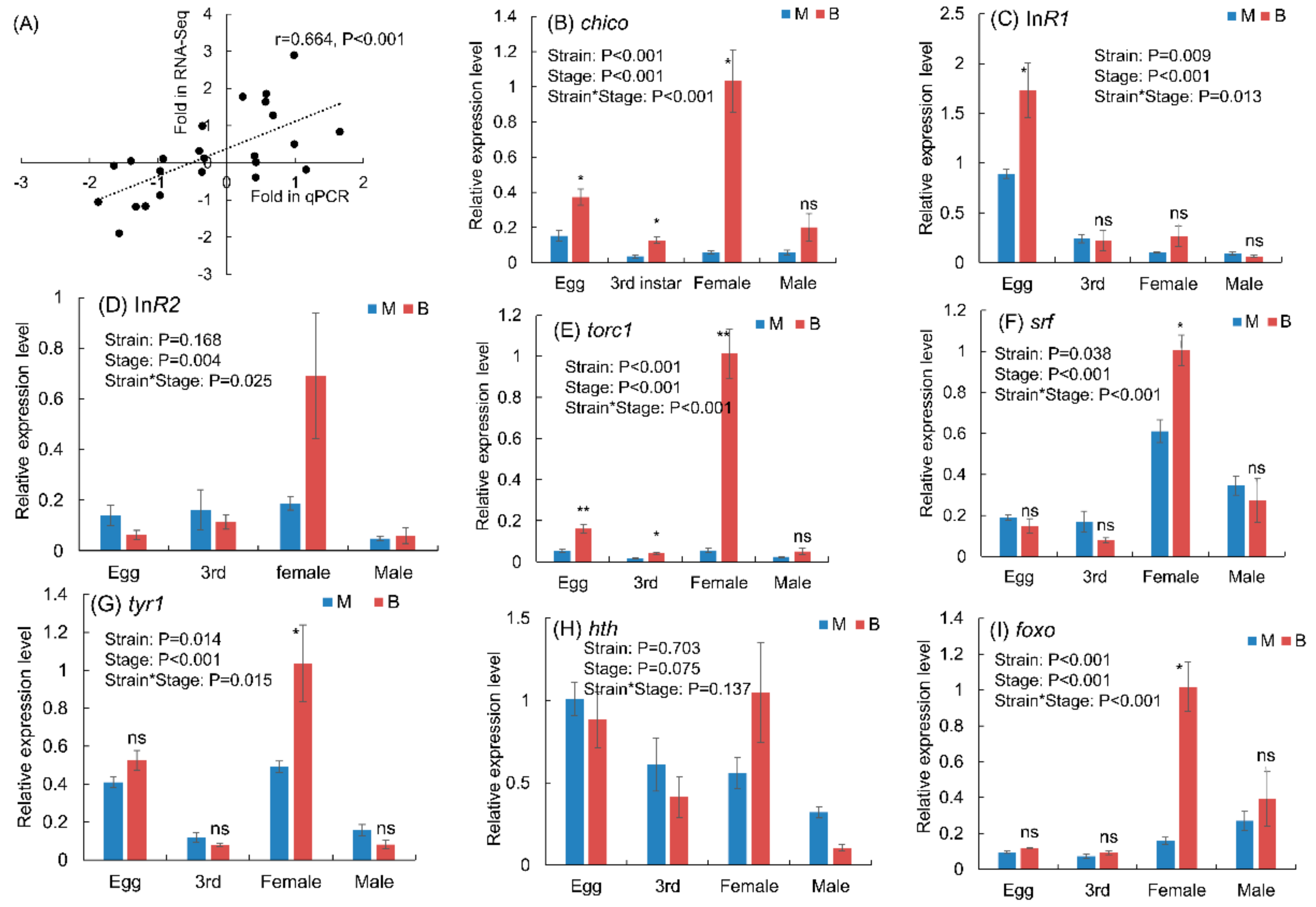
3.5. Expression Levels of Genes in Macropterous and Brachypterous Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simpson, S.J.; Sword, G.A.; Lo, N. Polyphenism in insect. Curr. Biol. 2011, 21, R738–R749. [Google Scholar] [CrossRef] [PubMed]
- Braendle, C.; Friebe, I.; Caillaud, M.C.; Stern, D.L. Genetic variation for an aphid wing polyphenism is genetically linked to a naturally occurring wing polymorphism. Proc. R. Soc. B 2005, 272, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, X.; Zhang, Y.; Coates, B.; Zhou, X.; Cheng, D. Bacterial symbionts, Buchnera, and starvation on wing dimorphism in English grain aphid, Sitobion avenae (F.) (Homoptera: Aphididae). Front. Physiol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, K.; Tojo, S.; Nagata, T. Immigration of the brown planthopper, Nilaparvata lugens, exhibiting various responses to density in relation to wing morphism. Entomol. Exp. Appl. 1985, 38, 101–108. [Google Scholar] [CrossRef]
- Peng, J.; Zhang, C.; An, Z.F.; Yu, J.L.; Liu, X.D. Genetic analysis of wing-form determination in three species of rice planthoppers (Hemiptera: Delphacidae). Acta Entomol. Sin. 2012, 55, 971–980. [Google Scholar]
- Zera, A.J.; Innes, D.J.; Saks, M.E. Genetic and environmental determinants of wing polymorphism in the waterstrider Limnoporus canaliculatus. Evolution 1983, 37, 513–522. [Google Scholar] [CrossRef][Green Version]
- Xu, H.J.; Xue, J.; Lu, B.; Zhang, X.C.; Zhuo, J.C.; He, S.F.; Ma, X.F.; Jiang, Y.Q.; Fan, H.W.; Xu, J.Y.; et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 2015, 519, 464–467. [Google Scholar] [CrossRef]
- Vellichirammal, N.N.; Madayiputhiya, N.; Brisson, J.A. The genomewide transcriptional response underlying the pea aphid wing polyphenism. Mol. Ecol. 2016, 25, 4146–4160. [Google Scholar] [CrossRef]
- Aukema, B. Wing-length determination in two wing-dimorphic Calathus species (Coleoptera: Carabidae). Hereditas 1990, 113, 189–202. [Google Scholar] [CrossRef]
- Ghanim, M.; Dombrovsky, A.; Raccah, B.; Sherman, A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (S.). Insect Biochem. Mol. Biol. 2006, 36, 857–868. [Google Scholar] [CrossRef]
- Grantham, M.E.; Brisson, J.A. Extensive differential splicing underlies phenotypically plastic aphid morphs. Mol. Biol. Evol. 2018, 35, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Denno, R.F.; Roderick, G.K. Population biology of planthoppers. Ann. Rev. Entomol. 1990, 35, 489–520. [Google Scholar] [CrossRef]
- Padgham, D. The influence of host-plant on the development of the adult brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae), and its significance in migration. B. Entomol. Res. 1983, 73, 117–128. [Google Scholar] [CrossRef]
- An, Z.F.; Yu, J.L.; Peng, J.; Zhang, C.; Liu, X.D. Differentiation of wing forms in pure macropterous and brachypterous lineages is less subject to photoperiod in rice planthoppers (Hemiptera: Delphacidae). Acta Entomol. Sin. 2014, 57, 1306–1314. [Google Scholar]
- Mori, K.; Nakasuji, F. Effects of day length and density on development and wing form of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Appl. Entomol. Zool. 1991, 26, 557–561. [Google Scholar] [CrossRef]
- Matsumura, M.; Suzuki, Y. Direct and feeding-induced interactions between two rice planthoppers, Sogatella furcifera and Nilaparvata lugens: Effects on dispersal capability and performance. Ecol. Entomol. 2003, 28, 174–182. [Google Scholar] [CrossRef]
- Olvido, A.E.; Elvington, E.S.; Mousseau, T.A. Relative effects of climate and crowding on wing polymorphism in the southern ground cricket, Allonemobius socius (Orthoptera: Gryllidae). Fla. Entomol. 2003, 86, 158–164. [Google Scholar] [CrossRef]
- Nakao, S.; Chikamori, C. Temperature-dependent wing dimorphism in a Japanese strain of tobacco thrips, Frankliniella fusca (Thysanoptera: Thripidae). Appl. Entomol. Zool. 2013, 48, 337–343. [Google Scholar] [CrossRef]
- Lin, X.; Xu, Y.; Jiang, J.; Lavine, M.; Lavine, L.C. Host quality induces phenotypic plasticity in a wing polyphonic insect. Proc. Natl. Acad. Sci. USA 2018, 115, 7563–7568. [Google Scholar] [CrossRef]
- Rios Martinez, A.F.; Costamagna, A.C. Effects of crowding and host plant quality on morph determination in the soybean aphid, Aphis glycines. Entomol. Exp. Appl. 2018, 166, 53–62. [Google Scholar] [CrossRef]
- Bertuso, A.G.; Morooka, S.; Tojo, S. Sensitive periods for wing development and precocious metamorphosis after precocene treatment of the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 2002, 48, 221–229. [Google Scholar] [CrossRef]
- Yamada, S. Dual wing-form determination mechanism in the brown planthopper, Nilaparvata lugens Stål (Homoptera: Delphacidae). J. Soc. Appl. Entomol. Zool. 1991, 26, 590–592. [Google Scholar] [CrossRef]
- Brisson, J.A.; Stern, D.L. The pea aphid, Acyrthosiphon pisum: An emerging genomic model system for ecological, developmental and evolutionary studies. BioEssays 2006, 28, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, K.; Tojo, S. Effects of juvenile hormone and rearing density on wing dimorphism and oocyte development in the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 1986, 32, 585–590. [Google Scholar] [CrossRef]
- Chen, Y.; Verheggen, F.J.; Sun, D.; Wang, Z.; Francis, F.; He, K.L. Differential wing polyphenism adaptation across life stages under extreme high temperatures in corn leaf aphid. Sci. Rep. 2019, 9, 8744. [Google Scholar] [CrossRef]
- Muller, C.; Williams, I.S.; Hardie, J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol. Entomol. 2001, 26, 330–340. [Google Scholar] [CrossRef]
- Brisson, J.A.; Davis, G.K.; Stern, D.L. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum). Evol. Develop. 2007, 9, 338–346. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Xu, X.; Li, Z.; Li, Y.; Song, D.; Yu, T.; Zhu, F.; Zhang, Q.; Zhou, X. Gene expression profiling in winged and wingless cotton aphids, Aphis gossypii (Hemiptera: Aphididae). Int. J. Biol. Sci. 2014, 10, 257–267. [Google Scholar] [CrossRef]
- Hu, D.B.; Luo, B.Q.; Li, J.; Han, Y.; Jiang, T.R.; Liu, J.; Wu, G.; Hua, H.X.; Xiong, Y.F.; Li, J.S. Genome-wide analysis of Nilaparvata lugens nymphal responses to high-density and low-quality rice hosts. Insect Sci. 2013, 20, 703–716. [Google Scholar] [CrossRef]
- Brisson, J.A.; Ishikawa, A.; Miura, T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol. Biol. 2010, 19, 63–73. [Google Scholar] [CrossRef]
- Veale, A.J.; Foster, B.J.; Dearden, P.K.; Waters, J.M. Genotyping-by-sequencing supports a genetic basis for wing reduction in an alpine New Zealand stonefly. Sci. Rep. 2018, 8, 16275. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, G.A.; Oliphant, A.; Dearden, P.K.; Veale, A.J.; Ellen, C.W.; Waters, J.M. Comparative transcriptomic analysis of a wing-dimorphic stonefly reveals candidate wing loss genes. EvoDevo 2019, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Bao, Y.Y.; Li, B.L.; Cheng, Y.B.; Peng, Z.Y.; Liu, H.; Xu, H.J.; Zhu, Z.R.; Lou, Y.G.; Cheng, J.A.; et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 2010, 5, e14233. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.J.; Zhang, C.X. Insulin receptors and wing dimorphism in rice planthoppers. Phil. Trans. R. Soc. B 2017, 372, 20150489. [Google Scholar] [CrossRef] [PubMed]
- Roff, D.A. The genetic basis of wing dimorphism in the sand cricket, Gryllus firmus and its relevance to the evolution of wing dimorphisms in insects. Heredity 1986, 57, 221–231. [Google Scholar] [CrossRef]
- Zera, A.J.; Deo, R.F. Physiology and ecology of dispersal polymorphism in insects. Ann. Rev. Entomol. 1997, 42, 207–230. [Google Scholar] [CrossRef] [PubMed]
- Gatehouse, A.G. Behavior and ecological genetics of wind-borne migration by insects. Ann. Rev. Entomol. 1997, 42, 475–502. [Google Scholar] [CrossRef]
- Morooka, S.; Tojo, S. Maintenance and section of strains exhibiting specific wing form and body colour under high density conditions in the brown planthopper. Nilaparvata lugens (Homoptera: Delphacidae). Appl. Entomol. Zool. 1992, 27, 445–454. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–665. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Heidelberg, Germany, 6–10 August 1999; pp. 138–148. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Yao, Y.; Jin, M.; Li, Q. Characterization of the Distal-less gene homologue, NlDll, in the brown planthopper, Nilaparvata lugens (Stål). Gene 2014, 535, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Nakasuji, F. Genetic analysis of the wing-form determination of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Res. Popul. Ecol. 1990, 32, 279–289. [Google Scholar] [CrossRef]
- Vellichirammal, N.N.; Zera, A.J.; Schilder, R.J.; Wehrkamp, C.; Riethoven, J.J.M.; Brisson, J.A. De novo transcriptome assembly from fat body and flight muscles transcripts to identify morph-specific gene expression profiles in Gryllus firmus. PLoS ONE 2014, 9, e82129. [Google Scholar] [CrossRef]
- Matsumura, M. Genetic analysis of a threshold trait: Density-dependent wing dimorphism in Sogatella furcifera (Horváth) (Hemiptera: Delphacidae), the whitebacked planthopper. Heredity 1996, 76, 229–237. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Asadi, N.B.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Liu, L.J.; Zheng, H.Y.; Jiang, F.; Guo, W.; Zhou, S.T. Comparative transcriptional analysis of asexual and sexual morphs reveals possible mechanisms in reproductive polyphenism of the cotton aphid. PLoS ONE 2014, 9, e99506. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Xia, W.; Li, J.; Sheng, S.; Lei, L.; Zhao, S. Transcriptome profiling and digital gene expression analysis of Fallopia multiflora to discover putative genes involved in the biosynthesis of 2,3,5,4’-tetrahydroxy stilbene-2-O-β-D-glucoside. Gene 2014, 547, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Engqvist-Goldstein, A.E.; Drubin, D.G. Actin assembly and endocytosis: From yeast to mammals. Ann. Rev. Cell Dev. Biol. 2003, 19, 287–332. [Google Scholar] [CrossRef]
- Arasada, R.; Schleicher, M. Actin Cytoskeleton. In Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Mahmud, F.S. Alary polymorphism in the small brown planthopper Laodelphax striatellus (Homoptera: Delphacidae). Entomol. Exp. Appl. 1980, 28, 47–53. [Google Scholar] [CrossRef]
- Du, J.; Wang, Q.; Cheng, X. Histological study on the wing dimorphism of the brown planthopper Nilaparvata lugens (Stål). Acta Entomol. Sin. 1997, 40 (Suppl.), 135–138. [Google Scholar]
- Bohni, R.; Riesgo-Escovar, J.; Oldham, S.; Brogiolo, W.; Stocker, H.; Andruss, B.F.; Beckingham, K.; Hafen, E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 1999, 97, 865–875. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Thompson, B.J. Mal/SRF is dispensable for cell proliferation in Drosophila. PLoS ONE 2010, 5, e10077. [Google Scholar] [CrossRef][Green Version]
- Lyulcheva, E.; Taylor, E.; Michael, M.; Vehlow, A.; Tan, S.; Fletcher, A.; Krause, M.; Bennett, D. Drosophila Pico and its mammalian ortholog lamellipodin activate serum response factor and promote cell proliferation. Dev. Cell 2008, 15, 680–690. [Google Scholar] [CrossRef]
- Mochida, O. A strain producing abundant brachypterous adults in Nilaparvata lugens (Hompotera: Delphacidae). Entomol. Exp. Appl. 1975, 18, 465–471. [Google Scholar] [CrossRef]


| Gene | Primer | Sequence (5′-3′) | Size |
|---|---|---|---|
| Serum response factor (srf) | srf-F | GTTCGCCGCCTTCCAAT | 145 bp |
| srf-R | TGACGCCACCAGTAGCA | ||
| hth | hth-F | CAGAAGAAGCGCGGAAT | 167 bp |
| hth-R | CGCCTCGCATTGATAAAC | ||
| Insulin receptor 1 (InR1) | InR1-F | TGCCATCAAGACTGTCAAT | 152 bp |
| InR1-R | GCTCCATCACCACATAGG | ||
| Insulin receptor 2 (InR2) | InR2-F | CTTGCCGAACAGCCTTAC | 150 bp |
| InR2-R | GGGTCGTTTAGTGGGTCT | ||
| foxo | foxo-F | CGCCTGTAACCACCAAA | 199 bp |
| foxo-R | CTCGTGCTTGATGACCTC | ||
| Target of rapamycin complex 1 (torc1) | Torc1-F | GGCTACAGGGATGTCAAAG | 162 bp |
| Torc1-R | GCATCACCAGCGTTTTATG | ||
| Insulin receptor substrate gene (chico) | Chico-F | GCTGGAGTCGTTCTTCTAC | 148 bp |
| Chico-R | CAAGTCCTTCTGCGAGTTC | ||
| Tyramine receptor 1 (tyr1) | tyr1-F | AGGAGGACTCGGTAACTG | 182 bp |
| tyr1-R | GGTGGCTGGCATTATCAT | ||
| 18S RNA | 18S-F | CGCTACTACCGATTGAA | 132 bp |
| 18S-R | GGAAACCTTGTTACGACTT |
| Item | Short-Winged Strain | Long-Winged Strain | ||||
|---|---|---|---|---|---|---|
| Egg | 3rd Instar | Adult | Egg | 3rd Instar | Adult | |
| Total raw reads | 54,721,540 | 55,915,498 | 54,415,048 | 57,603,588 | 56,380,930 | 55,801,748 |
| Total clean reads | 51,363,210 | 52,020,498 | 51,285,176 | 54,061,654 | 52,684,738 | 52,726,358 |
| Total clean nucleotides (nt) | 4,622,688,900 | 4,681,844,820 | 4,615,665,840 | 4,865,548,860 | 4,741,626,420 | 4,745,372,220 |
| Q20 percentage | 97.87% | 97.78% | 97.95% | 97.83% | 97.80% | 97.98% |
| Total number of contig | 115,221 | 104,974 | 93,064 | 113,494 | 101,678 | 101,760 |
| Total length of contig (nt) | 51,970,647 | 45,085,737 | 40,546,907 | 52,387,287 | 45,157,354 | 42,697,500 |
| Mean length of contig (nt) | 451 | 429 | 436 | 462 | 444 | 420 |
| Total number of unigene | 72,598 | 66,433 | 60,612 | 71,586 | 66,103 | 64,619 |
| Total length of unigene (nt) | 67,908,204 | 52,260,520 | 46,158,712 | 68,344,039 | 52,676,037 | 48,676,881 |
| Mean length of unigene (nt) | 935 | 787 | 762 | 955 | 797 | 753 |
| Distinct clusters of unigene | 19,705 | 14,068 | 11,570 | 19,325 | 13,226 | 12,965 |
| Distinct singletons of unigene | 52,893 | 52,365 | 49,042 | 52,261 | 52,877 | 51,654 |
| Stage | Type of Ontology | GO Term | Term_ID | Log10 Corrected p-Value | Uniqueness | Number of DEGs |
|---|---|---|---|---|---|---|
| Egg | Biological process | Cell fate determination | GO:0001709 | −6.3675 | 0.423 | 27 |
| Regulation of cell projection organization | GO:0031344 | −5.8210 | 0.498 | 27 | ||
| Morphogenesis of a branching structure | GO:0001763 | −5.4486 | 0.391 | 25 | ||
| Branching morphogenesis of an epithelial tube | GO:0048754 | −5.4486 | 0.323 | 25 | ||
| Regulation of neurogenesis | GO:0050767 | −5.3706 | 0.243 | 26 | ||
| Cell fate commitment | GO:0045165 | −3.6383 | 0.378 | 37 | ||
| Open tracheal system development | GO:0007424 | −3.5086 | 0.389 | 31 | ||
| Respiratory system development | GO:0060541 | −3.5086 | 0.379 | 31 | ||
| Regulation of cell size | GO:0008361 | −3.0000 | 0.607 | 16 | ||
| Negative regulation of cytoskeleton organization | GO:0051494 | −2.8894 | 0.501 | 17 | ||
| Regulation of cell growth | GO:0001558 | −2.8894 | 0.500 | 17 | ||
| Regulation of microtubule-based process | GO:0032886 | -2.5272 | 0.669 | 15 | ||
| Determination of muscle attachment site | GO:0016204 | −2.5272 | 0.453 | 15 | ||
| Taxis | GO:0042330 | −2.3242 | 0.966 | 33 | ||
| Cytoskeleton organization | GO:0007010 | −1.9488 | 0.664 | 49 | ||
| Mushroom body development | GO:0016319 | −1.9458 | 0.426 | 16 | ||
| Negative regulation of cellular component organization | GO:0051129 | −1.9374 | 0.525 | 18 | ||
| Animal organ morphogenesis | GO:0009887 | −1.8901 | 0.312 | 59 | ||
| Regulation of multicellular organismal process | GO:0051239 | −1.8380 | 0.392 | 30 | ||
| Regulation of developmental process | GO:0050793 | −1.6552 | 0.295 | 35 | ||
| Regulation of protein complex disassembly | GO:0043244 | −1.5176 | 0.476 | 15 | ||
| Tube development | GO:0035295 | −1.3578 | 0.353 | 49 | ||
| Cellular component | Site of polarized growth | GO:0030427 | −4.0414 | 0.833 | 19 | |
| Growth cone | GO:0030426 | −4.0414 | 0.770 | 19 | ||
| Astral microtubule | GO:0000235 | −3.6990 | 0.577 | 15 | ||
| Aster | GO:0005818 | −3.6990 | 0.621 | 15 | ||
| Spindle microtubule | GO:0005876 | −3.6990 | 0.592 | 15 | ||
| Spindle | GO:0005819 | −3.5850 | 0.607 | 18 | ||
| Adherens junction | GO:0005912 | −3.1938 | 0.692 | 19 | ||
| Fusome | GO:0045169 | −3.1427 | 0.759 | 15 | ||
| Cytoplasmic microtubule | GO:0005881 | −3.1427 | 0.566 | 15 | ||
| Neuron part | GO:0097458 | −2.1506 | 0.828 | 24 | ||
| Apical part of cell | GO:0045177 | −1.8884 | 0.834 | 19 | ||
| Apicolateral plasma membrane | GO:0016327 | −1.7804 | 0.703 | 10 | ||
| Subapical complex | GO:0035003 | −1.7804 | 0.580 | 10 | ||
| Cell cortex part | GO:0044448 | −1.7352 | 0.572 | 11 | ||
| Molecular function | Microtubule binding | GO:0008017 | −2.4473 | 0.633 | 15 | |
| Phosphate transmembrane transporter activity | GO:1901677 | −2.0022 | 0.584 | 14 | ||
| Organophosphate ester transmembrane transporter activity | GO:0015605 | −2.0022 | 0.521 | 14 | ||
| Carbohydrate kinase activity | GO:0019200 | −1.8655 | 0.705 | 10 | ||
| Hexokinase activity | GO:0004396 | −1.6906 | 0.706 | 8 | ||
| Organic phosphonate transmembrane transporter activity | GO:0015604 | −1.6029 | 0.597 | 12 | ||
| Catalytic activity | GO:0003824 | −1.4090 | 0.926 | 689 | ||
| Transferase activity | GO:0016740 | −1.4046 | 0.813 | 260 | ||
| 3rd instar nymph | Biological process | Metabolic process | GO:0008152 | −1.8339 | 1 | 593 |
| Cellular component | No significantly enriched terms | |||||
| Molecular function | Catalytic activity | GO:0003824 | −5.3363 | 0.889 | 778 | |
| Antioxidant activity | GO:0016209 | −4.6536 | 0.696 | 30 | ||
| Oxidoreductase activity, acting on peroxide as acceptor | GO:0016684 | −5.5171 | 0.588 | 30 | ||
| Transaminase activity | GO:0008483 | −2.1337 | 0.563 | 19 | ||
| Hexokinase activity | GO:0004396 | −1.3505 | 0.578 | 8 | ||
| Transferase activity, transferring nitrogenous groups | GO:0016769 | −2.1337 | 0.563 | 19 | ||
| Peroxidase activity | GO:0004601 | −3.6198 | 0.588 | 24 | ||
| Adult | Biological process | Metabolic process | GO:0008152 | −2.5045 | 0.568 | 658 |
| Oxidation-reduction process | GO:0055114 | −4.3915 | 0.023 | 132 | ||
| Cellular component | Cytoplasm | GO:0005737 | −1.7203 | 1 | 242 | |
| Molecular function | Antioxidant activity | GO:0016209 | −1.6189 | 0.254 | 27 | |
| Oxidoreductase activity, acting on peroxide as acceptor | GO:0016684 | −2.2262 | 0.233 | 27 | ||
| Oxidoreductase activity | GO:0016491 | −3.699 | 0.297 | 170 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Liu, X.-D. Transcriptomic Analysis Suggests Genes Expressed Stage-Independently and Stage–Dependently Modulating the Wing Dimorphism of the Brown Planthopper. Genes 2020, 11, 19. https://doi.org/10.3390/genes11010019
Zhang C, Liu X-D. Transcriptomic Analysis Suggests Genes Expressed Stage-Independently and Stage–Dependently Modulating the Wing Dimorphism of the Brown Planthopper. Genes. 2020; 11(1):19. https://doi.org/10.3390/genes11010019
Chicago/Turabian StyleZhang, Chao, and Xiang-Dong Liu. 2020. "Transcriptomic Analysis Suggests Genes Expressed Stage-Independently and Stage–Dependently Modulating the Wing Dimorphism of the Brown Planthopper" Genes 11, no. 1: 19. https://doi.org/10.3390/genes11010019
APA StyleZhang, C., & Liu, X.-D. (2020). Transcriptomic Analysis Suggests Genes Expressed Stage-Independently and Stage–Dependently Modulating the Wing Dimorphism of the Brown Planthopper. Genes, 11(1), 19. https://doi.org/10.3390/genes11010019




