Abstract
The SWEET family, which includes transcripts of a cohort of plant hexose and sucrose transporters, is considered key to improving crop stress tolerance and yield through its role in manipulating the carbohydrate partitioning process. The functions and regulatory roles of this gene family are variable among different species; thus, to determine these roles, more species-specific information is needed. Brassica rapa displays complicated regulation after a whole-genome triplication (WGT) event, which provides enormous advantages for use in genetic studies, thus it is an ideal model for exploring the functional and regulatory roles of SWEETs from a genetic perspective. In this study, the results of a homology search and phylogenetic relationship analysis revealed the evolutionary footprint of SWEETs among different plant taxa, which showed that plant SWEETs may have originated from Clade II and then expanded from vascular plants. The amino acid sequence characteristics and an analysis of the exon-intron structure of BrSWEETs duplicates clarified that SWEETs retention occurred after a WGT event in B. rapa. An analysis of the transcriptional levels of BrSWEETs in different tissues identified the expression differences among duplicated co-orthologs. In addition, qRT-PCR indicated that the BrSWEETs’ co-orthologs were varied in their stress responses. This study greatly enriches our knowledge of SWEETs in the B. rapa species, which will contribute to future studies on the Brassica-specific regulatory pathways and to creating genetic innovations.
1. Introduction
Higher plants have evolved well for the ability to employ solar energy to convert CO2 into organic carbon in photosynthetic source leaf tissues, and this carbon then travels to various heterotrophic sink organs (such as seeds, roots, and fruits) [1]. The efficient distribution (translocation and partitioning) of sugars between a source–sink system is governed by sugar transporters, and it is crucial for sink organ development and positive feedback to source tissues to ensure sufficient energy allocation and to maintain a trade-off between the different parts of plants [1,2]. Therefore, sugar transporters usually act as bridges, and they are fundamentally pivotal to the cellular exchange of carbon and energy in multicellular organisms to perform further biological functions. Research has shown that membrane-localized sugar transporters in plants can be categorized into MFSs (the major facilitator superfamily) and SWEET (Sugars Will Eventually be Exported Transporter, PFAM: PF03083) family [3]. The homologs of the SWEET family among the higher plants are structurally characterized by seven transmembrane domains (TMDs) harboring two MtN3/PQ-loop domains connected by a loop, and they are phylogenetically divided into four clades with mono- and disaccharide transport activity by a uniport mechanism [4,5]. Their functions are essential for various activities in plants, such as phloem loading [6], pollen development [7], nectar secretion [8], seed filling [9,10,11], freezing tolerance [12], pathogen resistance [13,14,15], and others, and they act by mediating the translocation and partitioning of hexoses and/or sucrose.
SWEETs homologs have not only been found in prokaryotes (semiSWEETs) but have also reportedly appeared in all the eukaryotic kingdoms ranging from fungi to plants and animals [4,16,17]. Plant SWEETs were initially identified as sugar effluxers with glucose or sucrose transport activities in Arabidopsis and rice [5]. Thereafter, additional homologs of SWEETs from different plant varieties were explored and their functions were determined to improve crop survival and yield potential. For example, in the model plant Arabidopsis, plasma membrane (PM)-localized AtSWEET11 and AtSWEET12 of the clade III sucrose transporters were characterized by their role in the preparation of sucrose apoplastic phloem loading [18]. A double mutation (Atsweet11/12) blocks assimilation exudation, which results in a shorter root, abnormal vascular structure and lower freezing tolerance [6,18]. In addition, AtSWEET11/12/15 are specifically expressed in developing seeds to transfer sugars from the seed coat to the embryo, and the triple mutant (Atsweet11/12/15) resulted in severe seed defects [19]. Like other members of clade III, AtSWEET9 has a demonstrated function as an essential sucrose efflux transporter in nectar production, and its orthologs are functionally conserved as nectary-specific transporters in core eudicots [8]; AtSWEET10 is involved in the stress response, and AtSWEET13 and AtSWEET14 function within the anthers by tangling with the gibberellic acid response [20,21]. Apart from the sucrose transporters in Clade III, AtSWEET1 [5] and AtSWEET2 in clade I, AtSWEET4 [22] and AtSWEET8 [23,24] in clade II, and AtSWEET16 [12] and AtSWEET17 [25] in Clade IV, have been specifically explored for their significant functions in various biological activities through the manipulation of sugar transport. Parallel with the gradual growth of SWEETs research in Arabidopsis, the further study of SWEETs homologs in other plants, including rice [5,24,26,27], tomatoes [11,28,29], grapes [30,31], Medicago truncatula [32], etc., have been reported and rigorously updated. Therefore, given the versatile regulators and obvious yield potential of SWEETs, more detailed and in-depth studies in more abundant species are needed.
The Brassica rapa plant provides a well-established model of the “U” triangle in the Brassica genus, which is characterized by broad genetic and morphological diversity that allowed for the domestication of this genus to produce leafy vegetables, vegetable oils, turnip roots, turnip greens, turnip tops, and fodder turnip [33,34,35]. The release of the B. rapa and B. oleracea genomes confirmed an additional γ whole-genome triplication (WGT) event in the Brassica species after they diverged from Arabidopsis thaliana approximately 9–28 million years ago [34,35,36,37,38,39]. In comparing the ~27,000 genes in the A. thaliana genome, the retained genes (~42,000) in the B. rapa genome are considerably fewer than the theoretical value from a simple WGT event. Approximately ~70%, ~46%, and 36% of genes remained in the sub-genomes of the least fractionated (LF), medium fractionated (MF1), and most fractionated (MF2) individuals, respectively, and they were distributed into 71 GBs (genetic blocks) comparing 24 conserved ancestral GBs in A. thaliana [34,40]. Previous reports have suggested that the reshuffle process, namely whole-genome duplication/triplication (WGD/WGT), subsequently intertwined the fractionation and re-diploidization that were ubiquitous in the plant kingdom, which had different impacts on the gene sets/family [41,42]. Structural and functional evidence has suggested that having multiple copies of SWEET genes in eukaryotes allows for diversification in metabolic regulation. However, the puzzle of the molecular characteristics of SWEETs in B. rapa remains unresolved.
With the aim of exploring the SWEET gene family in B. rapa (BrSWEETs), we investigated the evolutionary footprint of SWEETs in different plant taxa by homology search and phylogenetic relationship. Subsequently, the SWEETs were retained in B. rapa after the WGT event were analyzed using basic information, including its distribution on the chromosome, sub-genome, and 24 GB, and then it was characterized in detail by multiple sequence alignment, phylogenetic relationships, conserved domains, and amino acid motifs, along with exon-intron structure through comparisons with its homologs in A. thaliana. After that, the BrSWEETs’ transcriptional levels in different B. rapa tissues were investigated to illuminate the duplicated paralog expression. In addition, the WGD-duplicated SWEET’s paralogous expression under abiotic stress conditions was analyzed to understand the stress response pattern in B. rapa. Our fundamental work here will contribute to the growing body of knowledge centered on understanding the role of SWEET gene family expansion, and it may be valuable for target gene selection and further molecular function characterization to improve crop yields by modifying stress resistance.
2. Materials and Methods
2.1. Identification and Characterization of SWEET Genes in B. rapa
Homologous SWEETs in B. rapa were revealed in the BRAD database [43] according to a syntenic matchup with SWEET isoforms in A. thaliana, and they were subsequently confirmed by local BLASTP alignment (E-value ≤ 1e−5 and identity ≥ 60%) [44] and PFAM domain analysis (https://pfam.xfam.org). The position of each identified member on the ancestral karyotypes, genetic blocks, and sub-genomes (LF, MF1, and MF2), along with the exon-intron structure was verified by searching the BRAD database (B. rapa genome v1.5, accession Chinese cabbage Chiifu-401-42, http://brassicadb.org/brad/index.php). The MCScanX (Multiple Collinearity Scan toolkit) (http://chibba.pgml.uga.edu/mcscan2/; parameter settings of match_score: 50, match_size: 5, gap_score: −3, and E-value: 1e−5) was employed to identify duplication events for the SWEETs families in A. thaliana and B. rapa. A TMHMM server (V.2.0) (http://www.cbs.dtu.dk/services/TMHMM/) was used to determine the transmembrane helices (TMs). The conserved domain and motif analysis were subsequently performed using online CDD tools (Conserved Domains Database) (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and the MEME program (http://meme-suite.org/tools/meme).
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
For the homologous SWEETs’ analyses in B. rapa and A. thaliana, a multiple sequence alignment of the amino acid sequence was performed with the MAFFT program (v7.245) [45] and was demonstrated by TEXshade (www.pharmazie.uni-kiel.de/chem/Prof Beitz/biotex.html) [46]. The seven TM domains were assigned using a sequence alignment based on the AtSWEET1 structure indicated by Tao et al. [4]. The phylogenetic relationship was inferred using the NJ (neighbor-joining) method [47] based on the optimal model (JTT+G) with the support of 1000 bootstrap replications with a MEGA program [48].
Additionally, to investigate the footprint of SWEET genes in different plant groups, the homologs were retrieved from the PLAZA database by BLASTP search strategy (parameter cutoff setting: E-value ≤ 1e−10 and identity ≥ 60%) [44]. Only the candidate homologs with two MtN3/PQ-loop domains harboring seven TMs structure were further used to perform the phylogenetic analysis. The ML (maximum likelihood) phylogenetic trees were constructed using PhyML-3.1 software [49], after multiple sequence alignments with the MAFFT program (v7.245) [45] by setting up the parameters as follows: 1000 bootstrap replications; NJ/BioNJ as the initial tree; the optimal model (JTT+G) determined by the Protest program (v3.2) [50] according to BIC (Bayesian Information Criterion); and everything else according to default settings [51]. The output phylogenetic tree with the domain structures of each member was finally edited using the ITOL tool (http://itol.embl.de).
2.3. Expression Pattern in Different Tissues
The expression pattern of co-orthologous SWEETs pairs was analyzed at the transcript level by retrieving the Illumina RNA-Seq data that have been reported previously (Table S4) [52]. The values of FPKM from different tissues including two batches of 7-week-old root (root1, root2) and leaf (leaf1, leaf2); the same stages for the stems and flowers, the silique 15 days after pollination, and a callus generated by tissue culture; were log2-normalized and displayed using the Multi-Experiment Viewer (MeV 4.8) program (http://mev.tm4.org). After that, Pearson correlation coefficient analyses were performed by R program.
2.4. Plant Materials, Growth Conditions, and Stress Treatment
Chinese cabbage seeds (cultivar Chiifu-401-42) were surface-sterilized with 70% ethanol for 1 min, vortexed in a 10% bleach solution 1500 rpm for 15 min, and then thoroughly washed with sterilized reverse-osmosis water 3–5 times before soaking overnight for two days in dark conditions at 24 °C. The germinated seeds were grown in half-strength Hoagland’s solution (pH = 6.0) in plastic containers and kept in the phytotron at the Nanjing Agricultural University at 70% relative humidity and 100 μmol m−2s−1 light intensity (light at 24 °C for 16 h, and darkness at 18 °C for 8 h). Five-leaved seedlings were accustomed to stress treatments by transferring the plants to chambers under the same conditions mentioned above but at different temperatures for the cold (4 °C) and hot (38 °C) treatments and with the replenishment of exogenous substances for the salt (200 mM NaCl) treatment. Samples with three biological replicates were taken after treatment at 0 h (CK), 1 h, 2 h, 4 h, 6 h, 8 h, and 10 h and immediately dropped in liquid nitrogen, and then they were stored at −80 °C for RNA isolation.
2.5. RNA Isolation and qRT-PCR
The extractions of the total RNAs and the syntheses of the first-strand complementary DNA (cDNA) were performed separately using a Total RNA Kit and an RNA TIANScript cDNA Synthesis Kit (samples; Tiangen Biotech, Beijing) according to the manufacturer’s guidance. The qRT-PCR was performed in a volume of 20 μL (containing 10 μL of SuperMix, 0.4 μL of each primer, 1 μL of the template (×10 diluted cDNAs), and 7.2 μL of sterile distilled water). The condition set-up was as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, 55 °C for 15 s, and 72 °C for 10 s in the ABI StepOne Plus real-time PCR system. The relative gene expression analysis was counted using the 2−ΔΔCt methods by using ACTIN and UBQ as reference genes [53] with the three independent replicates. All the primer pairs were designed with the Primer program (v5.0) (http://www.broadinstitute.org/ftp/pub/software/Primer5.0/) and they are listed in Table S5.
3. Results
3.1. The Evolutionary Footprint of SWEETs in Plants
A comparative analysis of the molecular function and structural features between semiSWEETs in prokaryotes and SWEETs in eukaryotes illustrated that SWEETs expanded their functional capacity by enriching the structural features in eukaryotes [4]. Furthermore, mounting evidence shows that the extension of the SWEET gene family allows for diversification in metabolic regulation and specification in gene spatio-temporal expression [52,54,55,56,57]. Here, with the aim of investigating the footprint of the SWEET gene family in more widely distributed plant species, 486 homologs of SWEETs were retrieved using a BLASTP search (E-value ≤ 1e−10 and identity ≥ 60%) and MtN3/PQ-loop domain identification from 27 sequenced plant species in a different taxonomic group (Table S1). The retrieved result showed that the SWEETs’ gene set was extended from land plants, and its isoform numbers are species-specific, ranging from six to 52 (Figure 1A). A statistical analysis on the transmembrane domain (TMs) suggested that many SWEET candidate genes have a certain sequence similarity by homologous search, but fewer than seven TMs also extended from land plants and tended to have more complex structural features from flowering plants. Since functionally verified structural features of SWEET proteins with two triple-helical bundles (THBs) are essential for functional sugar-translocating pores in eukaryotic organisms [4,58], the candidates (410 sequences) with two MtN3 domains were selected to further construct a phylogenetic relationship. Those sequences were phylogenetically divided into four groups (Figure S1). An orthologous analysis according to the phylogenetic relationship found that plant SWEETs may originate from Clade II, and they may have expanded from the vascular plant with the evolution of the vascular system, since more groups (Clade III and Clade IV) appeared from Selaginella moellendorffii. Clade I appeared soon afterward, from the flowering plant Amborella trichopoda (Figure 1B). Additionally, although SWEET genes set and functional differentiation expanded after land plants, they were seemingly barely related to the cyclic rounds of the genome duplication event (WGD or WGT) in the plant kingdom [59].
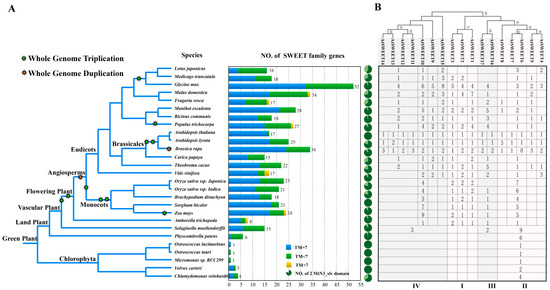
Figure 1.
Molecular footprint and phylogenetical classification of SWEETs in the plant kingdom. (A) Identification and distribution of homologous SWEETs in different plants and (B) functional classification based on maximum likelihood (ML) phylogenetic analysis by Figure S1.
3.2. SWEETs Homologs in the B. rapa Genome
The genome release of B. rapa confirmed that, other than sharing the three paleopolyploidy events (γ, β, and α) with the genome of A. thaliana, B. rapa suffered an additional WGT event after the divergence from A. thaliana 13–17 million years ago [34,43,60,61]. The diploidization process to follow involved genome reshuffling and gene losses, which have different effects on the retention of the gene family set. To analyze the duplication and retention pattern of SWEETs in B. rapa, the orthologs or co-orthologs of SWEETs in A. thaliana were investigated. Here, a total of 33 BrSWEET genes were first identified in the B. rapa genome according to their syntenic relationship and a BLASTp search with the best-matched hits for A. thaliana homologs (Table S2) [5]. Each member was named on the basis of their orthologous relationship with A. thaliana and with co-orthologs in different sub-genomes by adding an LF, MF1, or MF2 suffix (Figure 2A). When mapping the SWEETs of A. thaliana and B. rapa onto 24 GBs of ancestral crucifer karyotype, all of them are mapped on 14 of the 24 GBs and they shared the selfsame GBs for orthologous genes distributed over eight ancestral crucifer karyotypes and expanding copies of AK1/3/5/6/7/8 in B. rapa (Figure 2B). By comparing them to the neighboring syntenic genes (10 genes on either side) in the flanking region, a higher proportion of BrSWEETs’ genes were retained for those homologs with two or three copies (Figure 2C), and more members were kept in the sub-genomes of MF1 and MF2 (Figure 2D).
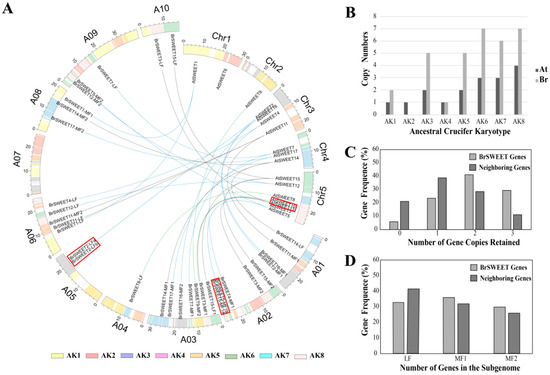
Figure 2.
Basic information of the SWEET genes in the Arabidopsis thaliana and Brassica rapa genome. (A) Assignment of the SWEET genes to the chromosomes and 24 gene blocks (GBs) of thaliana and B. rapa; (B) SWEETs numbers on the inferred ancestral crucifer karyotype; (C) gene frequencies of retained homologous copies in the syntenic region; and (D) three sub-genomes with SWEETs in B. rapa. The red box labeled gene pairs in (A) expanded by tandem duplication and the fractionation of SWEETs homologs in different sub-genome are listed in Table S3. Different colored lines indicate the orthologous SWEETs (green for single copy, blue for two copies, and black for three copies) between A. thaliana and B. rapa.
After that, with the aim of understanding the species-specific amplification of SWEET genes in B. rapa better, basic information, including the phylogenetic relationships; copy number variations of orthologs; duplication event types; and their corresponding amino acid sequence alignment, TMs, conservative motifs, domains, and the exon-intron structures of homologs between A. thaliana and B. rapa were investigated. From an overall perspective, the result of a multiple sequence alignment of amino acid sequences illustrated that TM4 is less conservative for all the sequences (Figure 3B). All the BrSWEETs proteins were strictly conserved for the key functional sites, which validated that they were essential for structural formation, with sites such as Tyr57, Trp176, Val188, and four conserved prolines (P23, P43, P145, and P162) in AtSWEET1 [4], excepting isoforms with obvious fragment deletion (Figure S2). An analysis of the conservative motif and domain suggested that all the members of Clade III have longer protein sequences, which embed an extra motif 7 near the position of TM4, an extra motif 8 or motif 10 near C-terminal region, and its composed domains are closer to MtN3 than the PQ loop according to the sequence similarity retrieval prediction.
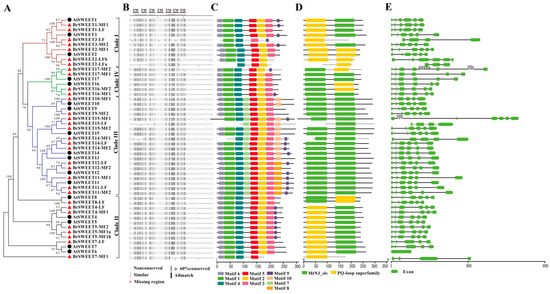
Figure 3.
Sequence feature analysis of SWEETs in A. thaliana and B. rapa. (A) NJ-phylogenetic relationship, (B) sequence alignment of amino acid, (C) conserved motif, (D) conserved domain, and (E) exon-intron structure. The seven transmembrane™ domains were assigned a sequence alignment based on the AtSWEET1 structure indicated by Tao et al. [4].
A further analysis on the phylogenetic relationship, copy number variation, and duplication event type for SWEETs in B. rapa suggested that the case can be divided into three types as follows: (1) Removed, as with AtSWEET6: the only isoform with six TMs and without introns in A. thaliana loses its orthologs from the B. rapa genome; (2) single, such as tandemly duplicated gene pairs of AtSWEET10/13, AtSWEET8, and AtSWEET9, all of which left only one orthologous copy in the B. rapa genome (BrSWEET8-LF, BrSWEET9-MF2, and BrSWEET10-MF1), but kept their uniform structural features of seven TMs, motif distribution, two MtN3/PQ-loop domains, and exons/introns with orthologs in A. thaliana; (3) expanded, the remaining members are expanded with the Brassica-specific WGT event by two (BrSWEET1/2/4/5/7/12/16/17) or three (BrSWEET3/11/14/15) copies except for an extra tandem duplication event for BrSWEET2-LFa/LFb and BrSWEET5-MF1a/MF1b, respectively (Figure 3).
For the expanded members, the amino acid sequence features and gene structure suggested that those co-orthologs had different fragments missing, although they still remained in the B. rapa genome. For the members with two copies that were duplicated by WGT, except for the co-orthologs (BrSWEET4-LF and BrSWEET4-MF1) of AtSWEET4, BrSWEET1-LF, BrSWEET12-MF2, and BrSWEET16-MF2 that is missing motif 9. BrSWEET7-MF1 is missing motif 2/3, and BrSWEET17-MF2 is missing motif 3, when compared to their corresponding orthologs in A. thaliana. For tandemly duplicated co-orthologs of BrSWEET2-LFa and BrSWEET2-LFb, both have an obviously truncated sequence in the N and/or C-terminus of the flanking region, accompanying the missing conservative motifs, domain, and intron-exon structures at the corresponding region. For another pair of tandem duplicates, BrSWEET5-MF1a and BrSWEET5-MF1b, together with BrSWEET5-MF2, there was perfect structural consistency with the AtSWEET5 ortholog for the motifs, domain, and intron-exon structure. For the members with three copies duplicated by WGT, who were co-orthologs of AtSWEET3 and AtSWEET15, only one remained with structural consistency. The orthologs of AtSWEET11 and AtSWEET14 maintained three and two perfect structurally consistent regions, respectively, with the exception of BrSWEET14-MF1, which deleted the fragment on the N-terminus of the flanking region. Summarily, SWEETs did expand their family sets (194.12%) in the B. rapa genome, through integrating the WGT and tandem duplication event according to the identification of homologous genes, but they only duplicated co-orthologs of BrSWEET4, BrSWEET14, BrSWEET5, and BrSWEET11, which were strictly conserved with their orthologs in A. thaliana.
3.3. In-Silico Assessment of Transcript Impact within Co-Orthologous BrSWEETs
Since the BrSWEETs in B. rapa have rarely been studied, the transcript level of log2 transformed fragments per kilobase of the exon model per million mapped reads (FPKM) from different tissues including root, stem, leaf, flower, and silique [52], was employed to assess the transcript impact within amplified co-orthologous genes (Figure 4) with the aim of determining the expression pattern change for the retained SWEETs after amplification by a Brassica-specific WGT event or tandem duplication. The transcriptional abundance within co-orthologs suggested that most of the WGT-amplified co-orthologous BrSWEETs were prone to maintaining a very similar expression pattern, and they rarely showed obvious tissue-specific expression patterns. For instance, co-orthologs with three copies of BrSWEET14 are comparatively highly expressed in the tissues of the root and flower, and the flower and silique, while BrSWEET15 is primarily expressed in the flower and silique. Co-orthologs with two copies of BrSWEET4 are both highly expressed in flowers, and BrSWEET1, BrSWEET12, and BrSWEET17 are expressed in multiple tested tissues with differential transcript abundance. The convergence in the expression profile within those WGT-amplified co-orthologous SWEET genes suggested that some of them may coordinate their expression towards an abundant final product. Interestingly, for two pairs of tandemly duplicated SWEETs that were structurally characterized, BrSWEET2-LFa and BrSWEET2-LFb had the obvious essential domain/motif deficiency mentioned above, but their transcripts were detectable in some tissues. BrSWEET5-MF1a and BrSWEET5-MF1b displayed their complete structural features but their transcripts were almost undetectable in all the investigated tissues. There was a phenomenon in which there was an incompletely conserved domain/motif but a detectable transcript for co-orthologs, not only for tandemly duplicated members but also WGT-amplified members, which suggested that the strictly conserved domain was probably not directly related to the transcript. In addition, only BrSWEET3-MF2 was found to have root-specific expression, and BrSWEET5-MF2 is exclusively expressed in the silique, although three co-orthologous copies were retained and their structural conservation was characterized differently. In addition, the PCC analysis for the detectable transcript members suggested that WGT-amplified co-orthologous SWEETs are transcriptionally correlated with others but not tandem-duplicated members.
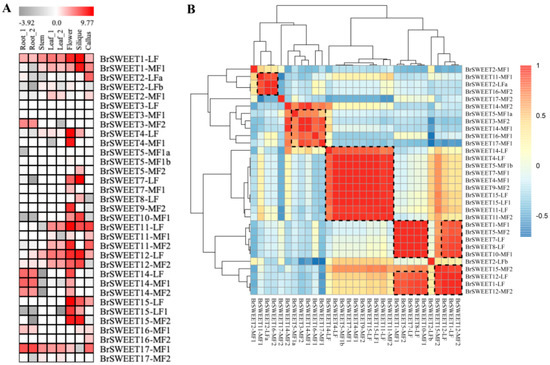
Figure 4.
Heat map (A) and Pearson correlation coefficient (PCC) analysis (B) of the SWEET gene transcriptional levels in different tissues of Brassica rapa. The dotted black box indicates the members with transcriptional correlation. Blocks with colors (A) indicate low (dark) to high (red) normalized transcript accumulation and blocks, with colors in (B) indicating a negative PCC correlation (−1) to positive PCC correlation (1).
3.4. Expanded Co-Orthologous Gene Expression in Response to Abiotic Stress
Generally, the duplicate genes retained after the fractionation process are supposed to produce advantageous or functional profitably [54], and they have particularly been hypothesized to act as standby members in the genome for stress survival by somehow responding to extreme environments. Sugar transporter SWEETs usually modulate the stress response osmotically and/or via sugar metabolism signaling by interfering with sugar allocation [55,62]. Therefore, to determine whether abiotic stress affects the WGT-amplified paralogous SWEETs genes in B. rapa, salt, cold, and heat stress experiments were performed on the WGT-duplicated co-orthologs of BrSWEET1, BrSWEET11, BrSWEET12, and BrSWEET17—which are highly expressed in leaf tissues, as indicated by the in-silico investigation above (Figure 5). The expression patterns of two BrSWEET1 co-orthologs suggested that BrSWEET1-LF and BrSWEET1-MF1 are both significantly up-regulated after 2 h of salt and cold treatment, but only BrSWEET1-MF1 was dominantly expressed in response to heat stress at the 2 h and 10 h time points, indicating that they are both transcriptionally detectable but have different expression responses to various stresses. For three co-orthologs of BrSWEET11, BrSWEET11-LF slightly responded to salt and cold treatment, and it was barely expressed in the heat treatment; BrSWEET11-MF1 was strongly induced under all stress treatments, but BrSWEET11-MF2 was only dominantly expressed under heat treatment after 8 h. In addition, two co-orthologs of BrSWEET12 are both induced under different stresses. Their expression pattern suggested that BrSWEET11-LF and the two co-orthologues of BrSWEET12 probably have a broader response to stress environments compared to BrSWEET11-MF2. Additionally, BrSWEET17-MF1 was dominantly expressed in response to different stress treatments, but BrSWEET17-MF2 were barely detected.

Figure 5.
Expression patterns of co-orthologs of SWEETs under salt, cold, and heat stress treatments in B. rapa by RT-qPCR. Error bars represent ± SE (n = 3). The one-way analysis of variance was calculated by Duncan’s new multiple range test, n = 3; different letters above the bars indicate significant differences (p < 0.05).
4. Discussion
Higher plants have updated their way of transporting the photosynthetic assimilates, not only through open plasma membranes (PMs), which are connected by the plasmodesmata bridge, but also through enclosed membranes harboring sugar transporters to balance sugar export and import relying on demand and supply [1,56,57,63]. The identification of SWEET proteins not only fills the missing step for the apoplastic pathway of the phloem loading and unloading process but also clarifies the confusion about the sugar efflux-involved sink organ’s development and nutrition communication between plants and pathogens. Therefore, both its molecular function and regulatory mechanism have been explored for biomass improvement and environmental adaption, which depend on allocation deficiency.
In the current study, an orthologous analysis of SWEETs from different plant groups according to the phylogenetic relationship suggested that higher plant SWEETs probably originated from Clade II and expanded from vascular plants with the evolution of the vascular system, since more groups (Clade III and Clade IV) appeared from S. moellendorffii and Clade I appeared soon afterward from flowering plants in A. trichopoda (Figure 1B). Previous publications have documented that Clade III transports sucrose, and Clade I, II, and IV predominantly transport hexose [6,18,32]. Furthermore, PM-localized SWEETs with sucrose transport capacity are hypothesized to move sucrose out of leaves involving the long-distance transport of assimilation product by way of apoplastic phloem loading in the plant vascular system. Therefore, the expansion of SWEETs’ functions in vascular plants probably extended their lifestyle by updating the distribution passage of assimilation products.
After that, 33 BrSWEET genes were identified from the B. rapa genome, and co-orthologs of SWEETs in A. thaliana were labeled by adding the sub-genome of the LF, MF1, or MF2 as a suffix (Figure 2A). Compared to the 34 SWEETs identified by Miao L et al. [64], BrSWEET3-MF1 (gene ID: Bra029090) is not thought to belong to the BrSWEETs family, because of the absence of any of MTs or MtN3/PQ-loop and because it barely qualified for phylogenetic analysis. The distribution of SWEETs in A. thaliana and B. rapa on 24 GBs of ancestral crucifer karyotypes indicated that all of them are mapped on 14 of 24 GBs and they shared the same GBs for co-orthologous SWEETs (Figure 2A). The expanded copies of BrSWEETs occurred at AK1/3/5/6/7/8 (Figure 2B), and a higher proportion of BrSWEETs genes were retained for homologs with two or three copies compared to the neighboring syntenic genes in the flanking region (Figure 2C). Furthermore, more members remained in the sub-genomes of MF1 and MF2 (Figure 2D). From an overall perspective of the identified SWEETs in B. rapa, TM4 is less conserved and all the members showed strict conservation of the key functional sites, except for isoforms with obvious fragment deletion. An analysis of the conserved motif and domain suggested that all the members in Clade III have longer protein sequences, which embed an extra motif 7 near the TM4 position; an extra motif 8 or motif 10 near the C-terminal region; and their composed domains are closer to MtN3 rather than the PQ-loop, according to the sequence similarity retrieval prediction. Further detailed analysis combining the copy number variation, duplication event type and sequence features suggested that AtSWEET6 lost its orthologs in the B. rapa genome; tandemly duplicated AtSWEET10 and AtSWEET13, leaving only one orthologous copy in the B. rapa genome; and most of the remaining amplified SWEETs in the B. rapa genome occurred by WGT or tandem duplication events, which left only one isoform with strictly conserved orthologs in A. thaliana, except for the co-orthologs of BrSWEET4, BrSWEET5, BrSWEET11, and BrSWEET14. Subsequently, the transcriptional level of BrSWEETs in different tissues illustrated that there was an incompletely conserved domain/motif but also a detectable transcript for co-orthologs; not only for tandemly duplicated members but also WGT-amplified members, indicating that the strictly conserved domain was probably not directly related to the transcript. Further analysis of the expression levels under salt, cold, and heat stress conditions for WGT-duplicated co-orthologs of BrSWEET1, BrSWEET11, BrSWEET12, and BrSWEET17, which are highly expressed in leaf tissues according to in-silico investigation, showed that BrSWEET11-LF and the two co-orthologs of BrSWEET12 probably provide broad responses to stressful environments compared to the others, which have specific responses to different stresses.
In summary, from the evolutionary footprint of the SWEETs, although the SWEETs gene set and functional differentiation expanded after land plants occurred, it was seemingly barely related to the cyclic rounds of genome duplication events (WGD or WGT) in the plant kingdoms. However, for the recent WGT event in the B. rapa genome, more WGT-duplicated SWEETs were retained for amplifying the SWEETs family set, and their transcripts are detectable. The convergence or divergence in the expression of co-orthologous SWEETs’ responses to biotic stress implies functional diversity. This study provided a detailed investigation of the SWEETs in B. rapa, and it also raises a number of questions. For example, the relationship is between TM4 and motif 7 and whether motif 7 is necessary for sucrose transport, since it was specifically present in Clade III. This early work on SWEETs in B. rapa will fundamentally assist in further molecular function characterizations and will help during explorations of genetic innovation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/722/s1, Figure S1: The phylogenetic relationship and MtN3 domains of SWEET genes from different species. The phylogenetic tree was completed using the maximum likelihood method with the optimal model (JTT+G) and 1000 bootstrap replications. The exon-intron structural information feature was added by ITOL (http://itol.embl.de), Figure S2: Multiple sequence alignment of SWEETs proteins in A. thaliana and B. rapa. The seven TM domains were assigned based on the AtSWEET1 structure indicated by Tao et al. [4]. The experimentally validated key functional sites were labeled with arrows; Table S1: Identified SWEET family genes in different plant kingdoms, Table S2: Identification and characteristics of SWEETs in A. thaliana and B. rapa, Table S3: Fractionation of SWEET homolog genes after triplication in B. rapa. The genes retained during fractionation are indicated with red 1s. The genes lost during fractionation are indicated with white 0s. The three B. rapa sub-genomes are indicated by LF, MF1, and MF2. The retained items indicate the number of B. rapa genes retained relative to those of A. thaliana within the 21-gene syntenic window, Table S4: Fragments per kilobase of the exon model per million mapped reads (FPKM) values of SWEETs in B. rapa; Table S5: B. rapa SWEETs primers used for qPCR.
Author Contributions
Performing the experiments and writing the paper, Y.W.; manuscript revision and approval, D.X. and C.Z.; contribution to the interpretation of the results and coordination of the study, X.H.; all the authors read and approved the final manuscript.
Funding
This research was funded by The Major Program of National Key Research and Development of China, grant number 2017YFD0101803, and The National Key R&D Program of China, grant number 2016YFD0101701.
Acknowledgments
This work was supported by grants from the Major Program of National Key Research and Development of China (2017YFD0101803) and the National Key R&D Program of China (2016YFD0101701). Additionally, we would like to thank Ying-Li, Shah and Sam’s help for polishing the English style.
Conflicts of Interest
All the authors declare no competing interests.
References
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.-L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Cheung, L.S.; Li, S.; Eom, J.S.; Chen, L.Q.; Xu, Y.; Perry, K.; Frommer, W.B.; Feng, L. Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 2015, 527, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Li-Qing, C.; Xiao-Qing, Q.; Bi-Huei, H.; Davide, S.; Sonia, O.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Sun, M.-X.; Huang, X.-Y.; Yang, J.; Guan, Y.-F.; Yang, Z.-N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 26, 83–91. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.Q.; Gase, K.; Kim, S.G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.H.; Qu, X.Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, J.M.; Kang, S.K.; Kim, S.G.; Park, C.M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef]
- Lu, Y.; Gehan, J.P.; Sharkey, T.D. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005, 138, 2280–2291. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Hir, R.L.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.H.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe Interact. 2014, 27, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.-S.; Chen, L.-Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.-Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Structure and function of semiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tao, Y.; Cheung, L.S.; Fan, C.; Chen, L.Q.; Xu, S.; Perry, K.; Frommer, W.B.; Feng, L. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature 2014, 515, 448–452. [Google Scholar] [CrossRef]
- Le Hir, R.; Spinner, L.; Klemens, P.A.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Gery, C.; et al. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP 2–4, a waterlogging-responsive erf from mentha, regulates bidirectional sugar transporter at SWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2018, 16, 221–233. [Google Scholar] [CrossRef]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.-F.; Huang, X.-Y.; Zhu, J.; Gao, J.-F.; Zhang, H.-X.; Yang, Z.-N. Ruptured pollen grain1, a member of the MtN3/Saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Zhao, J.; Huang, R.; Li, X.; Xiao, J.; Wang, S. Rice MtN3/Saliva/SWEET gene family: Evolution, expression profiling, and sugar transport. J. Integr. Plant Biol. 2014, 56, 559–570. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef]
- Feng, C.-Y.; Han, J.-X.; Han, X.-X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Ho, L.-H.; Klemens, P.A.; Neuhaus, H.E.; Ko, H.-Y.; Hsieh, S.-Y.; Guo, W.-J. SlSWEET1a is involved in glucose import to young leaves in tomato plants. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, L.; Ren, C.; Ren, F.; Wang, Y.; Fan, P.; Li, S.; Liang, Z. VvSWEET10 mediates sugar accumulation in grapes. Genes 2019, 10, 255. [Google Scholar] [CrossRef]
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [PubMed]
- Kryvoruchko, I.S.; Sinharoy, S.; Torres-Jerez, I.; Sosso, D.; Pislariu, C.I.; Guan, D.; Murray, J.; Benedito, V.A.; Frommer, W.B.; Udvardi, M.K. MtSWEET11, a nodule-specific sucrose transporter of Medicago truncatula. Plant Physiol. 2016, 171, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Artemieva, A.M. Genetic diversity and intraspecific phylogenetic relationships of Brassica rapa l. Species crops based on microsatellite analysis. Вестник ВОГиС (Рoссийская Федерация) 2008. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef]
- Ramchiary, N.; Yong, P.L. Genetics of brassica rapa L. In Genetics and Genomics of the Brassicaceae; Springer: New York, NY, USA, 2010; pp. 215–260. [Google Scholar]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Sarah, M. Dated molecular phylogenies indicate a miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 18724–18728. [Google Scholar] [CrossRef]
- Lukens, L.N.; Quijada, P.A.; Udall, J.; Pires, J.C.; Schranz, M.E.; Osborn, T.C. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 2004, 82, 665–674. [Google Scholar] [CrossRef]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef]
- Arias, T.; Pires, J.C. Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 2014, 101, 86–91. [Google Scholar] [CrossRef]
- Haibao, T.; Woodhouse, M.R.; Feng, C.; Schnable, J.C.; Pedersen, B.S.; Gavin, C.; Xiaowu, W.; Michael, F.; Chris, P. Altered patterns of fractionation and exon deletions in brassica rapa support a two-step model of paleohexaploidy. Genetics 2012, 190, 1563–1574. [Google Scholar] [CrossRef]
- Peer, Y.V.D. A mystery unveiled. Genome Biol. 2011, 12, 113. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Jian, W.; Lu, F.; Wang, X. Syntenic gene analysis between Brassica rapa and other brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef]
- Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). Cold Spring Harb. Protoc. 2007, 2007, 17. [Google Scholar] [CrossRef] [PubMed]
- Kazutaka, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 102–343. [Google Scholar] [CrossRef]
- Beitz, E. Texshade: Shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Stéphane, G.; Olivier, G. Phyml-a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Devisetty, U.K.; Covington, M.F.; An, V.T.; Lekkala, S.; Maloof, J.N. Polymorphism identification and improved genome annotation of Brassica rapa through deep RNA sequencing. G3 Genes Genet. 2014, 4, 2065–2078. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, N.-W.; Zhao, J.-J.; Bonnema, G.; Hou, X.-L. Validation of reference genes for real-time quantitative PCR normalisation in non-heading Chinese cabbage. Funct. Plant Biol. 2012, 39, 342–350. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 67, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Gamalei, Y. Structure and function of leaf minor veins in trees and herbs. Trees 1989, 3, 96–110. [Google Scholar] [CrossRef]
- Gamalei, Y. Phloem loading and its development related to plant evolution from trees to herbs. Trees 1991, 5, 50–64. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.Q.; Sosso, D.; Ducat, D.C.; Hou, B.H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef] [PubMed]
- Proels, R.K.; Huckelhoven, R. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 2014, 15, 858–864. [Google Scholar] [CrossRef]
- Couvreur, T.L.; Franzke, A.; Al-Shehbaz, I.A.; Bakker, F.T.; Koch, M.A.; Mummenhoff, K. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 2010, 27, 55–71. [Google Scholar] [CrossRef]
- Franzke, A.; Lysak, M.A.; Alshehbaz, I.A.; Koch, M.A.; Mummenhoff, K. Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 2011, 16, 108–116. [Google Scholar] [CrossRef]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. Cell Mol. Biol. 2015, 83, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, A.J.E. Different phloem-loading machineries correlated with the climate. Acta Bot. Neerl. 1992, 41, 121–141. [Google Scholar] [CrossRef]
- Miao, L.; Lv, Y.; Kong, L.; Chen, Q.; Chen, C.; Li, J.; Zeng, F.; Wang, S.; Li, J.; Huang, L. Genome-wide identification, phylogeny, evolution, and expression patterns of MtN3/Saliva/SWEET genes and functional analysis of BcNS in Brassica rapa. BMC Genom. 2018, 19, 174. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).