Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Analysis and Phylogenetic Tree of OsMORF Genes
2.2. Plant Material and Abiotic Stress Treatments
2.3. RNA Isolation and Quantitative Real-Time Reverse-Transcription PCR (qRT-PCR) Analysis
2.4. Yeast Two-Hybrid (Y2H) Analysis
3. Results
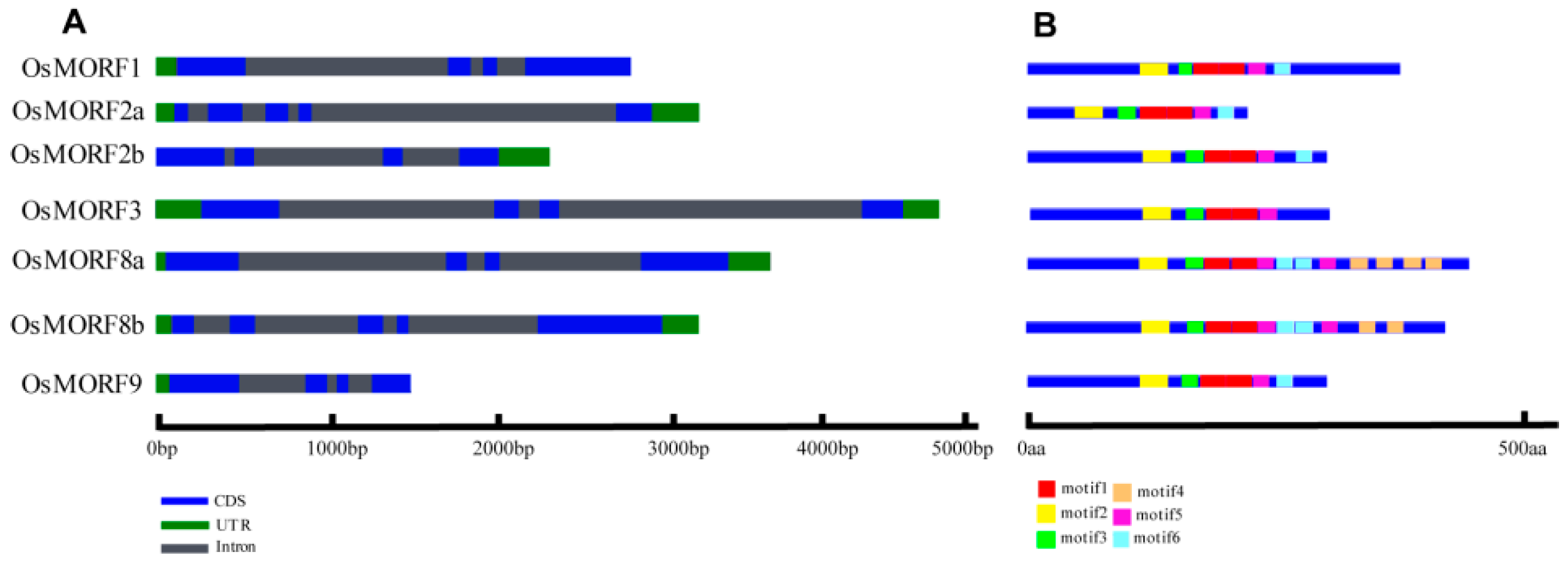
3.1. Sequence Analysis of the OsMORF Gene Family in Rice
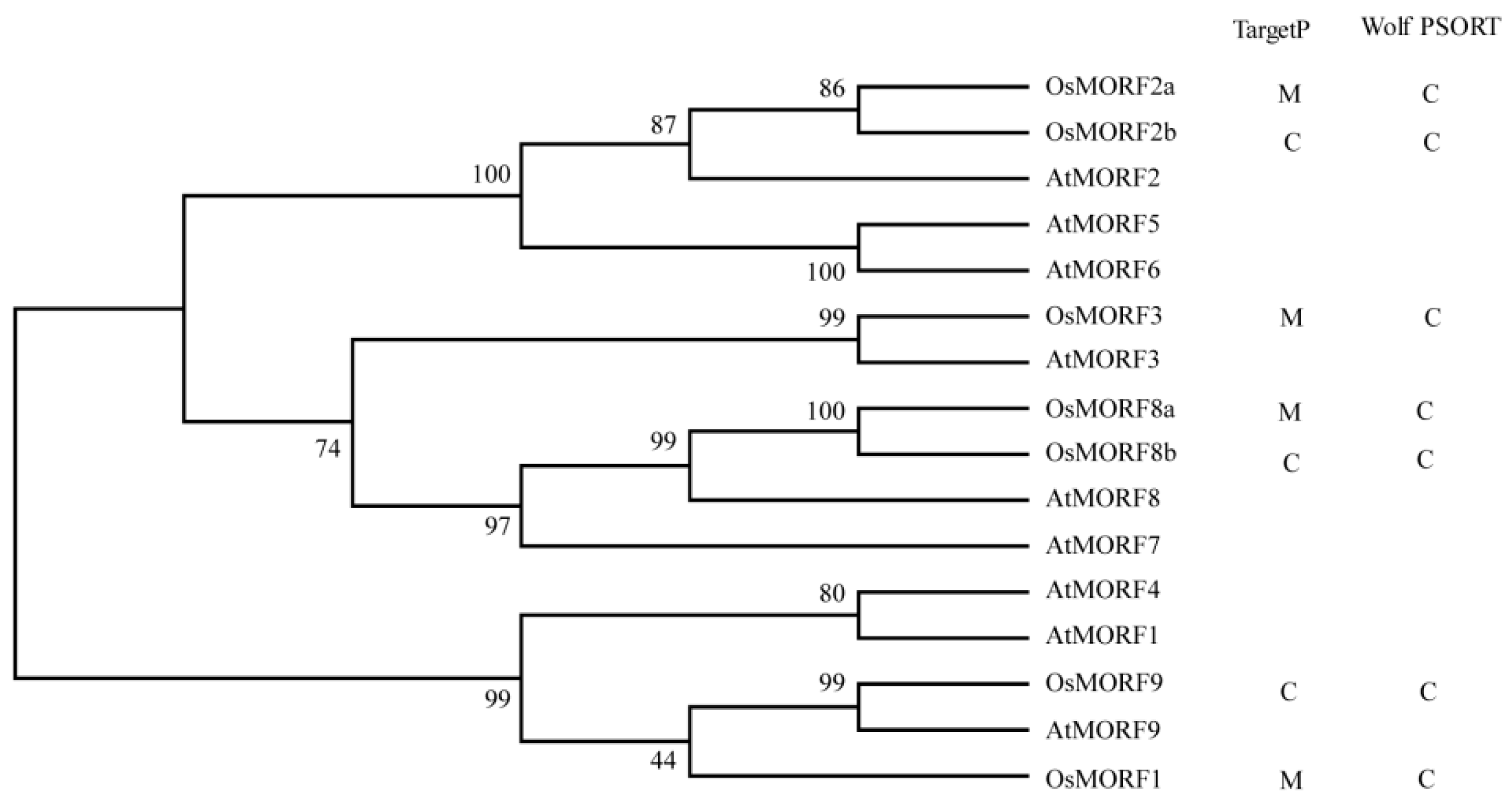
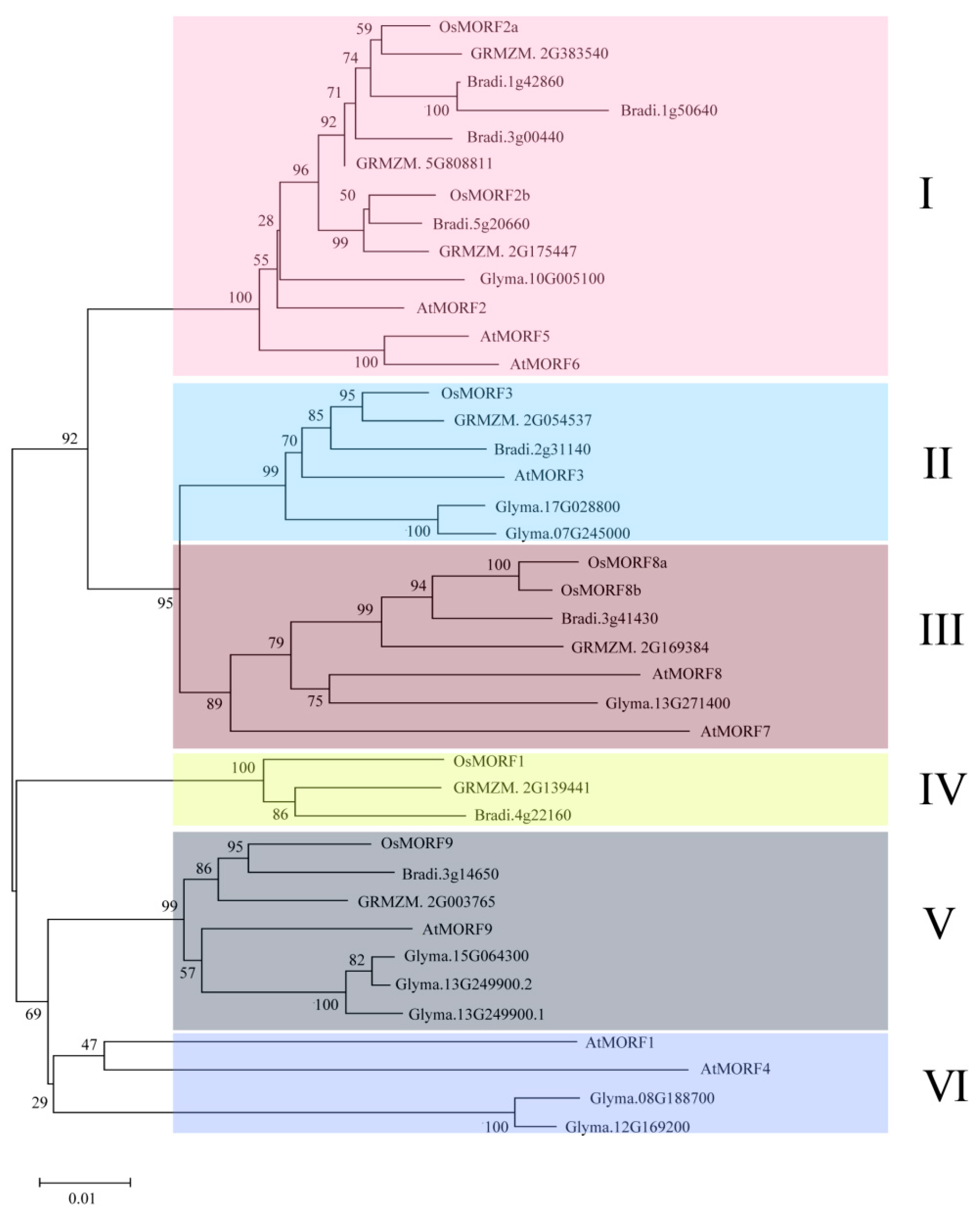
3.2. Phylogenetic Comparison of MORFs among Different Species
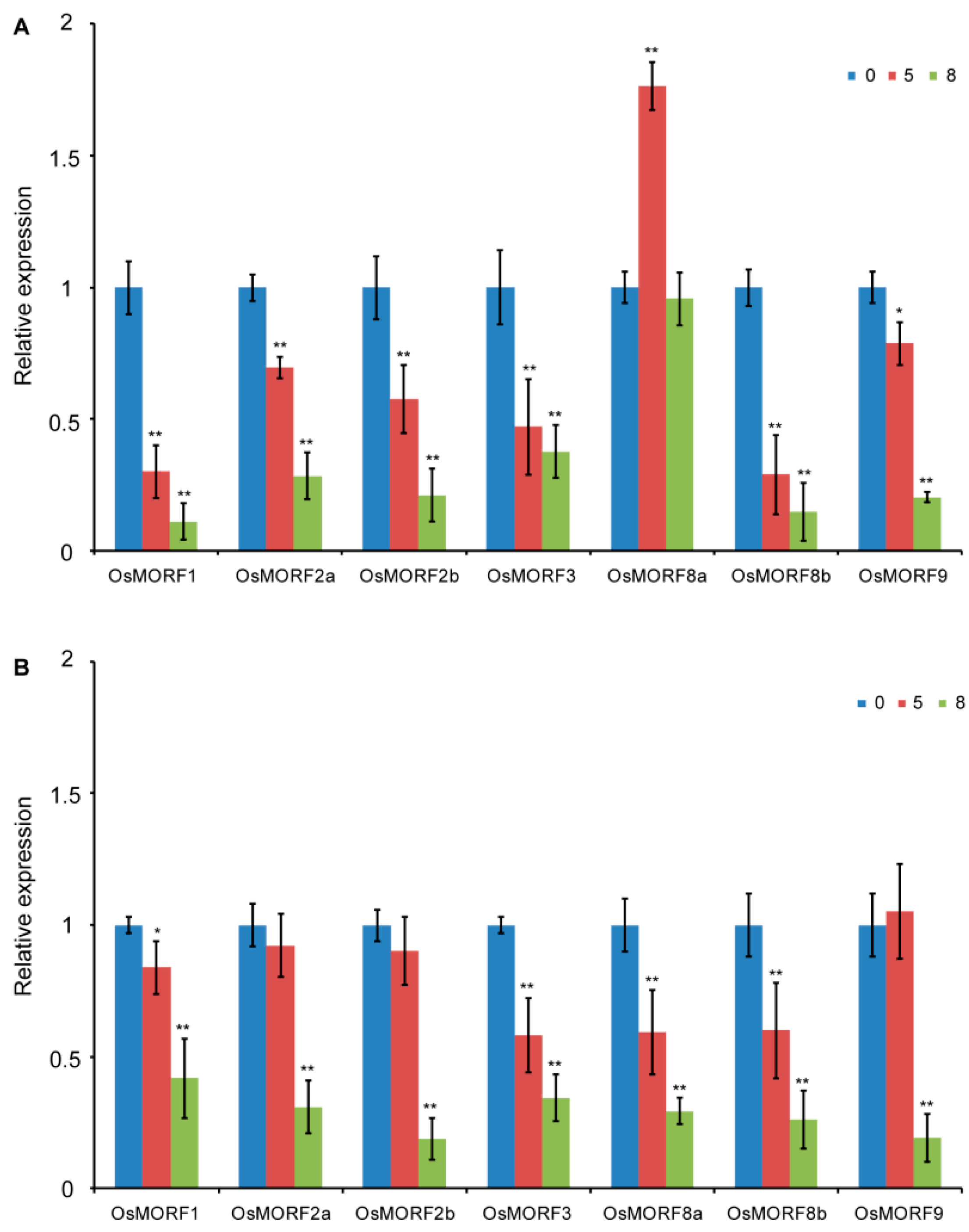
3.3. Expression Pattern Analysis of OsMORF Genes under Cold and Salt Stresses
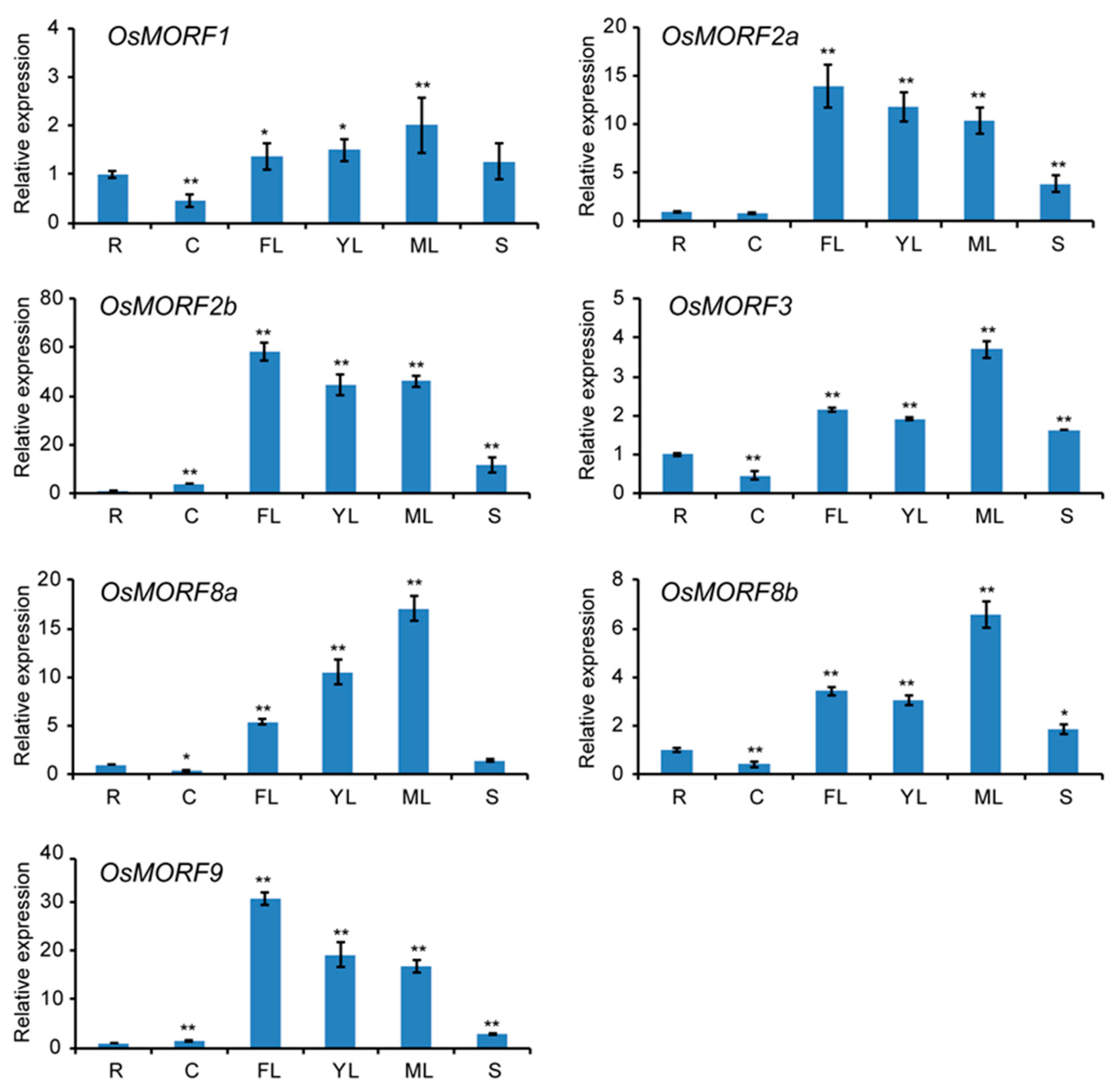
3.4. Expression Analysis of MORF Genes in Different Tissues
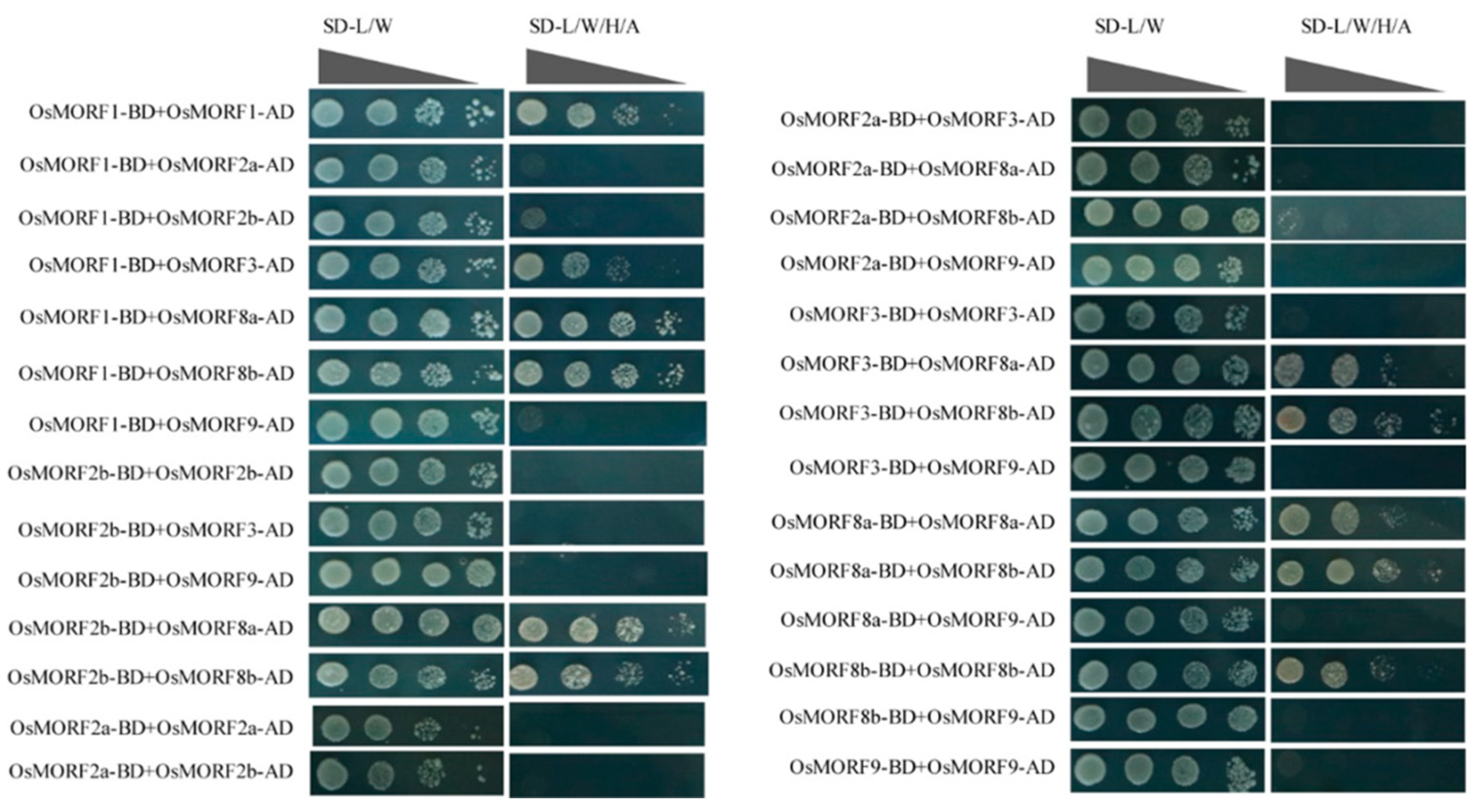
3.5. Analysis of Interactions between MORF Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chateigner-Boutin, A.L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Meng, S.; Su, W.; Bao, Y.; Lu, Y.; Yin, W.; Liu, C.; Xia, X. Genome-Wide Analysis of Multiple Organellar RNA Editing Factor Family in Poplar Reveals Evolution and Roles in Drought Stress. Int. J. Mol. Sci. 2019, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, Q.; Yin, P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018, 61, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Kugelmann, M.; Härtel, B.; Brennicke, A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5104–5109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Tang, W.; Hedtke, B.; Zhong, L.; Liu, L.; Peng, L.; Lu, C.; Grimm, B.; Lin, R. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. USA 2014, 111, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Boil. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-R.; Yang, J.-I.; Moon, S.; Ryu, C.-H.; An, K.; Kim, K.-M.; Yim, J.; An, G. RiceOGR1encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009, 59, 738–749. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.; Wu, J.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol. 2019, 221, 834–849. [Google Scholar] [CrossRef]
- Xiao, H.; Xu, Y.; Ni, C.; Zhang, Q.; Zhong, F.; Huang, J.; Liu, W.; Peng, L.; Zhu, Y.; Hu, J. A rice dual-localized pentatricopeptide repeat protein is involved in organellar RNA editing together with OsMORFs. J. Exp. Bot. 2018, 69, 2923–2936. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, Q.; Qin, X.; Xu, Y.; Ni, C.; Huang, J.; Zhu, L.; Zhong, F.; Liu, W.; Yao, G.; et al. Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 2018, 220, 878–892. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lan, J.; Huang, Y.; Cao, P.; Zhou, C.; Ren, Y.; He, N.; Liu, S.; Tian, Y.; Nguyen, T.; et al. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 2018, 69, 3949–3961. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Cai, M.; Zhang, J.; Li, Y.; Zhang, R.; Song, W.; Zhang, K.; Xiao, H.; Yue, B.; Zheng, Y.; et al. Functional divergence and origin of the DAG-like gene family in plants. Sci. Rep. 2017, 7, 5688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehrmann, A.; Härtel, B.; Glass, F.; Bayer-Császár, E.; Obata, T.; Meyer, E.; Brennicke, A.; Takenaka, M. Selective Homo- and Heteromer Interactions between the Multiple Organellar RNA Editing Factor (MORF) Proteins in Arabidopsis thaliana. J. Boil. Chem. 2015, 290, 6445–6456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Li, Z.-R.; Yu, Q.-B.; Ye, L.-S.; Cui, Y.-L.; Molloy, D.P.; Yang, Z.-N. MORF2 tightly associates with MORF9 to regulate chloroplast RNA editing in Arabidopsis. Plant Sci. 2019, 278, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A.; Takenaka, M. PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Boil. 2011, 8, 67–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentolila, S.; Heller, W.P.; Sun, T.; Babina, A.M.; Friso, G.; Van Wijk, K.J.; Hanson, M.R. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. USA 2012, 109, E1453–E1461. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, X.; Wang, Y.; Wu, J.; Gu, X.; Lu, T. The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol. Plant 2017, 10, 86–98. [Google Scholar] [CrossRef]
- Qiu, Z.; Zhu, L.; He, L.; Chen, D.; Zeng, D.; Chen, G.; Hu, J.; Zhang, G.; Ren, D.; Dong, G.; et al. DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytol. 2019, 222, 349–365. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Zhang, C.; Yates, G.; Bailey, M.; Brown, A.; Sadanandom, A. SUMO Is a Critical Regulator of Salt Stress Responses in Rice. Plant Physiol. 2016, 170, 2378–2391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Cai, W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 3277–3287. [Google Scholar] [CrossRef]
- Rodrigues, N.F.; Da Fonseca, G.C.; Kulcheski, F.R.; Margis, R. Salt stress affects mRNA editing in soybean chloroplasts. Genet. Mol. Boil. 2017, 40, 200–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Shen, L.; Ren, D.; Hu, J.; Chen, G.; Zhu, L.; Gao, Z.; Zhang, G.; Guo, L.; Zeng, D.; et al. Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice. Genes 2019, 10, 694. https://doi.org/10.3390/genes10090694
Zhang Q, Shen L, Ren D, Hu J, Chen G, Zhu L, Gao Z, Zhang G, Guo L, Zeng D, et al. Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice. Genes. 2019; 10(9):694. https://doi.org/10.3390/genes10090694
Chicago/Turabian StyleZhang, Qiang, Lan Shen, Deyong Ren, Jiang Hu, Guang Chen, Li Zhu, Zhenyu Gao, Guangheng Zhang, Longbiao Guo, Dali Zeng, and et al. 2019. "Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice" Genes 10, no. 9: 694. https://doi.org/10.3390/genes10090694
APA StyleZhang, Q., Shen, L., Ren, D., Hu, J., Chen, G., Zhu, L., Gao, Z., Zhang, G., Guo, L., Zeng, D., & Qian, Q. (2019). Characterization, Expression, and Interaction Analyses of OsMORF Gene Family in Rice. Genes, 10(9), 694. https://doi.org/10.3390/genes10090694










