Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Tissue Collection
2.2. RNA Extraction and cDNA Synthesis
2.3. Amplification of the Nucleotide Sequences of PcIAG
2.4. Sequence Analyses
2.5. Expression Pattern of PcIAG Detected by Quantitative Real-Time PCR (qPCR)
2.6. Western Blotting
2.7. Histology of the AG in P. clarkii
2.8. RNAi
3. Results
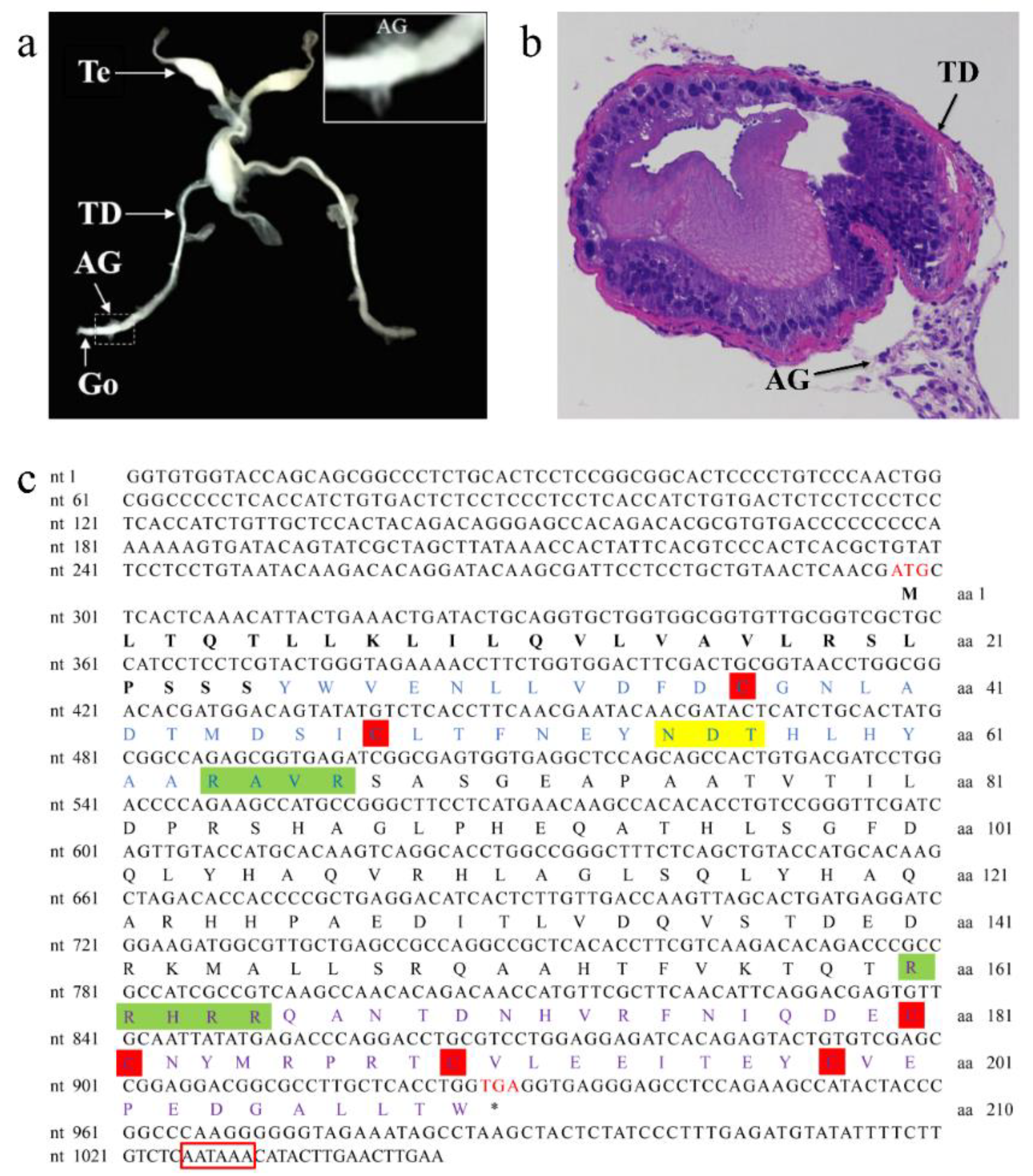
3.1. Characterization of the AG in P. clarkii and Sequence Analysis of PcIAG cDNA
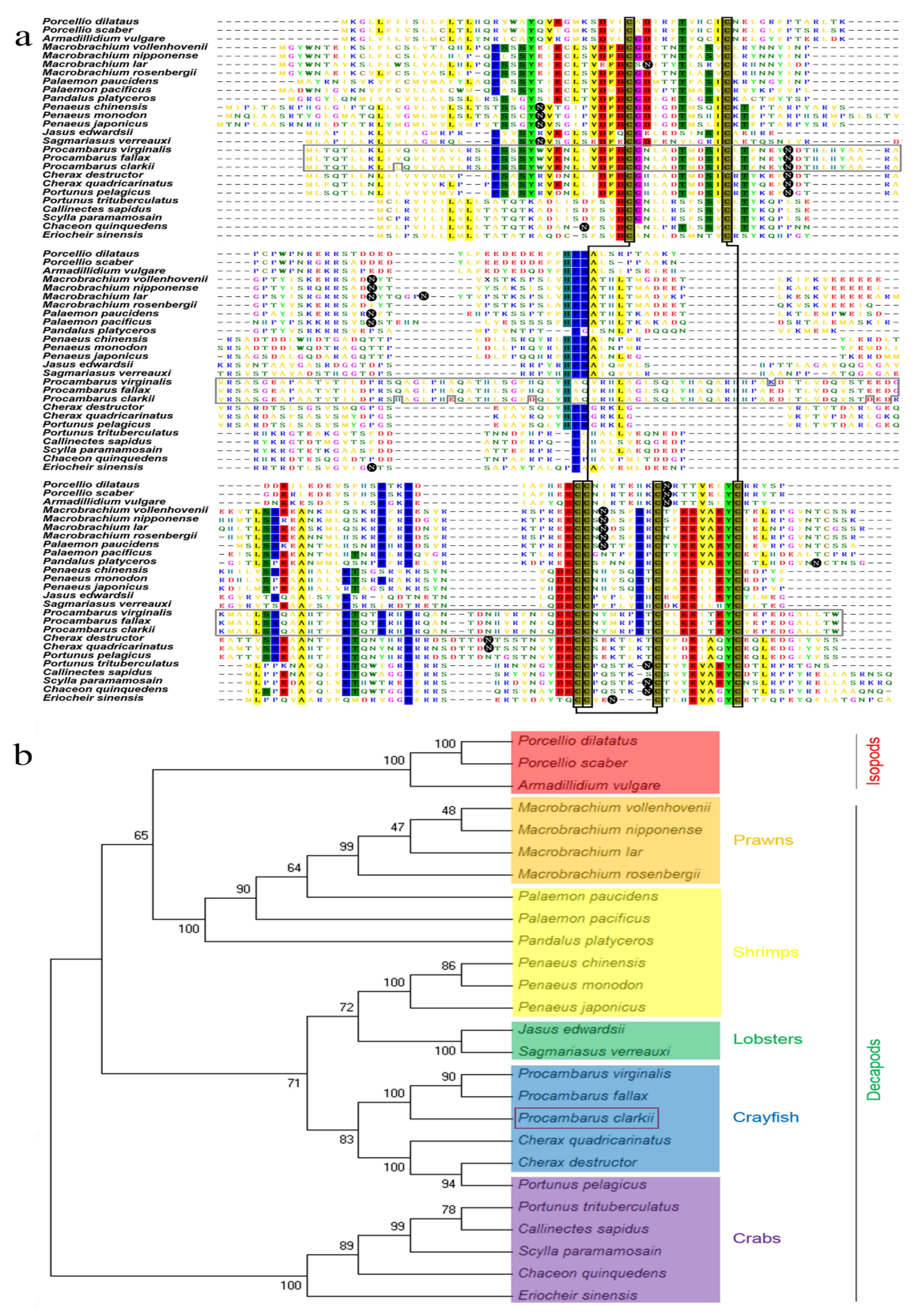
3.2. Multiple Alignment and Phylogenetic Tree Analysis of P. clarkii
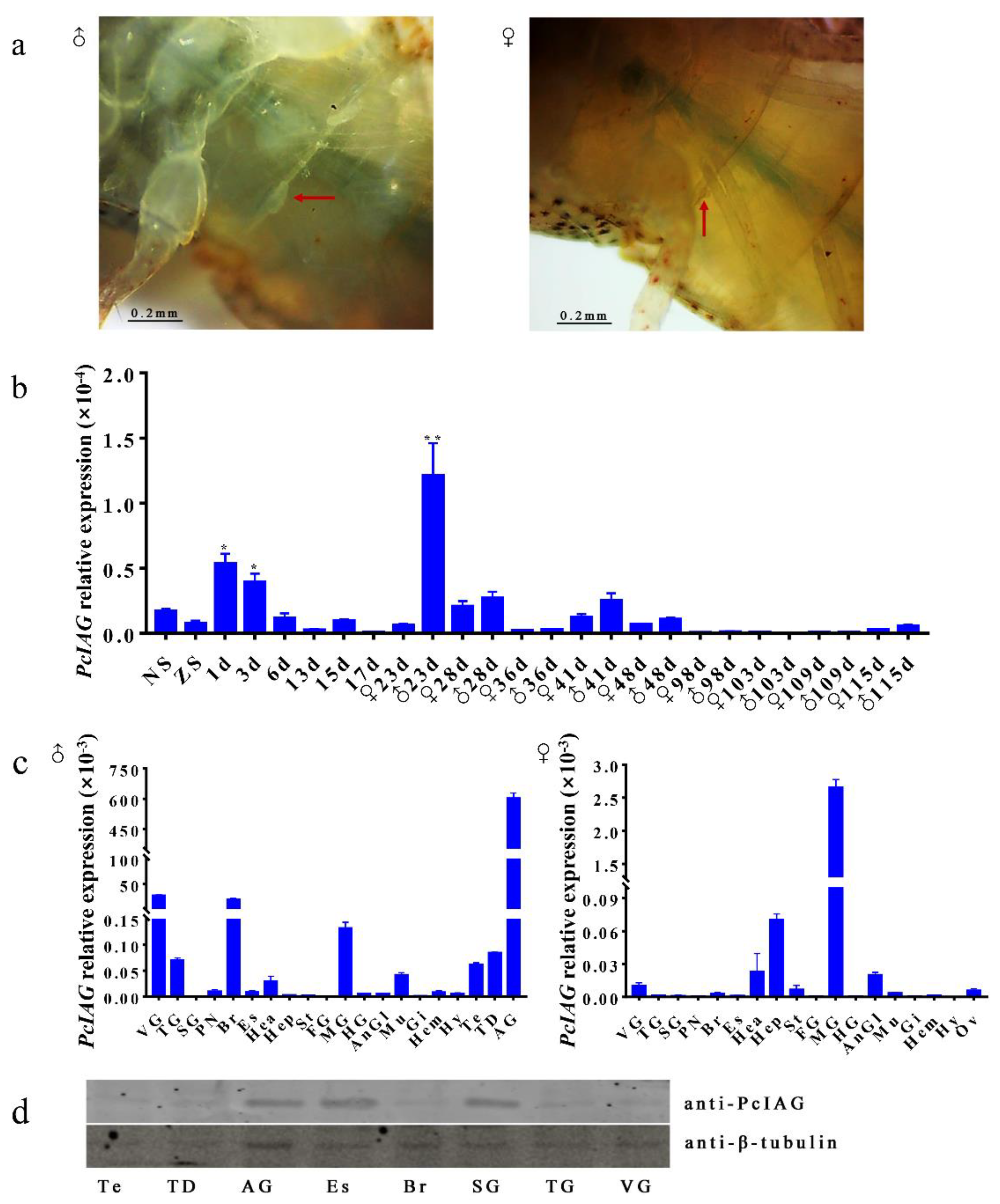
3.3. Expression Profile of PcIAG Gene in Different Developmental Stages of P. clarkii and Adult Crayfish
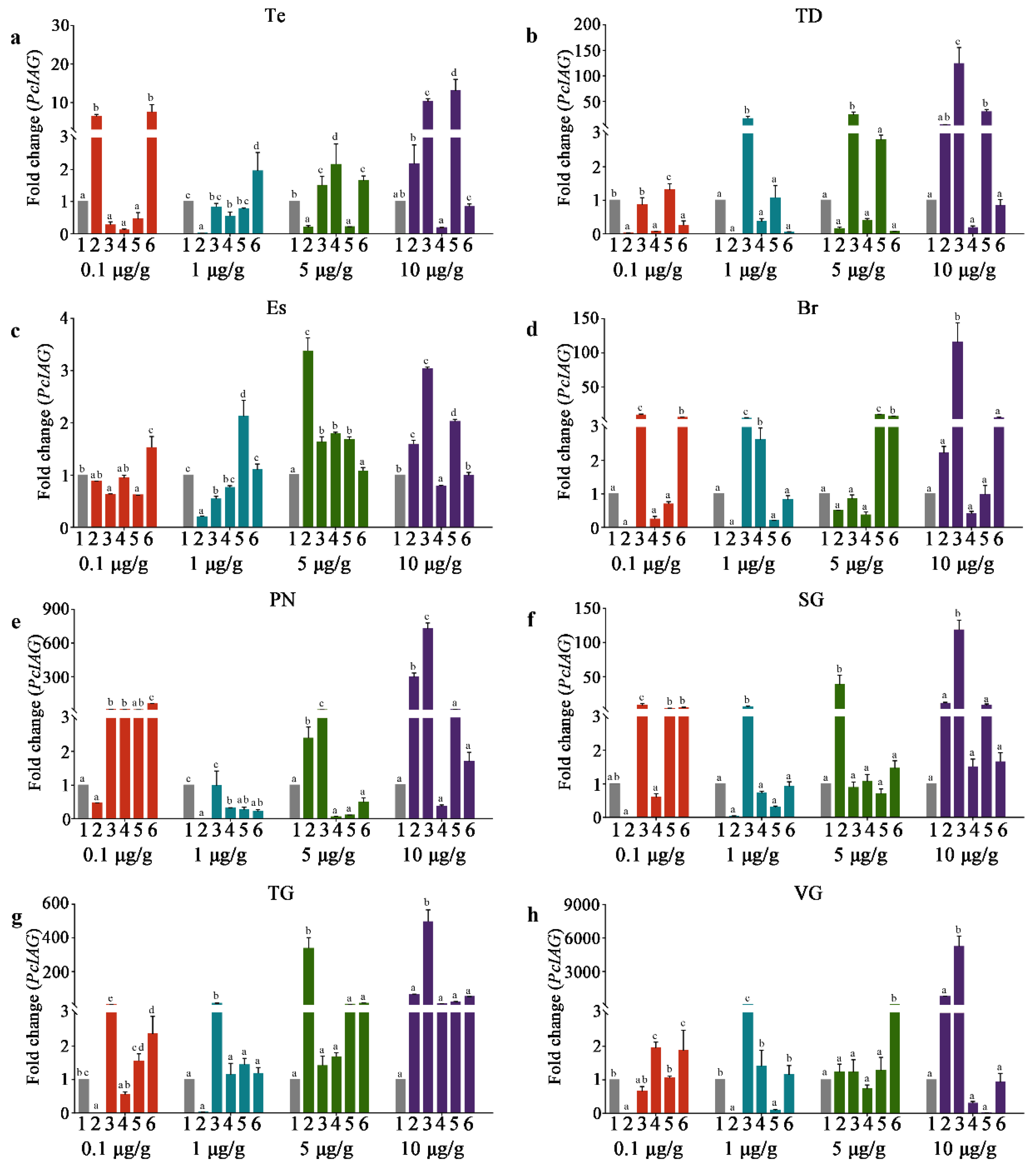
3.4. The PcIAG Gene Expression Profile in AG after RNAi
3.5. The PcIAG Gene Expression Profile in Other Tissues after RNAi
3.6. The Expression Profile of PcSxl after RNAi
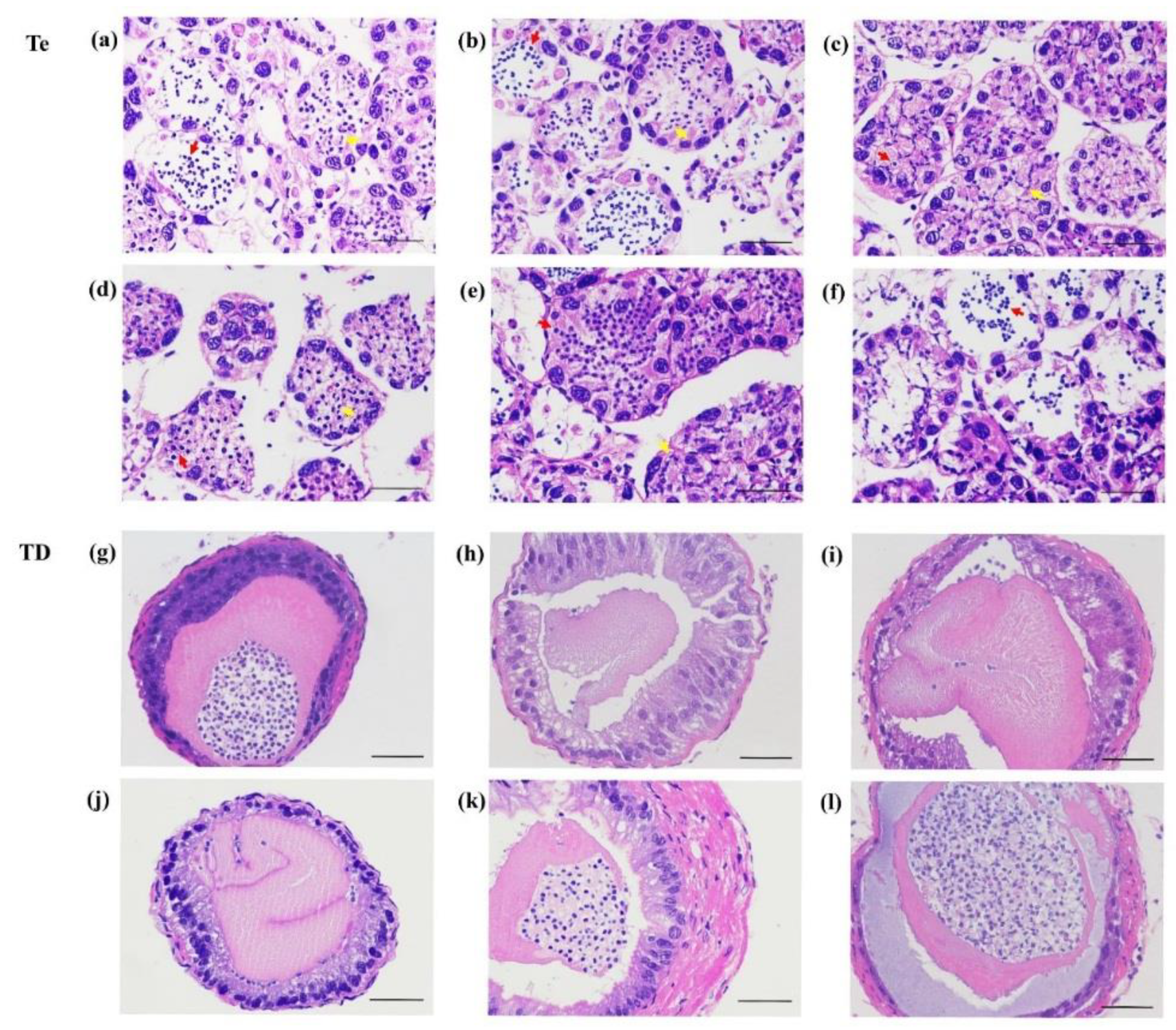
3.7. The Gonadal Development after Injection of dsRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harlıoğlu, M.M.; Farhadi, A.; Gür, S. Determination of sperm quality in decapod crustaceans. Aquaculture 2018, 490, 185–193. [Google Scholar] [CrossRef]
- Chandler, J.C.; Elizur, A.; Ventura, T. The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 2017, 825, 1–20. [Google Scholar] [CrossRef]
- Cui, Z.; Hui, M.; Liu, Y.; Song, C.; Li, X.; Li, Y.; Liu, L.; Shi, G.; Wang, S.; Li, F.; et al. High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 2015, 115, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Scalici, M.; Solano, E.; Gibertini, G. Karyological Analyses on the Australian Crayfish Cherax destructor (Decapoda: Parastacidae). J. Crustacean Biol. 2010, 30, 528–530. [Google Scholar] [CrossRef]
- Guo, Q.; Li, S.; Lv, X.; Xiang, J.H.; Sagi, A.; Manor, R.; Li, F.H. A putative insulin-like androgenic gland hormone receptor gene specifically expressed in male Chinese shrimp. Endocrinology 2018, 159, 2173–2185. [Google Scholar] [CrossRef] [PubMed]
- Okuno, A.; Hasegawa, Y.; Nagasawa, H. Purification and properties of androgenic gland hormone from the terrestrial isopod Armadillidium vulgare. Zoologicalence 1997, 14, 837–842. [Google Scholar] [CrossRef]
- Manor, R.; Weil, S.; Oren, S.; Glazer, L.; Afalo, E.D.; Ventura, T.; Chalia-Caspi, V.; Lapidot, M.; Sagi, A. Insulin and gender: An insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocr. 2007, 150, 326–336. [Google Scholar] [CrossRef]
- Li, F.J.; Bai, H.K.; Zhang, W.Y.; Fu, H.T.; Jiang, F.W.; Liang, G.X.; Jin, S.B.; Sun, S.M.; Qiao, H. Cloning of genomic sequences of three crustacean hyperglycemic hormone superfamily genes and elucidation of their roles of regulating insulin-like androgenic gland hormone gene. Gene 2015, 561, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Kubota, N.; Hojo, H.; Okada, A.; Kotaka, S.; Tsutsui, N.; Ohira, T. Direct evidence for the function of crustacean insulin-like androgenic gland factor (IAG): Total chemical synthesis of IAG. Bioorgan. Med. Chem. 2014, 22, 5783–5789. [Google Scholar] [CrossRef]
- Aizen, J.; Chandler, J.C.; Fitzgibbon, Q.P.; Sagi, A.; Battaglene, S.C.; Elizur, A.; Ventura, T. Production of recombinant insulin-like androgenic gland hormones from three decapod species: In vitro testicular phosphorylation and activation of a newly identified tyrosine kinase receptor from the Eastern spiny lobster, Sagmariasus verreauxi. Gen. Comp. Endocr. 2016, 229, 8–18. [Google Scholar] [CrossRef]
- Lawrence, A.; Green, S.; Chung, J.S. Isolation and tissue distribution of an insulin-like androgenic gland hormone (IAG) of the male red deep-sea crab. Chaceon Quinquedens Mar. Drugs 2017, 15, 241. [Google Scholar] [CrossRef] [PubMed]
- Levy, T.; Rosen, O.; Simons, O.; Savaya, A.A.; Sagi, A. The gene encoding the insulin-like androgenic gland hormone in an all-female parthenogenetic crayfish. PLoS ONE 2017, 12, e0189982. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.S.; Ye, H.H.; Huang, H.Y.; Yang, Y.N.; Gong, J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Deng, W.; Yang, K.L.; Wang, W.M. The expression of prophenoloxidase mRNA in red swamp crayfish, Procambarus clarkii, when it was challenged. Genomics 2012, 99, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Taketomi, Y. Ultrastructure of the androgenic gland of the crayfish, Procambarus clarkii. Cell Biol. Int. Rep. 1986, 10, 131–136. [Google Scholar] [CrossRef]
- Nagamine, C.; Knight, A. Masculinization of female crayfish, Procambarus clarki (Girard). Int. J. Invertebr. Reprod. 1987, 11, 77–87. [Google Scholar] [CrossRef]
- Taketomi, Y.; Murata, M.; Miyawaki, M. Androgenic gland and secondary sexual characters in the crayfish Procambarus clarkii. J. Crustacean Biol. 1990, 10, 492–497. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weils, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn. Macrobrachium Rosenb. Biol. Reprod. 2012, 86, 1–6. [Google Scholar] [CrossRef]
- Barki, A.; Karplus, I.; Manor, R.; Sagi, A. Intersexuality and behavior in crayfish: The de-masculinization effects of androgenic gland ablation. Horm. Behav. 2006, 50, 322–331. [Google Scholar] [CrossRef]
- Sagi, A.; Cohen, D. Growth, maturation and progeny of sex-reversed Macrobrachium rosenbergii males. World Aquacult 1990, 21, 87–90. [Google Scholar]
- Shi, L.L.; Yi, S.K.; Li, Y.H. Genome survey sequencing of red swamp crayfish Procambarus clarkii. Mol. Biol. Rep. 2018, 45, 799–806. [Google Scholar] [CrossRef]
- Li, Y.H.; Zheng, F.L.; Chen, H.Q.; Wang, H.Z.; Wang, L.Q.; Xu, D.Q. Cloning and sequence analysis of prophenoloxidase from haemocytes of the red swamp crayfish. Procambarus clarkii Agr. Sci. China. 2009, 8, 369–379. [Google Scholar] [CrossRef]
- Tan, H.Y.; Shao, G.M.; Kang, P.F.; Wang, Y.F. Full-length normalization subtractive hybridization analysis provides new insights into sexual precocity and ovarian development of red swamp crayfish. Procambarus clarkii. Aquac. 2017, 2017. 468, 417–427. [Google Scholar] [CrossRef]
- Geng, R.J.; Liu, H.; Wang, W.M. Differential expression of six Rnase2 and three Rnase3 paralogs identified in blunt snout bream in response to aeromonas hydrophila infection. Genes 2018, 9, 95. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Khalaila, I.; Rosen, O.; Sagi, A. Expression of an androgenic gland-specific insulin-like peptide during the course of prawn sexual and morphotypic differentiation. ISRN. Endocrinology 2011, 6, 476283. [Google Scholar]
- Okuno, A.; Hasegawa, Y.; Ohira, T.; Nagasawa, H. Characterization and cDNA cloning of androgenic gland hormone of the terrestrial isopod. Armadillidium vulgare. Biochem. Bioph. Res. Commun. 1999, 264, 419–423. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Qiao, K.; Wang, S.P.; Peng, H.; Shan, Z.G.; Wang, K.J. Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab. Scylla paramamosain. Aquac. 2014, 433, 325–334. [Google Scholar] [CrossRef]
- Scholtz, G.; Braband, A.; Tolley, L.; Reimann, A.; Mittmann, B.; Lukhaup, C.; Steuerwald, F.; Vogt, G. Ecology: Parthenogenesis in an outsider crayfish. Nature 2003, 421, 806. [Google Scholar] [CrossRef]
- Vogt, G.; Falckenhayn, C.; Schrimpf, A.; Schmid, K.; Hanna, K.; Panteleit, J.; Helm, M.; Schulz, R.; Lyko, F. The marbled crayfish as a paradigm for saltational speciation by autopolyploidy and parthenogenesis in animals. Biol. Open 2015, 4, 1583–1594. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.S.; Manor, R.; Sagi, A. Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: Implications for eyestalk regulation of IAG expression. Gen. Comp. Endocr. 2011, 173, 4–10. [Google Scholar] [CrossRef]
- Song, K.; Xu, T.S.; Zang, Y.N.; Serwadda, A.; Dai, T.H.; Ma, Y.C.; Shen, H.S. Insulin-like androgenic gland hormone gene in the freshwater Chinese mitten crab Eriocheir sinensis: cDNA cloning, expression pattern, and interaction with EsIGFBP7. Turk. J. Fish. Aquat. Sci. 2018, 18, 17–25. [Google Scholar] [CrossRef]
- Nef, S.; Verma-Kurvari, S.; Merenmies, J.; Vassalli, J.; Efstratiadis, A.; Accili, D.; Parada, L.F. Testis determination requires insulin receptor family function in mice. Nature 2003, 426, 291. [Google Scholar] [CrossRef]
- Li, F.J.; Bai, H.K.; Xiong, Y.W.; Fu, H.T.; Jiang, S.F.; Jiang, F.W.; Jin, S.B.; Sun, S.M.; Qiao, H.; Zhang, W.Y. Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gene. Gen. Comp. Endocrinol. 2015, 216, 152–160. [Google Scholar] [CrossRef]
- Garza-Torres, R.; Campos-Ramos, R.; Maeda-Martínez, A.M. Organogenesis and subsequent development of the genital organs in female and male Pacific white shrimp Penaeus (Litopenaeus) vannamei. Aquaculture 2009, 296, 136–142. [Google Scholar] [CrossRef]
- Hasuwa, H.; Kaseda, K.; Einarsdottir, T.; Okabe, M. Small interfering RNA and gene silencing in transgenic mice and rats. Febs. Let. 2002, 532, 227–230. [Google Scholar] [CrossRef]
- Dorsett, T.; Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Disco 2004, 3, 318–329. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, C.C.; Choy, K.W.; Du, Q.; Chen, J.; Wang, Q.; Li, L.; Chung, T.K.H.; Tang, T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538, 217–227. [Google Scholar] [CrossRef]
- Hutvágner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002, 12, 225–232. [Google Scholar] [CrossRef]
- Escobedo-Bonilla, C.M. Application of RNA Interference (RNAi) against Viral Infections in Shrimp: A Review. J. Antivir. Antiretrovir. 2011. [Google Scholar] [CrossRef]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.W.; Man, X.; Huang, X.; Wang, Y.; Song, Q.S.; Hui, K.M.; Zhang, H.W. Identification of a C-type lectin possessing both antibacterial and antiviral activities from red swamp crayfish. Fish Shellfish Immu. 2018, 77, 22–30. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Y.H.; Wang, Z.; Ren, Q. Novel myeloid differentiation factor 88, EsMyD88, exhibits EsTube-binding activity in Chinese mitten crab Eriocheir sinensis. Dev. Comp. Immunol. 2014, 47, 298–308. [Google Scholar] [CrossRef]
- Xue, S.X.; Yang, W.J.; Sun, J.S. Role of chymotrypsin-like serine proteinase in white spot syndrome virus infection in Fenneropenaeus chinensis. Fish Shellfish. Immunol. 2013, 34, 403–409. [Google Scholar] [CrossRef]
- Wargelius, A.; Ellingsen, S.; Fjose, A. Double–stranded RNA induces specific developmental defects in Zebrafish embryos. Biochem. Bioph. Res. Commun. 1999, 263, 156–161. [Google Scholar] [CrossRef]
- Lagos, D.; Ruiz, M.F.; Sánchez, L.; Komitopoulou, K. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 2005, 348, 111–121. [Google Scholar] [CrossRef]
- Glenner, H.; Thomsen, P.F.; Hebsgaard, M.B.; Sørensen, M.V.; Willerslev, E. Evolution. The origin of insects. Science 2006, 314, 1883–1884. [Google Scholar] [CrossRef]
- López-Cuadros, I.; García-Gasca, A.; Gomez-Anduro, G.; Escobedo-Freqoso, C.; Ller-Herrera, R.A.; Ibarra, A.M. Isolation of the sex-determining gene Sex-lethal (Sxl) in Penaeus (Litopenaeus) vannamei (Boone, 1931) and characterization of its embryogenic, gametogenic, and tissue-specific expression. Gene 2018, 668, 33–47. [Google Scholar] [CrossRef]
- Chen, P.; Xu, S.L.; Zhou, W.; Guo, X.G.; Wang, C.L.; Wang, D.L.; Zhao, Y.L. Cloning and expression analysis of a transformer gene in Daphnia pulex during different reproduction stages. Anim. Reprod. Sci. 2014, 146, 227–237. [Google Scholar] [CrossRef]
- Li, S.H.; Li, F.H.; Yu, K.J.; Xiang, J.H. Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 2018, 649, 1–7. [Google Scholar] [CrossRef]
- Chandler, J.C.; Fitzgibbon, Q.P.; Smith, G.; Elizur, A.; Ventura, T. Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: The first invertebrate sex-linked Dmrt. Dev. Biol. 2017, 430, 337–345. [Google Scholar] [CrossRef]
- Yu, Y.Q.; Ma, W.M.; Zeng, Q.G.; Qian, Y.Q.; Yang, J.S.; Yang, W.J. Molecular cloning and sexually dimorphic expression of two Dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agric. Res. 2014, 3, 181–191. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Qiao, H.; Zhang, W.Y.; Sun, S.M.; Jiang, S.F.; Gong, Y.S.; Xiong, Y.W.; Jin, S.B.; Fu, H.T. Molecular cloning and expression analysis of two sex-lethal homolog genes during development in the oriental river prawn, Macrobrachium nipponense. Genet. Mol. Res. 2013, 12, 4698–4711. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Han, S.; Fei, J.; Zhang, L.; Ray, J.W.; Wang, W.; Li, Y. Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii. Genes 2019, 10, 645. https://doi.org/10.3390/genes10090645
Shi L, Han S, Fei J, Zhang L, Ray JW, Wang W, Li Y. Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii. Genes. 2019; 10(9):645. https://doi.org/10.3390/genes10090645
Chicago/Turabian StyleShi, Linlin, Shuxin Han, Jiamin Fei, Long Zhang, Jonathan W Ray, Weimin Wang, and Yanhe Li. 2019. "Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii" Genes 10, no. 9: 645. https://doi.org/10.3390/genes10090645
APA StyleShi, L., Han, S., Fei, J., Zhang, L., Ray, J. W., Wang, W., & Li, Y. (2019). Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii. Genes, 10(9), 645. https://doi.org/10.3390/genes10090645



