Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Vector Construction and Plant Transformation
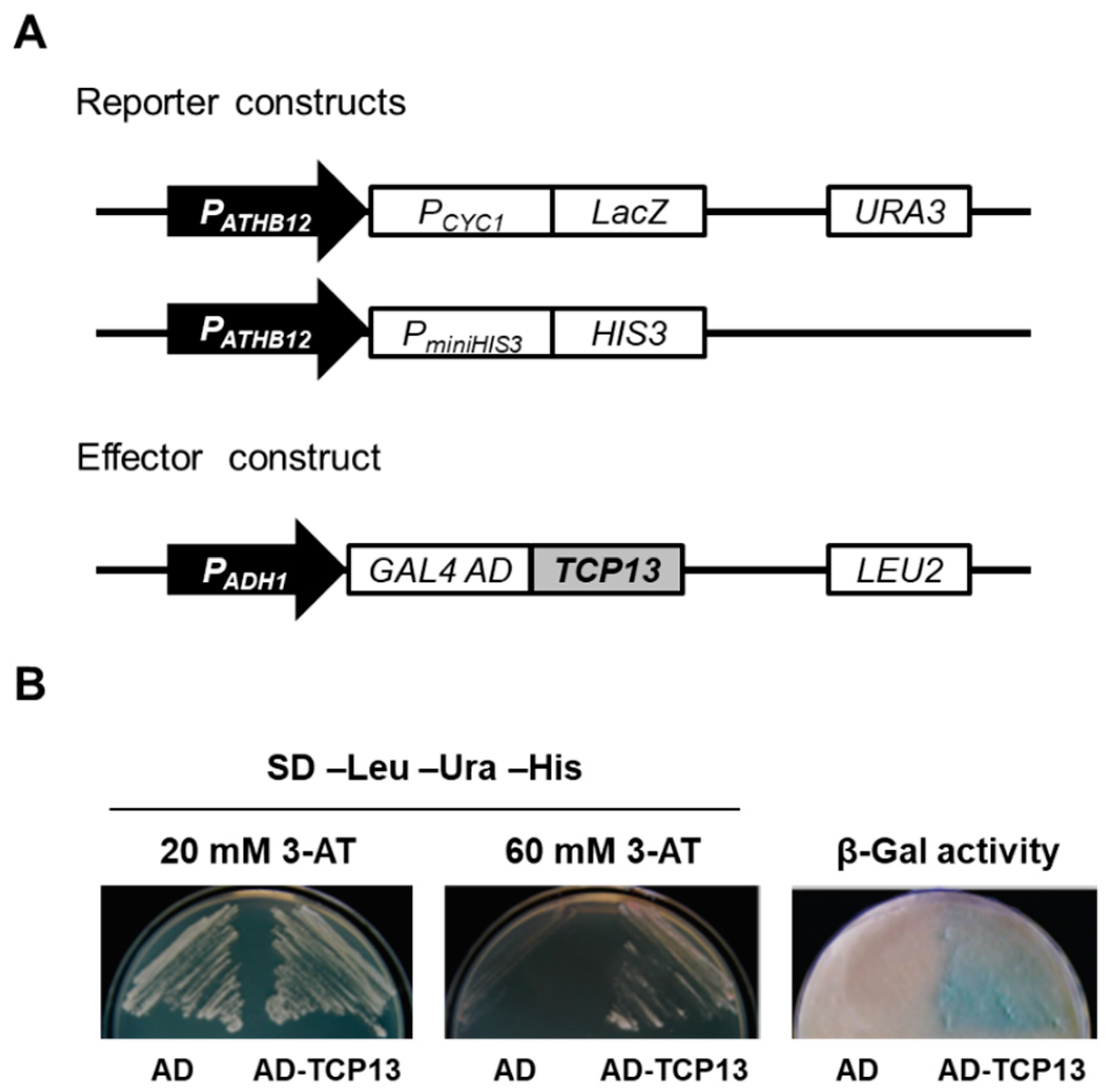
2.3. Yeast One-Hybrid (Y1H) Screening and Yeast Two-Hybrid (Y2H) Assays
2.4. Bimolecular Fluorescence Complementation (BiFC)
2.5. Microscopic Observation
2.6. Real-Time Quantitative PCR
2.7. GUS Assay
2.8. Chromatin Immunoprecipitation (ChIP)-qPCR
2.9. Luciferase Assay
3. Results
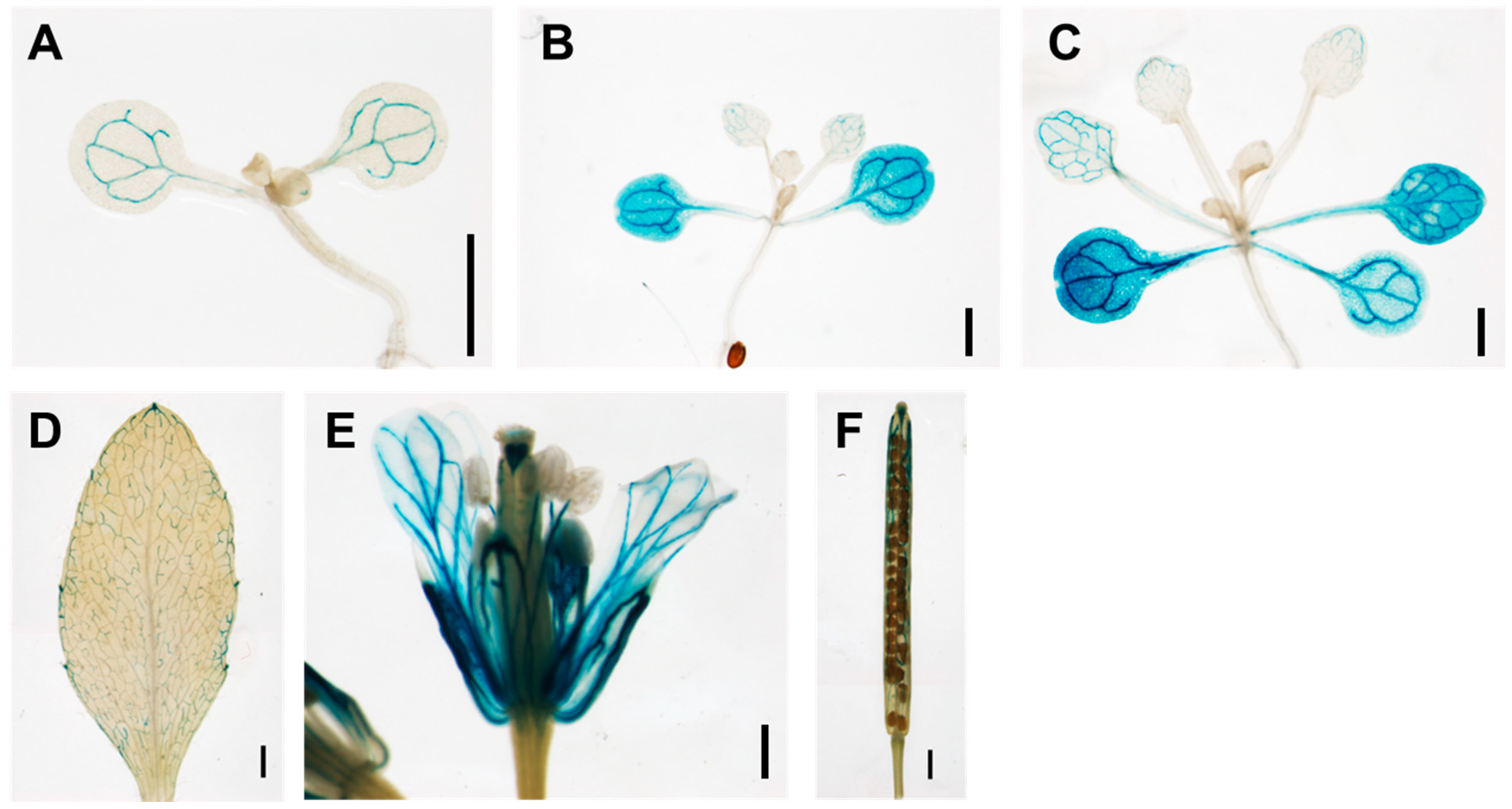
3.1. TCP13 Is an Upstream Regulator of ATHB12
3.2. Alterations of TCP13 Level Affect Leaf Morphology
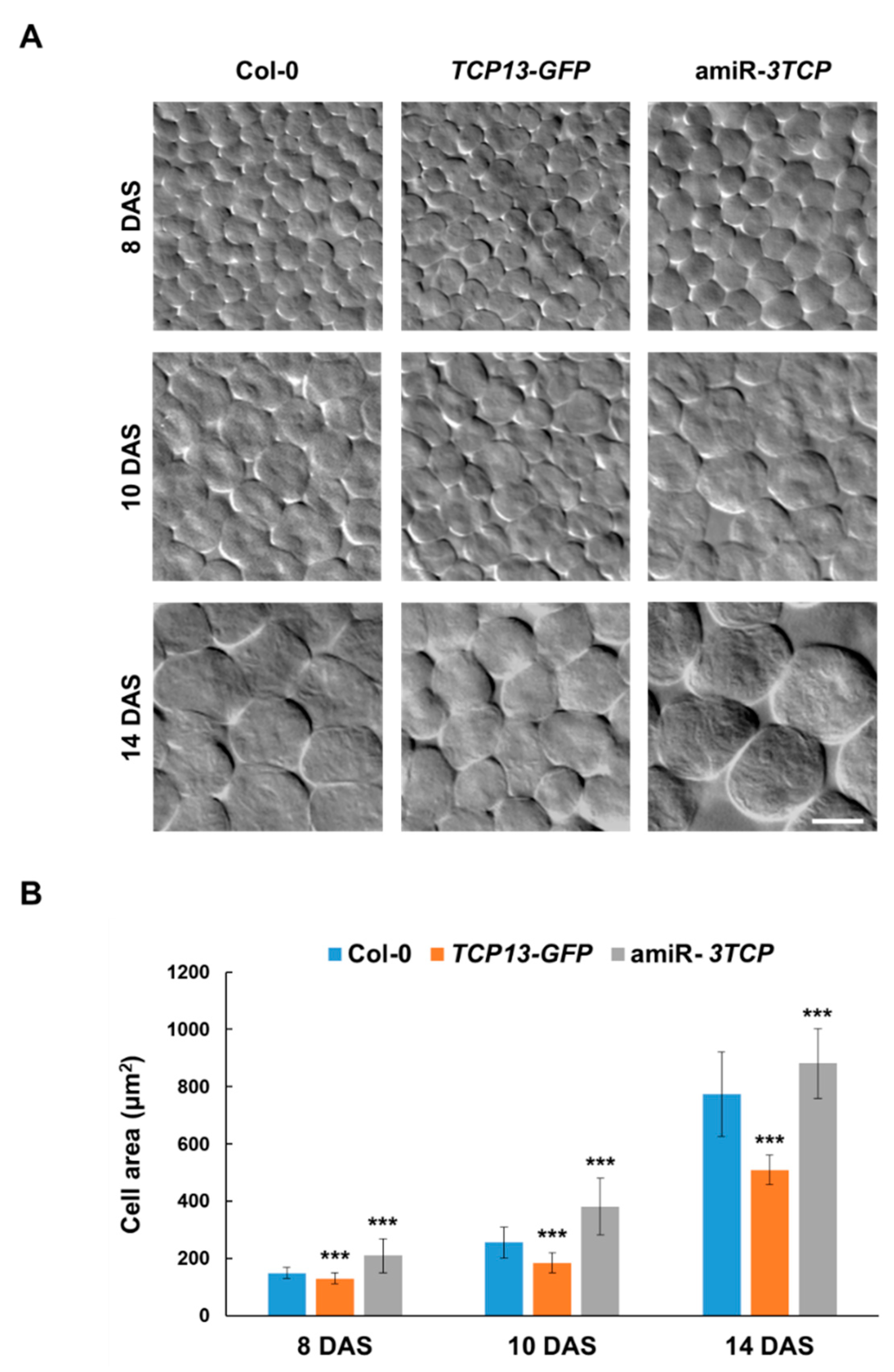
3.3. TCP13 Controls Leaf Growth Mainly by Repressing Cell Expansion
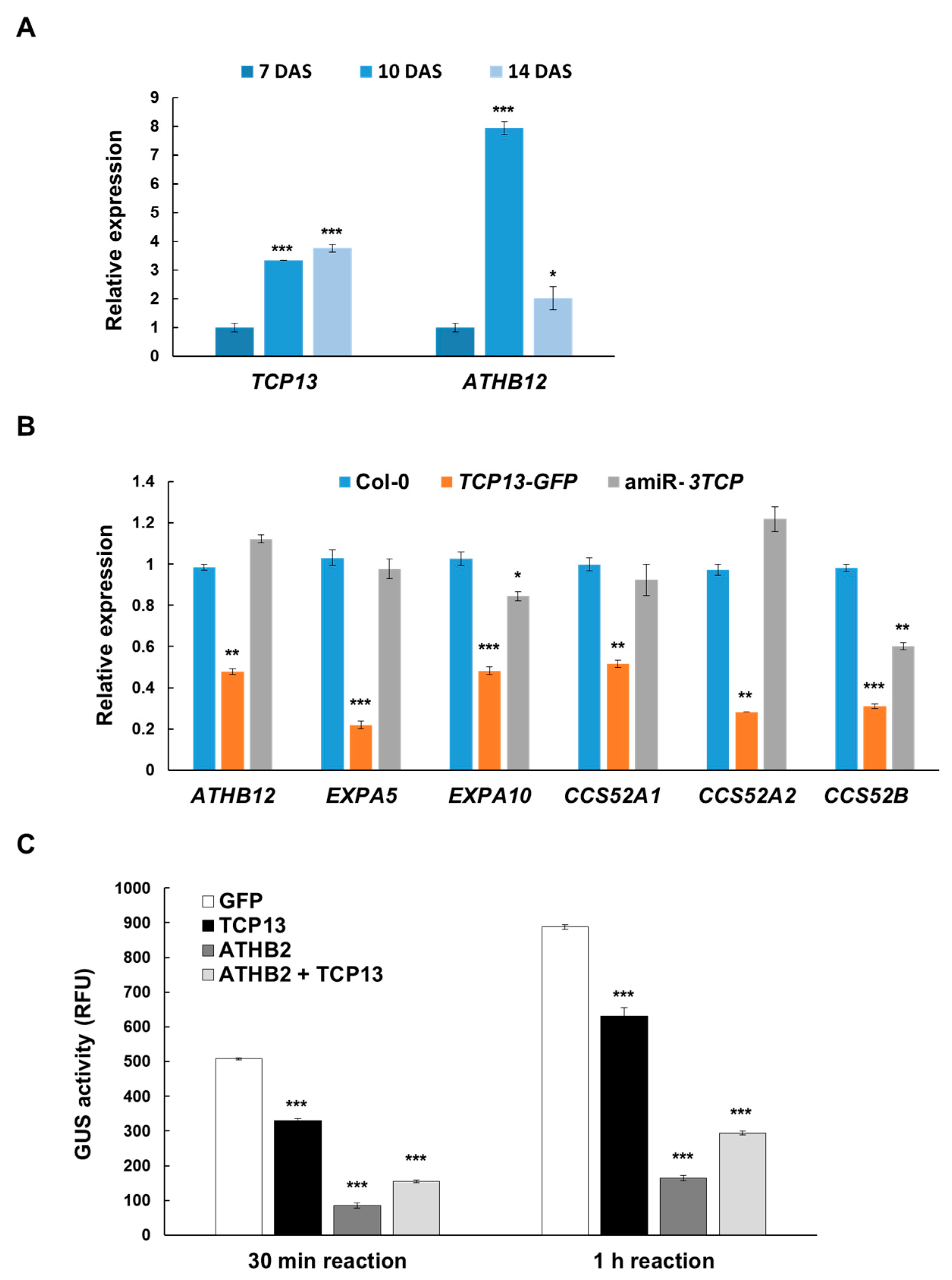
3.4. TCP13 Negatively Controls ATHB12 Expression
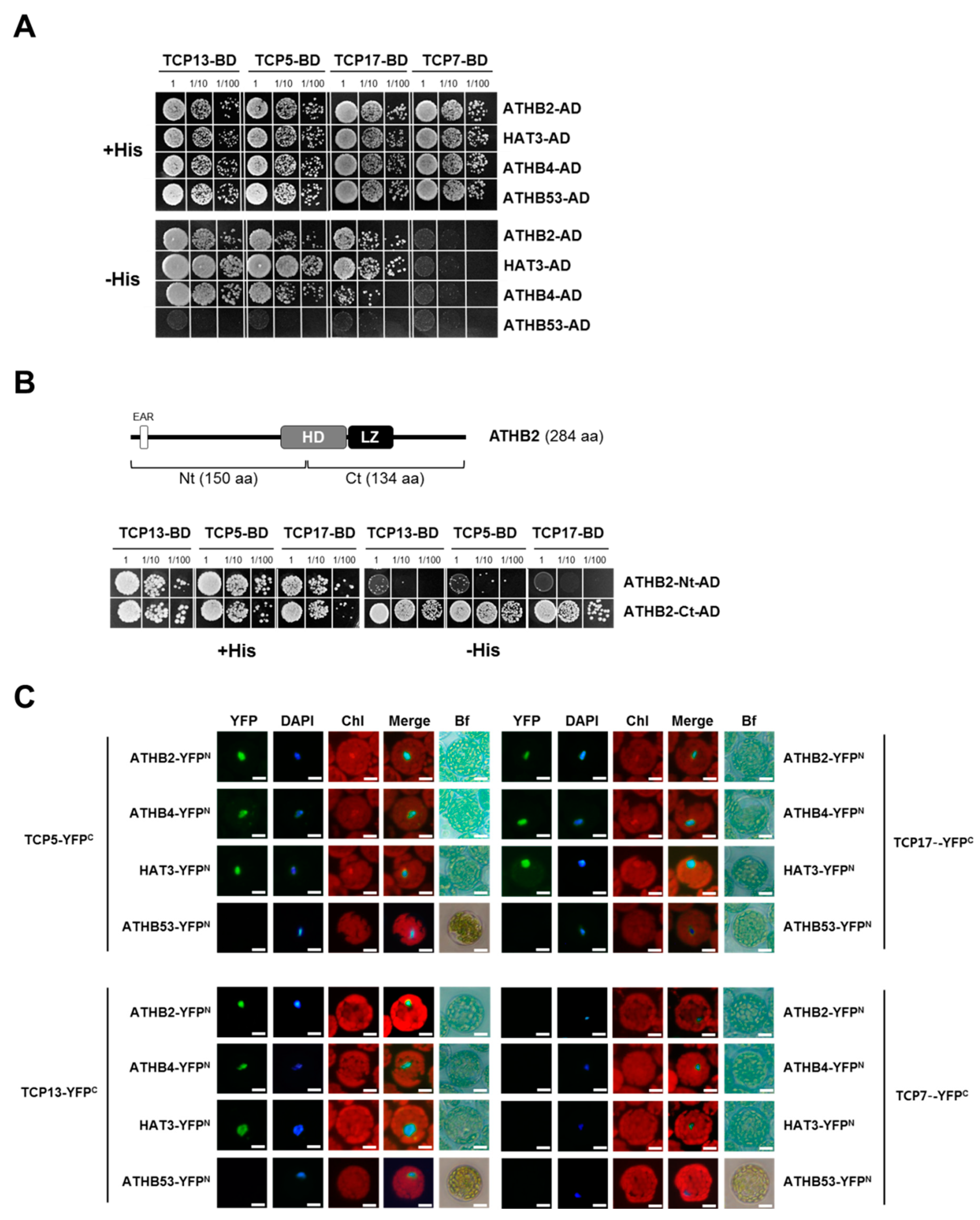
3.5. TCP13 Interacts with ATHB2
3.6. TCP13 Binds to the Promoter of ATHB12 in Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nelissen, H.; Gonzalez, N.; Inze, D. Leaf growth in dicots and monocots: So different yet so alike. Curr. Opin. Plant Biol. 2016, 33, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, Y.; Tsukaya, H. Behavior of leaf meristems and their modification. Front. Plant Sci. 2015, 6, 1060. [Google Scholar] [CrossRef] [PubMed]
- Sluis, A.; Hake, S. Organogenesis in plants: Initiation and elaboration of leaves. Trends Genet. 2015, 31, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Sarvepalli, K.; Das Gupta, M.; Challa, K.R.; Nath, U. Molecular cartography of leaf development—Role of transcription factors. Curr. Opin. Plant Biol. 2019, 47, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Selden, K.; Bediee, A.; Rolland, G.; Baumberger, N.; Noir, S.; Bach, L.; Lamy, G.; Granier, C.; Genschik, P. SIAMESE-RELATED1 is regulated posttranslationally and participates in repression of leaf growth under moderate drought. Plant Physiol. 2018, 176, 2834–2850. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Vanhaeren, H.; Inze, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef]
- Hur, Y.S.; Um, J.H.; Kim, S.; Kim, K.; Park, H.J.; Lim, J.S.; Kim, W.Y.; Jun, S.E.; Yoon, E.K.; Lim, J.; et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 2015, 205, 316–328. [Google Scholar] [CrossRef]
- Hake, S.; Smith, H.M.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The role of KNOX genes in plant development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef]
- Hay, A.; Tsiantis, M. KNOX genes: Versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef]
- Schena, M.; Davis, R.W. Structure of homeobox-leucine zipper genes suggests a model for the evolution of gene families. Proc. Natl. Acad. Sci. USA 1994, 91, 8393–8397. [Google Scholar] [CrossRef]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Johansson, H.; Hanson, J.; Engstrom, P.; Soderman, E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Steindler, C.; Matteucci, A.; Sessa, G.; Weimar, T.; Ohgishi, M.; Aoyama, T.; Morelli, G.; Ruberti, I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 1999, 126, 4235–4245. [Google Scholar] [PubMed]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Kunihiro, A.; Yamashino, T.; Nakamichi, N.; Niwa, Y.; Nakanishi, H.; Mizuno, T. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Merelo, P.; Ram, H.; Pia Caggiano, M.; Ohno, C.; Ott, F.; Straub, D.; Graeff, M.; Cho, S.K.; Yang, S.W.; Wenkel, S.; et al. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA 2016, 113, 11973–11978. [Google Scholar] [CrossRef]
- Lee, Y.H.; Chun, J.Y. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol. Biol. 1998, 37, 377–384. [Google Scholar] [CrossRef]
- Olsson, A.S.; Engstrom, P.; Soderman, E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef]
- Son, O.; Hur, Y.S.; Kim, Y.K.; Lee, H.J.; Kim, S.; Kim, M.R.; Nam, K.H.; Lee, M.S.; Kim, B.Y.; Park, J.; et al. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 2010, 51, 1537–1547. [Google Scholar] [CrossRef]
- Re, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Li, X.; Yang, R.; Gong, Y.; Chen, H. The Arabidopsis Mediator complex subunit MED19a is involved in ABI5-mediated ABA responses. J. Plant Biol. 2018, 61, 97–110. [Google Scholar] [CrossRef]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 2008, 20, 2293–2306. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr. Opin. Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Sarvepalli, K.; Nath, U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011, 67, 595–607. [Google Scholar] [CrossRef]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef]
- Challa, K.R.; Rath, M.; Nath, U. The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 2019, 15, e1007988. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Mitsuda, N.; Ikeda, M.; Takada, S.; Takiguchi, Y.; Kondou, Y.; Yoshizumi, T.; Fujita, M.; Shinozaki, K.; Matsui, M.; Ohme-Takagi, M. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 2145–2151. [Google Scholar] [CrossRef]
- Son, O.; Kim, S.; Hur, Y.S.; Cheon, C.I. Identification of the Raptor-binding motif on Arabidopsis S6 kinase and its use as a TOR signaling suppressor. Biochem. Biophys. Res. Commun. 2016, 472, 83–87. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Tsuge, T.; Tsukaya, H.; Uchimiya, H. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 1996, 122, 1589–1600. [Google Scholar]
- Rymen, B.; Coppens, F.; Dhondt, S.; Fiorani, F.; Beemster, G.T. Kinematic Analysis of Cell Division and Expansion. In Plant Developmental Biology Methods and Protocols; Hennig, L., Köhler, C., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 655, pp. 203–227. [Google Scholar]
- Lee, B.H.; Kwon, S.H.; Lee, S.J.; Park, S.K.; Song, J.T.; Lee, S.; Lee, M.M.; Hwang, Y.S.; Kim, J.H. The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol. Biol. 2015, 89, 529–538. [Google Scholar] [CrossRef]
- Saddic, L.A.; Huvermann, B.; Bezhani, S.; Su, Y.; Winter, C.M.; Kwon, C.S.; Collum, R.P.; Wagner, D. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 2006, 133, 1673–1682. [Google Scholar] [CrossRef]
- Nath, U.; Crawford, B.C.; Carpenter, R.; Coen, E. Genetic control of surface curvature. Science 2003, 299, 1404–1407. [Google Scholar] [CrossRef]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef]
- De Veylder, L.; Larkin, J.C.; Schnittger, A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011, 16, 624–634. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 2002, 29, 45–59. [Google Scholar] [CrossRef]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Shen, W.H.; Huang, H.; Dong, A. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 2012, 71, 99–107. [Google Scholar] [CrossRef]
- Tao, Q.; Guo, D.; Wei, B.; Zhang, F.; Pang, C.; Jiang, H.; Zhang, J.; Wei, T.; Gu, H.; Qu, L.J.; et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013, 25, 421–437. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, J.; Pang, C.; Yu, H.; Guo, D.; Jiang, H.; Ding, M.; Chen, Z.; Tao, Q.; Gu, H.; et al. The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 2015, 25, 121–134. [Google Scholar] [CrossRef]
- van Es, S.W.; Silveira, S.R.; Rocha, D.I.; Bimbo, A.; Martinelli, A.P.; Dornelas, M.C.; Angenent, G.C.; Immink, R.G.H. Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J. 2018, 94, 867–879. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hur, Y.-S.; Kim, J.; Kim, S.; Son, O.; Kim, W.-Y.; Kim, G.-T.; Ohme-Takagi, M.; Cheon, C.-I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes 2019, 10, 644. https://doi.org/10.3390/genes10090644
Hur Y-S, Kim J, Kim S, Son O, Kim W-Y, Kim G-T, Ohme-Takagi M, Cheon C-I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes. 2019; 10(9):644. https://doi.org/10.3390/genes10090644
Chicago/Turabian StyleHur, Yoon-Sun, Jiyoung Kim, Sunghan Kim, Ora Son, Woo-Young Kim, Gyung-Tae Kim, Masaru Ohme-Takagi, and Choong-Ill Cheon. 2019. "Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development" Genes 10, no. 9: 644. https://doi.org/10.3390/genes10090644
APA StyleHur, Y.-S., Kim, J., Kim, S., Son, O., Kim, W.-Y., Kim, G.-T., Ohme-Takagi, M., & Cheon, C.-I. (2019). Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes, 10(9), 644. https://doi.org/10.3390/genes10090644



