Gene Flow and Genetic Variation Explain Signatures of Selection across a Climate Gradient in Two Riparian Species
Abstract
1. Introduction
2. Materials and Methods
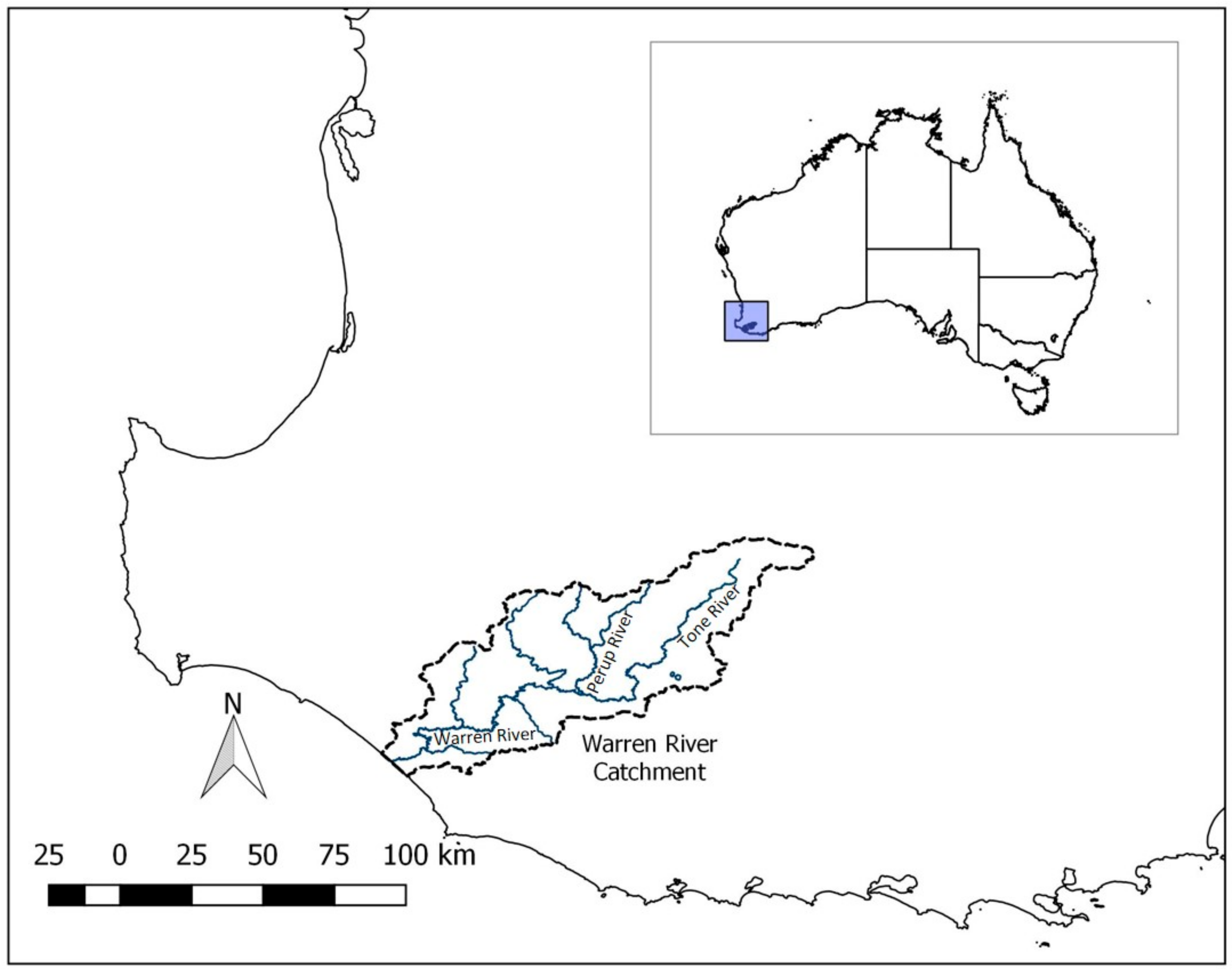
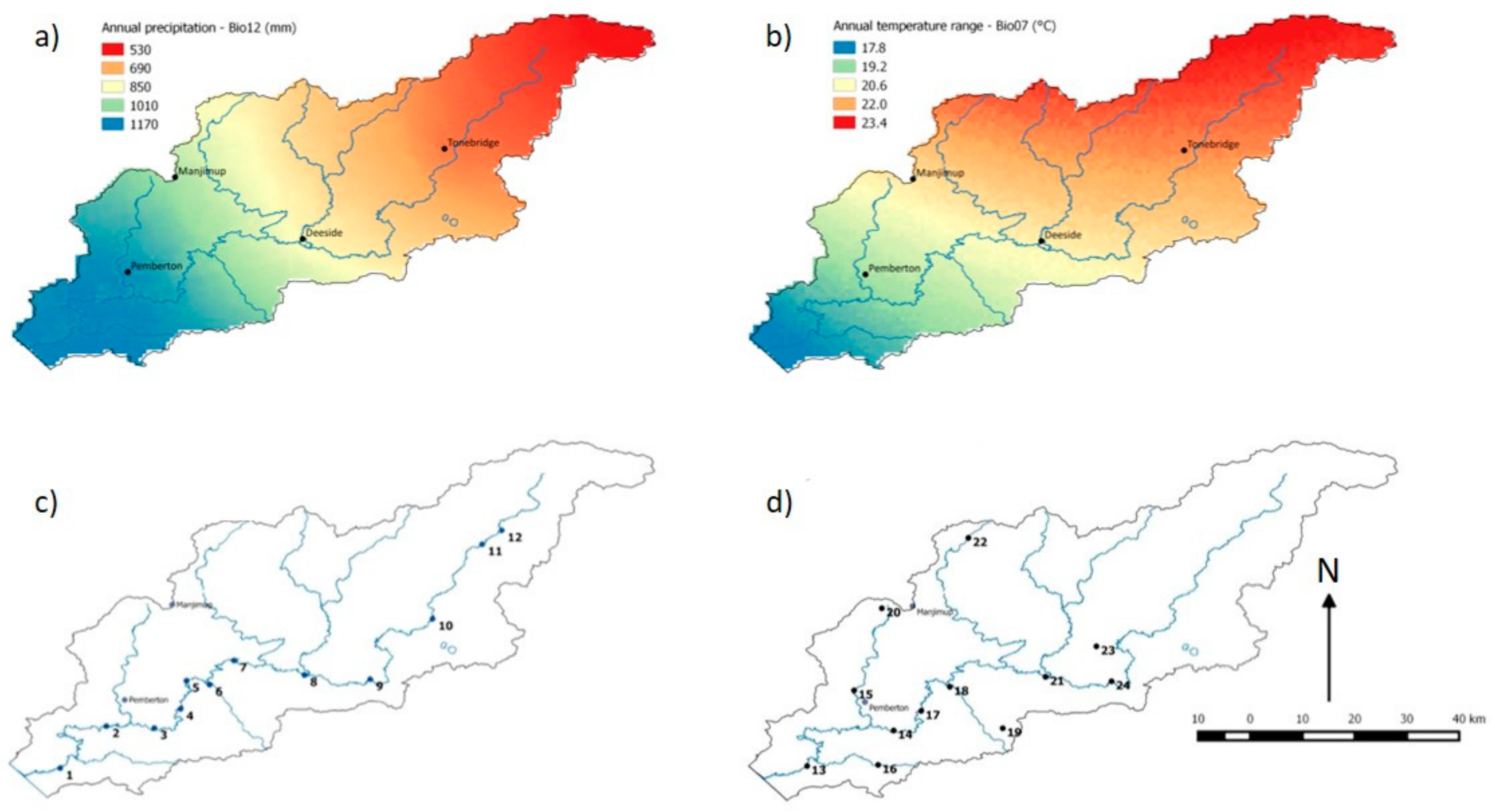
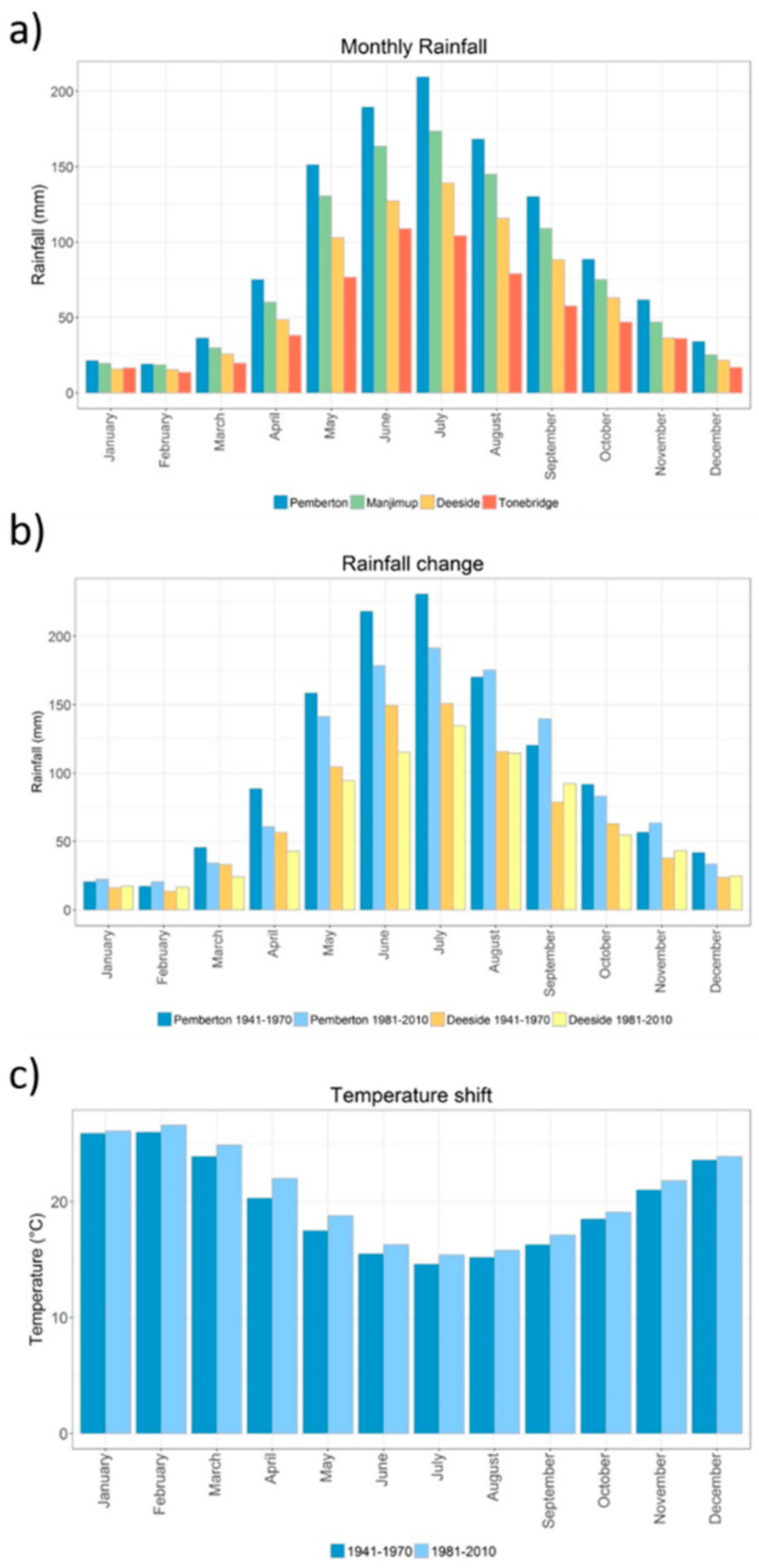
2.1. Study System and Sampling
2.2. Genomic Assay and Environmental Analysis
3. Results
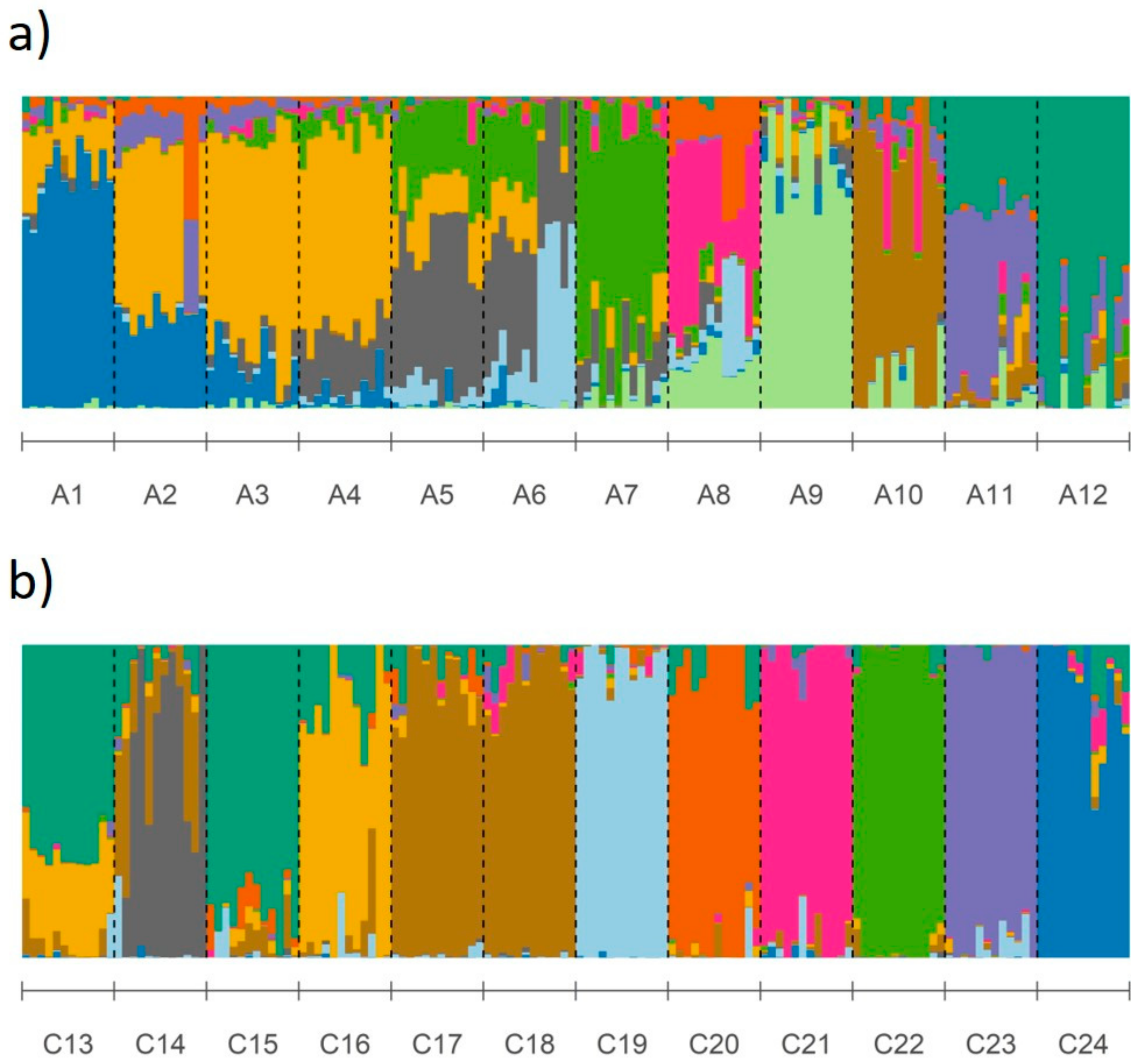
3.1. Genetic Differentiation and Population Structure
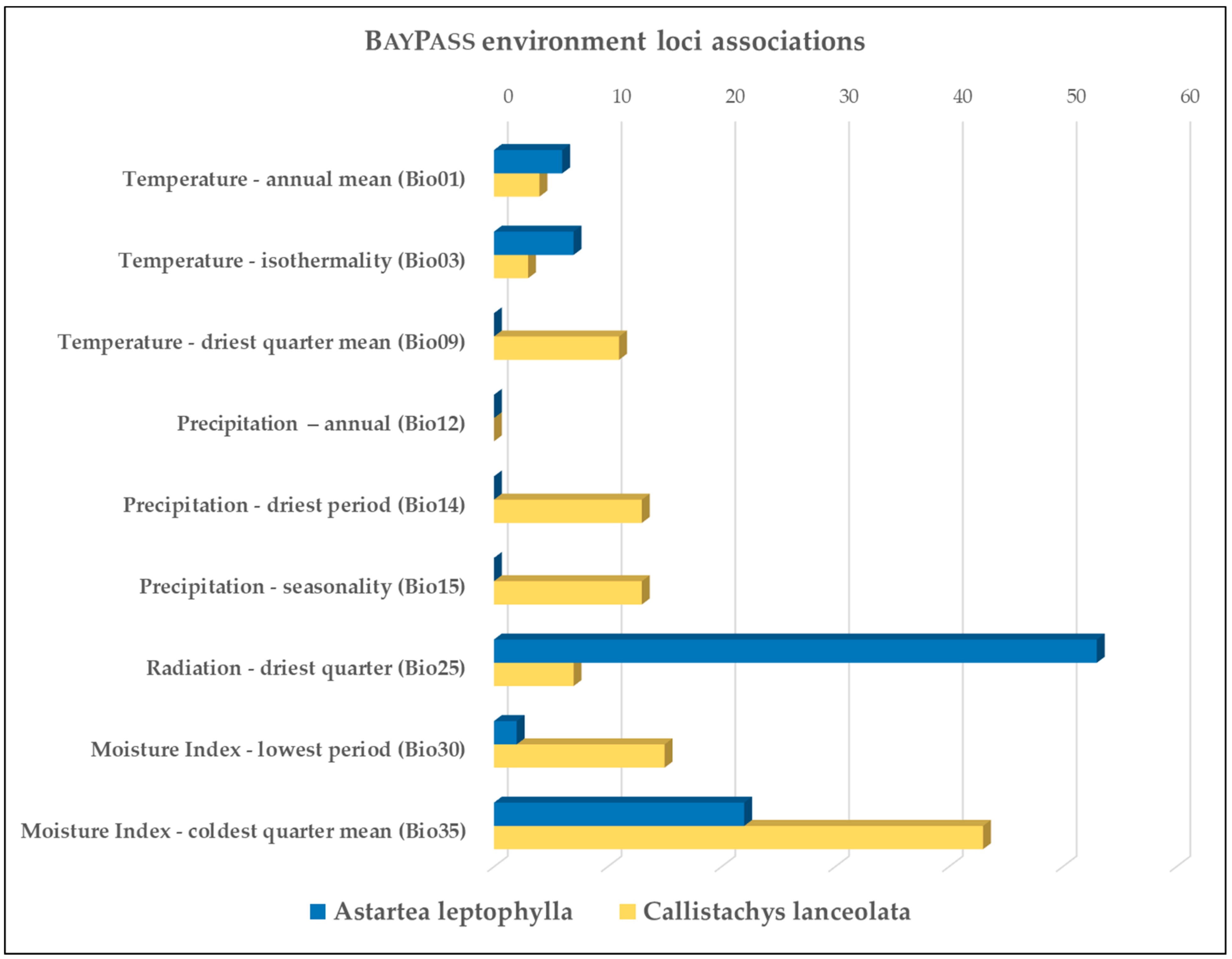
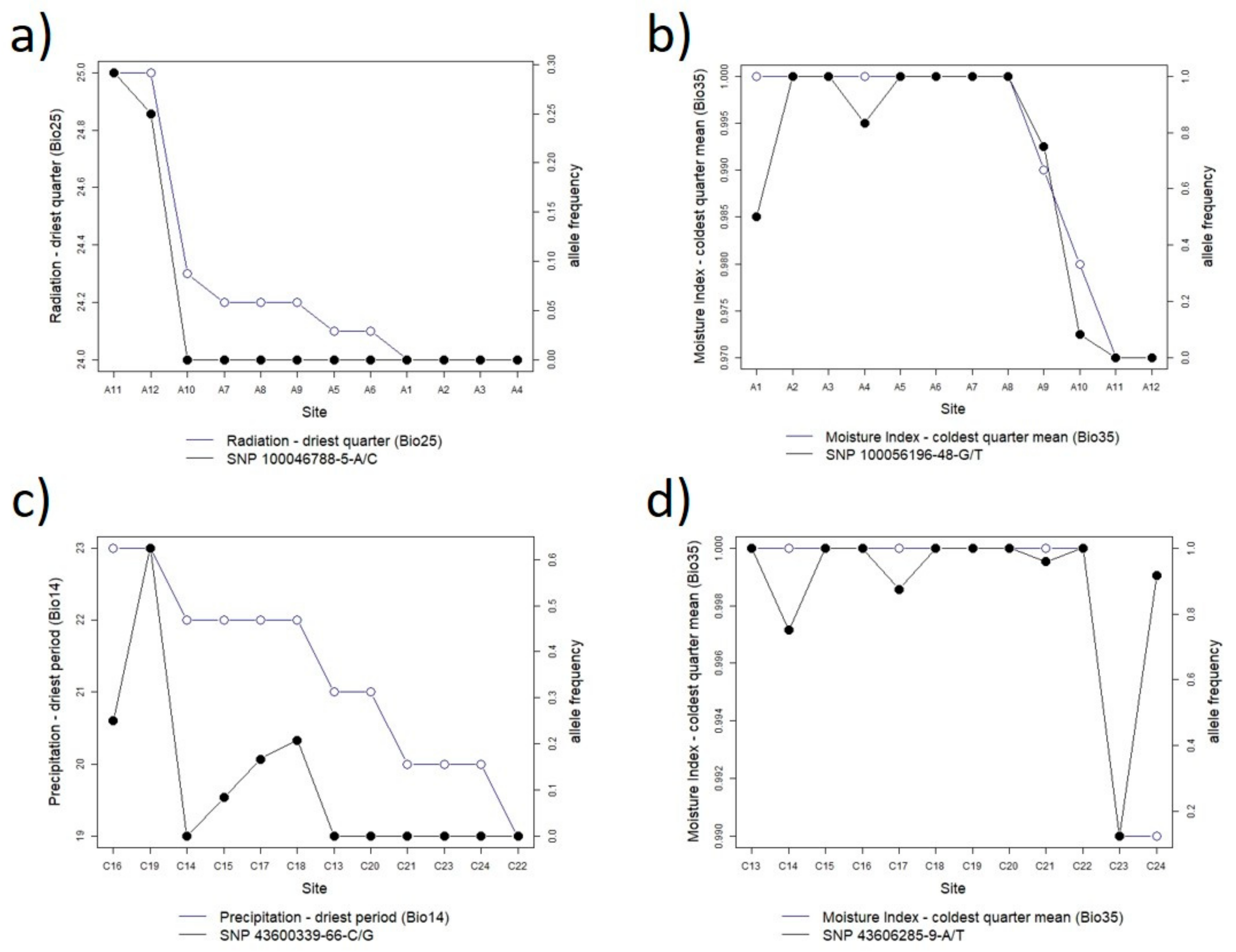
3.2. Environmental Associations
4. Discussion
4.1. Signals of Selection
4.2. Selection, Distribution and Gene Flow
4.3. Implications for Persistence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stocker, T.F.; Qin, D.; Plattner, G.-K.; Tignor, M.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; MidgleyStocker, P.M.; et al. (Eds.) Intergovernmental Panel on Climate Change Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013. [Google Scholar]
- Parmesan, C.; Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.B. Range Shifts and Adaptive Responses to Quaternary Climate Change. Science 2001, 292, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-C.; Hill, J.K.; Ohlemuller, R.; Roy, D.B.; Thomas, C.D. Rapid Range Shifts of Species Associated with High Levels of Climate Warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Nathan, R.; Horvitz, N.; He, Y.; Kuparinen, A.; Schurr, F.M.; Katul, G. Spread of North American wind-dispersed trees in future environments. Ecol. Lett. 2011, 14, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol. Appl. 2008, 1, 95–111. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Ikeda, D.H.; Max, T.L.; Allan, G.J.; Lau, M.K.; Shuster, S.M.; Whitham, T.G. Genetically informed ecological niche models improve climate change predictions. Glob. Chang. Biol. 2017, 23, 164–176. [Google Scholar] [CrossRef]
- Prober, S.M.; Byrne, M.; McLean, E.H.; Steane, D.A.; Potts, B.M.; Vaillancourt, R.E.; Stock, W.D. Climate-adjusted provenancing: A strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 2015, 3, 1–5. [Google Scholar] [CrossRef]
- Aitken, S.N.; Whitlock, M.C. Assisted Gene Flow to Facilitate Local Adaptation to Climate Change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Savolainen, O.; Lascoux, M.; Merilä, J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013, 14, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.; Schluter, D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008, 23, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 1985, 16, 393–430. [Google Scholar] [CrossRef]
- Nicotra, A.; Atkin, O.; Bonser, S.; Davidson, A.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.; Richards, C.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central–marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Hobbs, R.J.; Yates, C.J. Impacts of ecosystem fragmentation on plant populations: Generalising the idiosyncratic. Aust. J. Bot. 2003, 51, 471–488. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Ellstrand, N.C. Is gene flow the most important evolutionary force in plants? Am. J. Bot. 2014, 101, 737–753. [Google Scholar] [CrossRef]
- Tsumura, Y.; Uchiyama, K.; Moriguchi, Y.; Ueno, S.; Ihara-Ujino, T. Genome scanning for detecting adaptive genes along environmental gradients in the Japanese conifer, Cryptomeria japonica. Heredity 2012, 109, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Prunier, J.; Laroche, J.; Beaulieu, J.; Bousquet, J. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol. Ecol. 2011, 20, 1702–1716. [Google Scholar] [CrossRef] [PubMed]
- Namroud, M.-C.; Beaulieu, J.; Juge, N.; Laroche, J.; Bousquet, J. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol. Ecol. 2008, 17, 3599–3613. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, S.; Meirmans, P.G.; Aitken, S.N.; Ritland, K.; Isabel, N. The challenge of separating signatures of local adaptation from those of isolation by distance and colonization history: The case of two white pines. Ecol. Evol. 2016, 6, 8649–8664. [Google Scholar] [CrossRef] [PubMed]
- Rajora, O.P.; Eckert, A.J.; Zinck, J.W.R. Single-Locus versus Multilocus Patterns of Local Adaptation to Climate in Eastern White Pine (Pinus strobus, Pinaceae). PLoS ONE 2016, 11, e0158691. [Google Scholar] [CrossRef] [PubMed]
- Pais, A.L.; Whetten, R.W.; Xiang, Q.-Y.J. Ecological genomics of local adaptation in Cornus florida L. by genotyping by sequencing. Ecol. Evol. 2016, 7, 441–465. [Google Scholar] [CrossRef] [PubMed]
- Steane, D.A.; Potts, B.M.; McLean, E.; Collins, L.; Prober, S.M.; Stock, W.D.; Vaillancourt, R.E.; Byrne, M. Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt. Tree Genet. Genomes 2015, 11, 33. [Google Scholar] [CrossRef]
- Steane, D.A.; McLean, E.H.; Potts, B.M.; Prober, S.M.; Stock, W.D.; Stylianou, V.M.; Vaillancourt, R.E.; Byrne, M. Evidence for adaptation and acclimation in a widespread eucalypt of semi-arid Australia. Biol. J. Linn. Soc. 2017, 121, 484–500. [Google Scholar] [CrossRef]
- Steane, D.A.; Potts, B.M.; McLean, E.; Prober, S.M.; Stock, W.D.; Vaillancourt, R.E.; Byrne, M. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol. Ecol. 2014, 23, 2500–2513. [Google Scholar] [CrossRef] [PubMed]
- Lind, B.M.; Friedline, C.J.; Wegrzyn, J.L.; Maloney, P.E.; Vogler, D.R.; Neale, D.B.; Eckert, A.J. Water availability drives signatures of local adaptation in whitebark pine (Pinus albicaulis Engelm.) across fine spatial scales of the Lake Tahoe Basin, USA. Mol. Ecol. 2017, 26, 3168–3185. [Google Scholar] [CrossRef] [PubMed]
- Jump, A.S.; Hunt, J.M.; Peñuelas, J.; Martínez-Izquierdo, J.A.; Martínez-Izquierdo, J.A. Natural selection and climate change: Temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Mol. Ecol. 2006, 15, 3469–3480. [Google Scholar] [CrossRef] [PubMed]
- Eckert, A.J.; Maloney, P.E.; Vogler, D.R.; Jensen, C.E.; Mix, A.D.; Neale, D.B. Local adaptation at fine spatial scales: An example from sugar pine (Pinus lambertiana, Pinaceae). Tree Genet. Genomes 2015, 11, 42. [Google Scholar] [CrossRef]
- Seavy, N.E.; Gardali, T.; Golet, G.H.; Griggs, F.T.; Howell, C.A.; Kelsey, R.; Small, S.L.; Viers, J.H.; Weigand, J.F. Why Climate Change Makes Riparian Restoration More Important than Ever: Recommendations for Practice and Research. Ecol. Restor. 2009, 27, 330–338. [Google Scholar] [CrossRef]
- Wissmar, R.C.; Beschta, R.L. Restoration and management of riparian ecosystems: A catchment perspective. Freshw. Boil. 1998, 40, 571–585. [Google Scholar] [CrossRef]
- Perry, L.G.; Reynolds, L.V.; Beechie, T.J.; Collins, M.J.; Shafroth, P.B. Incorporating climate change projections into riparian restoration planning and design. Ecohydrology 2015, 8, 863–879. [Google Scholar] [CrossRef]
- Catford, J.A.; Naiman, R.J.; Chambers, L.E.; Roberts, J.; Douglas, M.; Davies, P. Predicting novel riparian ecosystems in a changing climate. Ecosystems 2013, 16, 382–400. [Google Scholar] [CrossRef]
- Capon, S.J.; Chambers, L.E.; Mac Nally, R.; Naiman, R.J.; Davies, P.; Marshall, N.; Pittock, J.; Reid, M.; Capon, T.; Douglas, M.; et al. Riparian Ecosystems in the 21st Century: Hotspots for Climate Change Adaptation? Ecosystems 2013, 16, 359–381. [Google Scholar] [CrossRef]
- Honnay, O.; Jacquemyn, H.; Nackaerts, K.; Breyne, P.; Van Looy, K. Patterns of population genetic diversity in riparian and aquatic plant species along rivers. J. Biogeogr. 2010, 37, 1730–1739. [Google Scholar] [CrossRef]
- Wubs, E.R.J.; Fraaije, R.G.A.; Groot, G.A.; Erkens, R.H.J.; Garssen, A.G.; Kleyheeg, E.; Raven, B.M.; Soons, M.B. Going against the flow: A case for upstream dispersal and detection of uncommon dispersal events. Freshw. Boil. 2016, 61, 580–595. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Indian Ocean Climate Initiative. Climate Variability and Change in South West Western Australia State Panel; Indian Ocean Climate Initiative: Panel, WA, USA, 2002; ISBN 1920687033. [Google Scholar]
- Hopley, T.; Byrne, M. Connectivity in riparian plants: Influence of vegetation type and habitat fragmentation overrides water flow. Oecologia 2018, 188, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Clim. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Bureau of Meterology; Australian Government Climate Data Online. Available online: http://www.bom.gov.au/climate/data (accessed on 12 November 2015).
- Rye, B.L. A revision of the south-western Australian genus Astartea (Myrtaceae: Chamelaucieae). Nuytsia 2013, 23, 189–269. [Google Scholar]
- Wheeler, J.; Marchant, N.; Lewington, M. Flora of the South West, Bunbury, Augusta, Denmark; UWA Publishing: Crawley, Australia, 2002; Volume 2. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Byrne, M.; Macdonald, B.; Francki, M. Incorporation of Sodium Sulfite into Extraction Protocol Minimizes Degradation of Acacia DNA. BioTechniques 2001, 30, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity Arrays Technology: A Generic Genome Profiling Technology on Open Platforms. In Evolutionary Genomics; Springer Science and Business Media LLC: Berlin, Germany, 2012; Volume 888, pp. 67–89. [Google Scholar]
- Gruber, B.; Georges, A. dartR: Importing and Analysing SNP and Silicodart Data Generated by Genome-Wide Restriction Fragment Analysis. 2019. Available online: https://rdrr.io/cran/dartR/ (accessed on 31 July 2019).
- R Core Team. R: A Language and Environment for Statistical Computing. 2016. Available online: https://www.gbif.org/en/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 31 July 2019).
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, 281. [Google Scholar] [CrossRef]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef]
- Naimi, B.; Hamm, N.A.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling. Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Gautier, M. Genome-Wide Scan for Adaptive Divergence and Association with Population-Specific Covariates. Genetics 2015, 201, 1555–1579. [Google Scholar] [CrossRef]
- Kass, R.E.; Raftery, A.E. Bayes Factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J.; Jombart, T. Hierfstat: Estimation and Tests of Hierarchical F-Statistics, R package version 0.04-22; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Francis, R.M. Pophelper: An R package and web app to analyse and visualise population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, S.; Jarvis, A. Regional heterogeneity and gene flow maintain variance in a quantitative trait within populations of lodgepole pine. Proc. R. Soc. B Boil. Sci. 2006, 273, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Lenormand, T. Gene flow and the limits to natural selection. Trends Ecol. Evol. 2002, 17, 183–189. [Google Scholar] [CrossRef]
- Chen, C.; Mitchell, S.E.; Elshire, R.J.; Buckler, E.S.; El-Kassaby, Y.A. Mining conifers’ mega-genome using rapid and efficient multiplexed high-throughput genotyping-by-sequencing (GBS) SNP discovery platform. Tree Genet. Genomes 2013, 9, 1537–1544. [Google Scholar] [CrossRef]
- White, H. Restoring Today’s Riparian Communities for Tomorrow’s Climate: Climate Adaptation in Riparian Ecosystems; University of Western Australia: Crawley, Australia, 2017. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hopley, T.; Byrne, M. Gene Flow and Genetic Variation Explain Signatures of Selection across a Climate Gradient in Two Riparian Species. Genes 2019, 10, 579. https://doi.org/10.3390/genes10080579
Hopley T, Byrne M. Gene Flow and Genetic Variation Explain Signatures of Selection across a Climate Gradient in Two Riparian Species. Genes. 2019; 10(8):579. https://doi.org/10.3390/genes10080579
Chicago/Turabian StyleHopley, Tara, and Margaret Byrne. 2019. "Gene Flow and Genetic Variation Explain Signatures of Selection across a Climate Gradient in Two Riparian Species" Genes 10, no. 8: 579. https://doi.org/10.3390/genes10080579
APA StyleHopley, T., & Byrne, M. (2019). Gene Flow and Genetic Variation Explain Signatures of Selection across a Climate Gradient in Two Riparian Species. Genes, 10(8), 579. https://doi.org/10.3390/genes10080579




