Abstract
Finding outlier loci underlying local adaptation is challenging and is best approached by suitable sampling design and rigorous method selection. In this study, we aimed to detect outlier loci (single nucleotide polymorphisms, SNPs) at the local scale by using Aleppo pine (Pinus halepensis), a drought resistant conifer that has colonized many habitats in the Mediterranean Basin, as the model species. We used a nested sampling approach that considered replicated altitudinal gradients for three contrasting sites. We genotyped samples at 294 SNPs located in genomic regions selected to maximize outlier detection. We then applied three different statistical methodologies—Two Bayesian outlier methods and one latent factor principal component method—To identify outlier loci. No SNP was an outlier for all three methods, while eight SNPs were detected by at least two methods and 17 were detected only by one method. From the intersection of outlier SNPs, only one presented an allelic frequency pattern associated with the elevational gradient across the three sites. In a context of multiple populations under similar selective pressures, our results underline the need for careful examination of outliers detected in genomic scans before considering them as candidates for convergent adaptation.
1. Introduction
Finding the genetic basis of local adaptation is a topic of crucial importance in evolutionary biology as it allows for the study of the mechanisms of natural selection [1,2]. However, there is still no consensus on how widespread local adaptation is as well as under which conditions it occurs. Studies based on reciprocal transplant and common garden experiments showed that plants can be locally adapted [3,4] and that local adaptation is more common for large populations. However, generalizing this phenomenon depends on the definition of local adaptation (relaxed versus a strict definition, [3]). Forest trees provide numerous examples of adaptive genetic divergence [5] as they generally combine a wide distribution across contrasting environmental conditions, large population sizes, and extensive levels of gene flow that may increase the rate of adaptation as well as high levels of genetic variation for fitness-related traits. However, examples that show that forest trees can genetically adapt at small geographical scales are limited [6]. The few studies that have looked at local adaptation in trees contemplating adaptive molecular divergence (e.g., [7,8,9,10]) or correlations between adaptive genetic variants and environmental variables [11,12,13] illustrate that the existence of spatially heterogeneous selective pressure may favor locally specialized genotypes despite extensive gene flow at the local scale.
In the present study, our objective was to shed new light on the genomic signatures of local adaptation in Aleppo pine (Pinus halepensis, Mill.). Aleppo pine has a circum-Mediterranean distribution that is both geographically fragmented and vast, spanning 3.5 million hectares [14]. This very drought resistant species is able to colonize and adapt to many habitats around the Mediterranean Basin as well as become invasive in southern hemisphere Mediterranean habitats [15,16]. Its environmental versatility lies both in its high plasticity [17,18] as well as, potentially, in its genetic adaptation to the Mediterranean region [19,20]. In Europe, paleoecological and fossil records as well as genetic data suggest that the Aleppo pine demographic history is characterized by different colonization and drift events, resulting in its genetic diversity following a longitudinal trend as many other species in the Mediterranean Basin [20,21]. This left Aleppo pine with a complex genetic pattern made of two main genetic clusters: an eastern/southern cluster that is more genetically diverse than the western cluster [20,22,23,24], with both being connected by population admixture [20,25]. The evolutionary history of Aleppo pine, combined with its ecological versatility in many Mediterranean habitats, constitute a good setting to test whether patterns of genetic divergence among heterogeneous environments hold across phylogeographic groups.
Ruiz Daniels et al. [20] previously investigated selection imprint in 44 populations of Aleppo pine covering the species range. Here, we focused on seven populations sampled along climatic gradients and aimed to detect convergent adaptation by combining the following. (1) A nested design of natural populations from distinct phylogeographic groups and contrasted environments, where we selected replicated populations in each site across two genetic clusters. Replicated populations were sampled at distinct elevations to reflect a combination of environments including differences in precipitation and temperature, two climatic variables known to affect the distribution and adaptation of this conifer [20,25,26,27,28,29]. Several studies have shown morphological and physiological adjustments of woody species along elevational gradients [30,31], which seem to therefore constitute relevant selection drivers. (2) A set of single nucleotide polymorphisms (SNPs) located in genomic regions selected to maximize outlier detection (see more details in the material and method section). The 294 SNPs genotyped in the seven populations fall within loci that were found to be under selection in pine species including Aleppo pine [19,20,24,32]. (3) A combination of methods that look at outlier loci based on environmental correlations or genetic differentiation. We took advantage of three recently developed statistical methods that correct for population structure: PCAdapt [33], Bayenv2 [34,35], and Baypass [36]. This attribute allows these methods to outperform methods that do not [37,38,39,40] and is a critical issue when there is evidence of shared history and gene flow among populations, as in Aleppo pine. The originality of the present work lies on the combination of these approaches to maximize the detection of outlier SNPs involved in convergent evolution in Aleppo pine.
2. Materials and Methods
2.1. Sampling
2.1.1. Plant Material and Climatic Data
Adult trees from three sites in three different countries spanning the two main phylogeographic clusters of Aleppo pine were sampled from population pairs or triplets: three French populations (356 individuals), two Italian populations (50 individuals), and two Spanish populations (70 individuals), totaling 476 individuals from seven populations (Figure 1a; see Tables S1 and S2 for more details). The sites are characterized by contrasted climatic conditions along altitudinal gradients. The aim of this sampling design was to maximize the differentiation between environments, while minimizing the differences in evolutionary history (gene flow should reduce differentiation at loci not under selection) [2]. This approach has been shown to be effective in comparative studies using simulated data [39] as well as when inferring outlier loci in empirical studies [12].
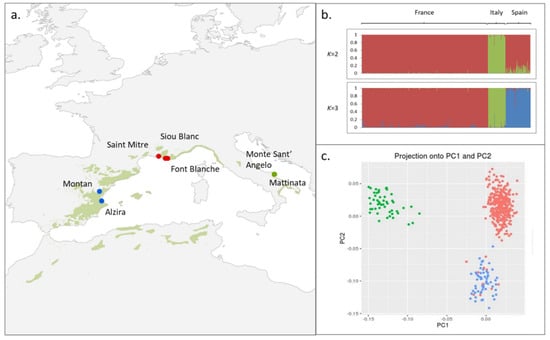
Figure 1.
Aleppo pine population structure. (a) Localization of the three populations superimposed on the natural range of Aleppo pine from EUFORGEN (green area). (b) Bayesian clustering performed in STRUCTURE for the seven populations. Population order from left to right is as follows: France (Font Blanche, Siou Blanc, Saint Mitre), Italy (Monte Sant’ Angelo, Mattinata), and Spain (Montan, Alzira). (c) Score plot of SNP data where each country is color coded (France: red, Italy: green, and Spain: blue). This plot displays the projections of the individuals onto the first PC (PC1) and the second PC (PC2).
For each population, the 19 bioclimatic variables for the period 1950–2000 (available from WorldClim [41]) were downloaded to explore SNP-environment associations (see Table S3). The accuracy of this coarse grain climatic dataset was checked with local climatic datasets for the climatic variables based on precipitation and temperature. Specifically, WorldClim variables for precipitation and temperature were compared with those from a functional phytoclimatic model based on raw data from meteorological stations [42] for the Spanish populations, and from local meteorological stations for the French populations. Furthermore, elevation was added as a potential driver of selection as it encompasses a combination of geophysical influences [43], bringing the total of environmental variables to 20 (see Table S2 for more details).
2.1.2. DNA Extraction, SNP Genotyping, and Gene Annotation
For every genotyped tree, 50 mg of needles were dried with silica gel and ground in a QIAwell (Qiagen, Venlo, the Netherlands) plate homogenized with a mixer mill MM300 (RETSH, Haan, Germany) under liquid nitrogen. DNA extractions were carried out with the Invisorb DNA plant HTS 96 kit (Invitek, Hayward, CA, USA) following the manufacturer’s instructions. DNA was quantified with Nanodrop 10,000 (Thermo Fisher Scientific, Wilmington, DE, USA).
Aleppo pine populations were successfully genotyped at 294 SNPs (conversion rate of 76.56%) using a 384-plex SNP assay with Illumina VeraCode technology as described in Pinosio et al. [44]. SNP genotypes are available at [45]. These SNPs originated from two sources. (i) 60% were identified from transcriptomic data coming from two Aleppo pine individuals with contrasting fire-response phenotypes [44]. A subset of these SNPs may be involved in adaption to fire as shown by the genetic association of some SNPs with serotiny [46]; and (ii) 40% were obtained by resequencing some loci originally developed in loblolly pine [47] as well as candidate genes from Grivet et al. [19,24]. Some of the loci involved in abiotic stress responses (e.g., cold, heat, drought, oxidative stresses) and phenology/photosystems have been shown to be under selection in pines, including that of Aleppo pine [19,20,24,32].
Loci for which SNPs were found putatively under selection using at least two different methods were annotated from homology with other plant species using Geneious version 6.1 [48] by searching against known loblolly pine EST contigs and the National Center for Biotechnology Information reference plant protein database.
2.2. Statistical Analysis
We used two Bayesian outlier approaches, Bayenv2 [34,35] and Baypass [36], as well as one method based on principal component analysis (PCA), implemented in the software PCAdapt [33]. Since methods for detecting outlier loci are based on underlying hypotheses that take into account different signals left by natural selection at the molecular level, there are inevitable discrepancies in the set of outlier loci [2,33,35,36,49]. Deciding which loci will be retained as candidates for further investigation is challenging, as is demonstrating that these outliers are candidates for convergent adaptation when detected across parallel elevational gradients.
2.2.1. Population Structure
To confirm that the population pairs/triplets were sampled from the same phylogeographic clusters, population genetic structure was estimated in two ways. First, pairwise FST was computed among all seven populations with Genepop v4.1 [50]. Second, the population genetic structure was inferred using the Bayesian clustering method STRUCTURE [51], with the following parameters: number of clusters (K) set from 1 to 10; number of iterations per K set to 10; number of steps set to 100,000 with a burn in period of 10,000 to minimize the effect of the starting configuration, and with an ancestry model of admixture. The convergence toward reliable allele frequency estimates in each genetic group and membership probabilities of individuals to a genetic group was assessed by checking that the alpha parameter was relatively constant, as indicated in the STRUCTURE manual.
2.2.2. Looking for Outliers Using PCA
The rationale of this method is that the FST index of genetic variation can also be viewed as the proportion of variance explained by principal components [52,53,54,55,56]. First, the number of principal components to be retained (K) was established by running the PCA analysis in PCAdapt [33] with a large enough number of components (10 in this case). The K displaying most of the cumulative explained variance was chosen based on a scree plot, which presents the percentage of variance explained by each principal component (PC) in decreasing order. The choice of K was also corroborated by plotting individuals on the first two components (called a score plot by PCAdapt) to verify if the level of clustering was consistent with the chosen value for K. To account for missing data, the correlation matrix between individuals was computed using only the markers available for each individual. The Mahalanobis distance was used to identify the outliers by measuring how distant a data point is from the multivariate space’s centroid (overall mean), considering the covariance structure of all of the data points in the sample. By default, alleles displaying MAF < 5% were removed in the PCAdapt analyses.
Both Manhattan and Quantile—Quantile plots were used to visualize the distribution of the data. The presence of outliers was confirmed by applying a false discovery rate (FDR), defined as the percentage of false positives among the list of candidate SNPs. The FDR was set at 10%, as recommended by Luu et al. [33] using the R package qvalue (R Core Team 2013) to compute the q-values based on the p-value distribution.
2.2.3. Looking for Outliers Using Bayesian Approaches
Bayesian approaches have been shown to perform well in situations where there is a non-uniform population structure [37,57]. The Baypass method is based on a very similar model as the Bayenv, with some improvements that allow for the better identification of significant outliers (see below). The two methodologies were used together with the intention of using the significance threshold for the XtX in Baypass to help define a cut-off point for the Bayenv outputs, for which standardized methods exist neither for the XtX statistics nor for the Bayes factors. The comparison aimed to also compare the outliers identified by the two methods.
- Bayenv
The method described in Coop et al. [34] and implemented in the Bayenv 2.0 package [35] estimates the population differentiation statistic XtX and evaluates the association of ecological variables with genetic marker differentiation. Bayenv2 first estimates the empirical pattern of covariance in allele frequencies between populations, and uses this as the null model to test each individual SNP for selection. To minimize the stochasticity in estimating the null model, three covariance matrices were produced from 100,000 iterations each, and the mean of these matrices (hereafter called mean covariance matrix) was then used. In order to test whether the mean covariance matrix represented the true variance of allele frequencies well across populations, it was compared to a pairwise FST matrix using a Mantel test in R (R Core Team 2013) with 1000 permutations.
This covariance matrix, which takes into account correlations due to population structure, was then used as the null model in two analyses aimed at detecting SNPs under selection. (1) Detecting outliers through differences in overall genetic differentiation as in classic outlier tests by estimating the XtX averages across multiple samples from the MCMC. Outlier XtX values from the main XtX distribution were considered as potentially under selection. Finally, these values were compared to the ones estimated using Baypass in order to ascertain significance. Indeed, in theory, Baypass and Bayenv2 both produce XtX statistics and can be easily compared against each other. However, Bayenv2 does suffer from a lack of pre-defined methods to establish outliers based on XtX statistics, which would have been difficult to do without the results from Baypass to use as a guideline. (2) Finding SNPs that show a significant correlation between an environmental variable and allele frequencies. This was carried out by comparing, via Bayes factors, a model that allows for a linear relationship between allele frequency from the 294 SNPs and the 20 environmental variables with a null model based on the covariance matrix. This approach was repeated three times for each combination of SNP-environmental variable tested for association to account for instability between independent runs [38]. The mean of the three runs was then used to infer the final Bayes factors (BF). The Kass and Raftery [58] criterion was then used to determine the probability of these SNPs under selection, with 2lnK values above six (BF > 20) classified as strong.
- Baypass
The Baypass method is based on a very similar model as Bayenv2, but is suggested to be an improvement in two aspects: (i) It has a higher accuracy in the estimation of the baseline (ancestral) allele frequency through the use of a hierarchical Bayesian model; and (ii) the use of simulated pseudo observed datasets (PODs) from the posterior predictive distribution in order to calibrate the XtX statistics and determine a cut-off point for detecting potentially significant outliers by applying a false discovery rate threshold.
Baypass [36] was run using the core model, and R (R Core Team 2013) was used to analyze and visualize outputs. First, the correlation matrix was computed and estimates of XtX differentiation were produced from the SNP data. The correlation matrix was then visualized as a hierarchical cluster tree in order to be compared to a neighbor joining tree of pairwise FST done in Genepop v4.1 [50]. Genetic distances between populations were computed using the R package APE [59]. Second, a POD was constructed using the simulate_baypass function in R with 1000 SNPs by estimating the posterior mean of the hyperparameters a_pi and b_pi, which specify the beta priors for Pi, the baseline (ancestral) frequency at each locus, and then simulating samples with these parameter values. The POD was then analyzed in the same way as the real data to produce values of XtX. The quantiles of these empirical XtX distributions were used to calibrate the XtX observed for each locus in the original data, and the 99% quantile of the XtX distribution from the POD analysis provided a 1% threshold XtX value (the default cut-off in Baypass). This threshold was then used to discriminate between outlier and neutral SNPs. Baypass is expected to outperform Bayenv2 because it uses the analyzed data to provide hyperparameters for the outlier detectors. However, as the two methods have not been used in conjunction, we ran them concomitantly to compare how these different features will impact outlier detection.
Baypass was also used to detect outlier SNPs showing a significant correlation between an environmental variable and allele frequencies. This was carried out by comparing a model that allows for a linear relationship between allele frequency and environmental variable with a null model using only the correlation matrix. For this purpose, Baypass was run using an importance sampling estimator (IS) model based on Coop et al. [34]. The empirical Bayesian p-value (eBPis) was calculated for each SNP-environmental relationship, allowing for evaluation of the support in favor of a non-null regression coefficient when the eBPis is above three [36]. As there is high variability among runs when using this feature in Bayenv2, it is advisable to run multiple analyses and then average the Bayes factors [38]. This is not the case with Baypass, where no such variability exists.
In the two Bayesian analyses, SNPs with a minimum allele frequency (MAF) below 5% were kept (contrary to the PCA-based method, where by default these SNPs are removed) as their removal is not recommended because some of these might be under selection.
3. Results
3.1. Population Structure
Pairwise population FST were all significant except between the two Italian populations (Table S4). Moreover, populations within each country were the least differentiated, while among countries, the French and Spanish populations were genetically closer to each other than to the Italian populations (Table S4). This result is in line with the Bayesian clustering that revealed an optimal grouping for K = 2 and K = 3 (Figure 1b and Figure S1), and as for K > 3, the structure signal was lost (data not shown) and the variance across the iterations increased (Figure S1). This grouping is in full agreement with the PCA analysis run with PCAdapt where the main proportion of explained variance lies mainly in two PCs; more specifically, PC1 separates the Italian populations from the French and Spanish populations (this scenario corresponds to K = 2 with STRUCTURE), while PC2 separates the French populations from the other two to a lesser degree (Figure 1c). The genetic closeness between the French and Spanish populations was also reflected by admixed individuals between the two countries as shown in Figure 1b,c. Furthermore, they shared other characteristics such as lower genetic diversity when compared to the Italian ones.
3.2. Looking for Outliers Using PCA
The scree plot clearly showed that most of the variation was accounted for at K = 3 (with the main proportion of explained variance for the two first PCs), and that it was unnecessary to use more PCs (Figure S2), as further confirmed by inspecting the scatterplot of the first two PCs (Figure 1c). The distribution of the p-values was visualized with a Manhattan plot and a QQ-plot, and then used to compute the q-values (Figure S3). Eleven outliers were detected (Table 1), setting q-values at a false discovery rate of 10%.

Table 1.
Summary statistics of the 14 outlier loci identified using the three outlier methods of the study.
3.3. Looking for Outliers Using Bayesian Approaches
- Bayenv
Confidence in the mean covariance matrix representing the true variance of allele frequencies across populations was assessed by the similarity between its heat map and that of pairwise FST values (Figure S4), that was found to be statistically significant according to the Mantel test (R2 = 0.801; p-value = 0.009).
The four top outliers from the XtX analysis were: SNP 4 (seq-9882-801), SNP 149 (seq-8671-529), SNP 316 (seq-10373-2483), and SNP 378 (seq-2_3941_01-381) (Table 1; Figure S5, left). When the Bayesian linear model was used with the 20 different environmental variables, four potential outlier SNPs were detected, classified as strong candidates for selection (20 < BF < 150) according to the Kass and Raftery (1995) criterion: SNP 169 (seq-0_10162_01-244), SNP 312 (seq-UMN_3408_01-293), SNP 316 (seq-10373-2483), and SNP 378 (seq-2_3941_01-381). Six environmental variables were involved in these correlations: elevation, BIO 2 (mean diurnal range), BIO 9 (mean temperature of driest quarter), BIO 12 (annual precipitation), BIO 16 (precipitation of wettest quarter), and BIO 19 (precipitation of coldest quarter) (Table S5).
- Baypass
Confidence in the correlation matrix , was first assessed by visually comparing the values among the populations (Figure S6). The correlation matrix can be viewed as a hierarchical cluster tree (Figure S7, left), where the relationships of the seven populations were similar to those found with a neighbor joining tree based on pairwise FST (Figure S7, right), with populations from the same country clustering together (the exception being the French population from Font Blanche in the hierarchical cluster based on that includes much more individuals, from five to ten times more than the other studied populations). Two outlier SNPs were observed: SNP 149 (seq-8671-529) and SNP 378 (seq-2_3941_01-381) (Table 1; Figure S5, right).
When Baypass was used to detect SNPs showing significant correlation with environmental variables based on eBPis > 3, 17 SNPs were detected (see Table S6). These were then compared to the ones in Bayenv2 and only the outliers detected using both methods for the same environmental variable were included as strong candidates for selection, leading to a total of three SNPs: SNP 169 (seq-0_10162_01-244), SNP 316 (seq-10373-2483), and SNP 378 (seq-2_3941_01-381) (Table 2).

Table 2.
SNPs that coincided in being outliers for the same environmental variables (Env.) for both Bayesian linear models performed with Bayenv 2 and Baypass.
3.4. Combined Results of All Outlier Tests
Eight SNPs were identified as outliers in at least two methods (Figure 2): SNP 4, SNP 149, SNP 169, SNP 258, SNP 269, SNP 281, SNP 316, and SNP 378.
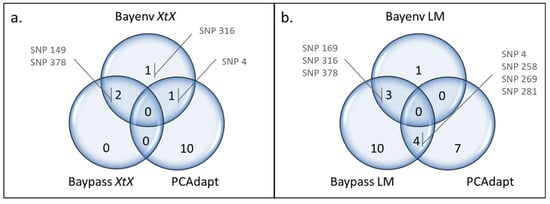
Figure 2.
Venn diagrams comparing the outliers detected using PCAdapt with those identified using (a) the XtX statistics from Bayenv2 and Baypass, and (b) the Bayesian linear model (LM) from Bayenv2 and Baypass. Only the SNPs found under selection in more than one method are indicated.
These SNPs belonged to six different loci, of which two could be annotated using the BLAST search (Table 3): seq-9882 (SNP 4 and SNP 258) corresponds to a PIN-like protein, while seq-10373-2483 (SNP 316) codes for a putative alpha-xylosidase (XYL1).

Table 3.
Annotation of the six loci where eight SNPs were found under selection with at least two methods.
Two outliers were detected with both Bayenv2 and Baypass making use of XtX statistics (SNP 149 and SNP 378), while Bayenv2 detected two additional ones (SNP 4 and SNP 316). Of the eleven outliers detected by PCAdapt, only SNP 4 was in common with the XtX outliers found in Bayenv2 (Figure 2a). Bayenv2 and Baypass revealed three outlier SNPs correlated with the same environmental variables (Figure 2b): SNP 169 was found in association with BIO9 (mean temperature driest quarter), SNP 316 with elevation, and SNP 378 with BIO12 (annual precipitation). The XtX and environmental association approaches revealed two outliers in common in Bayenv (SNP 316 and SNP 378; Table 1 and Table S5) and one in Baypass (SNP 378; Table 1 and Table S6). Out of the 11 SNPs detected with PCAdapt, four were in common with those detected by Baypass using environmental correlations (SNP 4, SNP 258, SNP 269, and SNP 281; Figure 2b), all associated with BIO12 (annual precipitation) (Table 4). Finally, SNP 316 detected with Bayenv2 based on the XtX statistics was also detected in environmental correlations with Bayenv2 and Baypass (Figure 2a,b).

Table 4.
SNPs found under selection by both PCAdapt and Baypass using the linear model to find SNPs associated with different environmental variables (Env.).
Two of the outlier SNPs potentially targeted by selection, SNP 4 and SNP 378, were remarkable in that they had allelic frequency patterns compatible with adaptation along the studied altitudinal gradients. The allelic frequency distribution of those top-candidate SNPs revealed several patterns: the allelic frequency distribution of SNP 4 indicated that allele A was at higher frequency in low elevation sites across the three countries, while its frequency was intermediate for the French site located at intermediate elevation (Figure 3).
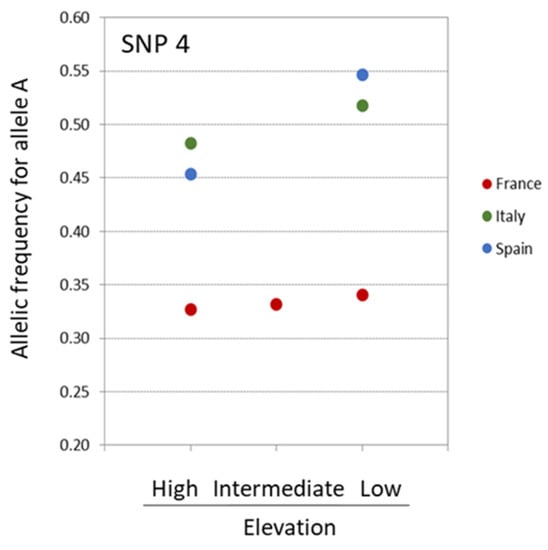
Figure 3.
Distribution of the frequency of allele A for SNP 4 detected under selection by PCAdapt, Bayenv2, and Baypass.
The allelic frequency distribution was somewhat similar for SNP 378 with the highest proportion of allele A associated with the site of highest altitude, while this proportion was lower for the intermediate and lowest altitude sites. However, this pattern was true only for the French replicates, with the Italian and Spanish replicates being monomorphic for the considered allele (Figure 4). Thus, only SNP 4 showed a pattern of allelic frequency across the three sets of populations compatible with convergent evolution under similar selection pressure linked to elevational gradient.
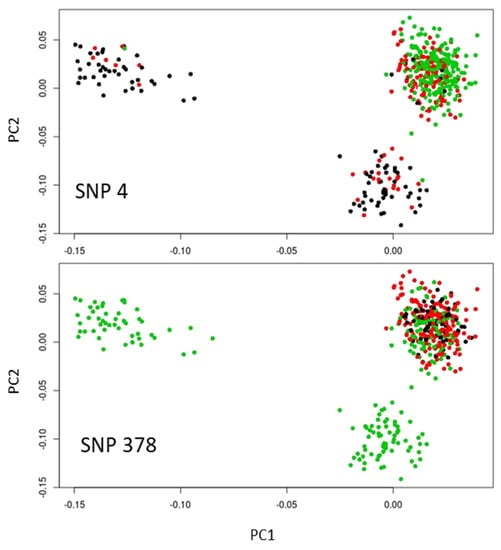
Figure 4.
Highlights on the score plot for the two main SNP outliers common to the outlier methods based on the XtX statistics (Bayenv2 and Baypass) and PCA (PCAdapt). Homozygotes for allele 1 and 2 are in green and black, respectively, while heterozygotes are in red. The populations are the same as in Figure 1c.
4. Discussion
Population structure analyses suggest that the Italian populations experienced a distinct evolutionary history when compared to the French and Spanish populations. This result is in agreement with the main genetic clusters previously defined: French and Spanish populations belong to the Western cluster of Aleppo pine and display lower genetic diversity when compared to the Italian populations that are part of the Eastern/Southern cluster [20]. Therefore, our sampling covered not only contrasting environmental conditions within population replicates (i.e., altitudinal gradient), but also distinct evolutionary genetic units. This sampling design allowed testing whether the pattern of genetic divergence among heterogeneous environments held between different genetic clusters. Sampling design (i.e., both sampling strategy and location) is one of the most influential factors when performing outlier detection tests in studies with simulated data [2,39]. Only recently have empirical analyses integrated an experimental approach based on replicated population pairs to increase the power of outlier detection [9,10].
Discovering genes potentially targeted by natural selection also relies on statistical frameworks that differ in the way they summarize complex data, and therefore could indicate different loci under selection. This outcome has been seen in both theoretical [39,57] and empirical [12,60,61] works including the present study. As of yet, there is no single widely accepted approach on how to combine statistical tests from multiple genome scan methods [40,49]. Even if the model specification is correct and the tests are properly calibrated, taking the intersection of the set of outlier SNPs leads to the identification of too few significant outliers, while taking the union leads to too many. On one hand, combining several well-calibrated tests can decrease the sensitivity to particular models and lead to more robust testing approaches [49]. On the other hand, by taking the intersection of SNPs, even if some have extreme p-values under some tests, we assume that the discarded loci are false-positives because the underlying assumptions of the respective tests are wrong. Finally, this conservative approach may lead to overlooking loci under weak selection. There are some indications that such SNPs exist in conifers [61]: local adaptation could result mainly from small, potentially undetectable, covarying shifts in frequency at many loci [2]. Looking for polygenic adaptation adds an extra layer of complexity when trying to disentangle the effects of demographic history and selection, and several methods have been recently developed to address this issue [62,63,64,65,66].
Locus annotation could be obtained only for one of the two top-candidate SNPs, SNP 4, which is located in a non-coding region of a PIN-like protein (PIN2 in Pinus tabuliformis; GenBank: AJP06341.1). This family of proteins act as auxin transporters and constitute key regulators in multiple developmental events ranging from embryogenesis through morphogenesis and organogenesis to growth responses to environmental stimuli [67]. The other top-candidate, SNP 378, which was detected by the two Bayesian linear models in association with annual precipitation in the French populations, was also detected independently using the Bayenv2 linear model in relation to the precipitation of the driest month in a study on the whole distribution range of Aleppo pine [20]. Altogether, these results point to SNP 378 as a very good candidate for selection and indicate that precipitation is a potential predominant driver of selection at two distinct spatial scales. The relevance of the precipitation regime for Aleppo pine has been emphasized in various disciplines such as ecophysiology [26,28,68], species distribution modeling [25], population genetics [20,27] and quantitative genetics [29].
The allelic frequency distribution of the two top-candidate SNPs highlights the importance of taking into account several aspects when interpreting the outputs of selection tests at a local scale: (i) the evolutionary history of the species (previous studies showed that Aleppo pine western populations are genetically depleted, [e.g. 20], which constitutes a limitation to detect polymorphic markers in the Spanish and French populations); (ii) the sampling size in terms of individuals per population (much more precise allelic frequencies could be obtained for the French population composed of 356 individuals compared to the Italian and Spanish populations comprising 50 and 70 individuals, respectively); and (iii) the sampling size in terms of replicated environmental gradients (three sites are somewhat limited to draw general conclusions, especially if some populations are monomorphic for the targeted SNPs). When detected, a convergent pattern of microevolution at local geographical scales may be attributed to parallel selective pressures at the locus, shared ancestral standing variation, or spread of the selected alleles via gene flow. Understanding the origin of convergent evolution can assist in dissecting the basis of local adaptation, and thereby in predicting adaptive response to selection [69,70]. In Aleppo pine, the selective pressure along an elevational cline could allow the species to cope with current and future climate changes characterized by higher temperature and lower precipitation in the Mediterranean Basin (International Panel on Climate Change, Fifth Assessment Report [71]). Moving uphill would allow individuals to be exposed to higher precipitation levels without latitudinal or longitudinal migration [72,73].
5. Conclusions
With the combination of the approaches used here (selection of putative candidate genes for genotyping, paired sampling technique, and multiple statistical approaches), eight SNPs were identified as outliers by at least two methods, of which only two (SNP 4 and SNP 378) had their allelic frequencies associated with elevational gradients, and one (SNP 4) across all population replicates. Looking for signal of convergent evolution at local or regional scales is an attractive and powerful approach to understand the molecular basis of adaptation. However, false positives could be linked not only to the statistical methods used, but also to the sampling. The outcome of our study cautions against considering outlier markers detected by genomic scans as automatic candidates for convergent adaptation in populations submitted to similar selective pressures.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/673/s1, Figure S1: Likelihood of K for each value of K estimated in STRUCTURE; Figure S2: Scree plot that displays the percentage of variance explained by each PC; Figure S3: Distribution of the empirical p-values obtained by PCAdapt visualized through a Manhattan plot and a QQ-plot; Figure S4: Heatmaps of the pairwise FST distance and the covariance matrix calculated in Bayenv2; Figure S5: The XtXs estimated from Bayenv2 and from Baypass; Figure S6: Correlation matrix comparing the values amongst the populations of the SNP dataset computed in Baypass; Figure S7: Correlation matrix visualized as a hierarchical cluster tree where the relationships between populations can be appreciated compared to a neighbor joining tree of pairwise FST; Table S1: Sampling details of the seven populations; Table S2: Environmental variables retrieved from WORDLCLIM for the seven populations; Table S3: List of bioclimatic variables BIO1–BIO19 available in WORDLCLIM; Table S4: Pairwise FST between the seven populations from France, Italy, and Spain; Table S5: Summary of the results of the Bayesian linear model performed in Bayenv2 with different bioclimatic variables; Table S6: Summary of the results of the Bayesian linear model performed in Baypass with different bioclimatic variables.
Author Contributions
Conceptualization, S.C.G-M., D.G., G.G.V., S.O-M., and B.F; Field work and data collection, S.C.G-M., D.G., G.G.V., S.O-M, A.P. and G.S.; SNP production, S.C.G-M., G.G.V., and D.G.; Data analysis, R.R.D. and R.S.T.; Contribution to data analysis, M.A.B. and D.G.; Original draft preparation, D.G. and R.R.D.; Review and editing, all the authors.
Funding
This project was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) (AdapCon CGL2011-30182-C02-00) by the project INIA-MAPAMA EG17-048 co-financed by FEADER (75%) according to EU Regulation 1305/2013 as well as by the ERA-Net BiodivERsA (projects LinkTree and TipTree). R.R.D. was supported by a FPI from the Spanish Ministry of Economy and Competitiveness; and R.S.T was funded through an Engineering and Physical Sciences Research Council (EPSRC) doctoral training grant (DTG) through the Bristol Centre for Complexity Science.
Acknowledgments
We would like to thank Fernando del Caño and Katharina Buddde (Spanish plots) as well as Celeste Labriola (Italian plots) for their field assistance, C. García-Barriga for DNA extraction, and Z. Lorenzo for help with SNP genotyping.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Savolainen, O.; Lascoux, M.; Merilä, J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013, 14, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Kelley, J.L.; Lotterhos, K.E.; Antolin, M.F.; Bradburd, G.; Lowry, D.B.; Poss, M.L.; Reed, L.K.; Storfer, A.; Whitlock, M.C. Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am. Nat. 2016, 188, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Leimu, R.; Fischer, M. A meta-analysis of local adaptation in plants. PLoS ONE 2008, 3, e4010. [Google Scholar] [CrossRef] [PubMed]
- Lascoux, M.; Glémin, S.; Savolainen, O. Local Adaptation in Plants; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 1–7. [Google Scholar]
- Savolainen, O.; Pyhäjärvi, T.; Knürr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- Scotti, I.; González-Martínez, S.C.; Budde, K.B.; Lalagüe, H. Fifty years of genetic studies: What to make of the large amounts of variation found within populations? Ann. For. Sci. 2016, 73, 69–75. [Google Scholar] [CrossRef]
- Audigeos, D.; Brousseau, L.; Traissac, S.; Scotti-Saintagne, C.; Scotti, I. Molecular divergence in tropical tree populations occupying environmental mosaics. J. Evol. Biol. 2013, 26, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, L.; Foll, M.; Scotti-Saintagne, C.; Scotti, I. Neutral and adaptive drivers of microgeographic genetic divergence within continuous populations: The case of the neotropical tree Eperua falcata (Aubl.). PLoS ONE 2015, 10, e0121394. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, L.; Postolache, D.; Lascoux, M.; Drouzas, A.D.; Källman, T.; Leonarduzzi, C.; Liepelt, S.; Piotti, A.; Popescu, F.; Roschanski, A.M.; et al. Local adaptation in European firs assessed through extensive sampling across altitudinal gradients in southern Europe. PLoS ONE 2016, 11, e0158216. [Google Scholar] [CrossRef]
- Roschanski, A.M.; Csilléry, K.; Liepelt, S.; Oddou-Muratorio, S.; Ziegenhagen, B.; Huard, F.; Ullrich, K.K.; Postolache, D.; Vendramin, G.G.; Fady, B. Evidence of divergent selection for drought and cold tolerance at landscape and local scales in Abies alba Mill. in the French Mediterranean Alps. Mol. Ecol. 2016, 25, 776–794. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.-P.; Rodríguez-Quilón, I.; Grivet, D.; Lepoittevin, C.; Sebastiani, F.; Heuertz, M.; Garnier-Géré, P.H.; Alía, R.; Plomion, C.; Vendramin, G.G.; et al. Molecular proxies for climate maladaptation in a long-lived tree (Pinus pinaster Aiton, Pinaceae). Genetics 2015, 199, 793–807. [Google Scholar] [CrossRef]
- Nadeau, S.; Meirmans, P.G.; Aitken, S.N.; Ritland, K.; Isabel, N. The challenge of separating signatures of local adaptation from those of isolation by distance and colonization history: The case of two white pines. Ecol. Evol. 2016, 6, 8649–8664. [Google Scholar] [CrossRef] [PubMed]
- Pluess, A.R.; Frank, A.; Heiri, C.; Lalagüe, H.; Vendramin, G.G.; Oddou-Muratorio, S. Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol. 2016, 210, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Fady, B. Pinus Halepensis and Pinus Brutia—Technical Guidelines for Genetic Conservation and Use for Aleppo and Brutia Pine: EUFORGEN—European forest genetic resources programme. 2003. Available online: http://www.euforgen.org/publications/publication/ipinus-halepensisi-and-ipinus-brutiai-technical-guidelines-for-genetic-conservation-and/ (accessed on 3 September 2019).
- Rouget, M.; Richardson, D.M.; Milton, S.J.; Polakow, D. Predicting invasion dynamics of four alien Pinus species in a highly fragmented semi-arid shrubland in South Africa. Plant Ecol. 2001, 152, 79–92. [Google Scholar] [CrossRef]
- Lavi, A.; Perevolotsky, A.; Kigel, J.; Noy-Meir, I. Invasion of Pinus halepensis from plantations into adjacent natural habitats. Appl. Veg. Sci. 2005, 8, 85–92. [Google Scholar] [CrossRef]
- Santos-Del-Blanco, L.; Climent, J.; González-Martínez, S.C.; Pannell, J.R. Genetic differentiation for size at first reproduction through male versus female functions in the widespread Mediterranean tree Pinus Pinaster. Ann. Bot. 2012, 110, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- de Luis, M.; Čufar, K.; Di Filippo, A.; Novak, K.; Papadopoulos, A.; Piovesan, G.; Rathgeber, C.B.K.; Raventós, J.; Saz, M.A.; Smith, K.T. Plasticity in dendroclimatic response across the distribution range of Aleppo pine (Pinus halepensis). PLoS ONE 2013, 8, e83550. [Google Scholar] [CrossRef] [PubMed]
- Grivet, D.; Sebastiani, F.; Alia, R.; Bataillon, T.; Torre, S.; Zabal-Aguirre, M.; Vendramin, G.G.; Gonzalez-Martinez, S.C. Molecular footprints of local adaptation in two Mediterranean conifers. Mol. Biol. Evol. 2011, 28, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Daniels, R.; Taylor, R.S.; Serra-Varela, M.J.; Vendramin, G.G.; González-Martínez, S.C.; Grivet, D. Inferring selection in instances of long-range colonization: The Aleppo pine (Pinus halepensis) in the Mediterranean Basin. Mol. Ecol. 2018, 27, 3331–3345. [Google Scholar] [CrossRef]
- Conord, C.; Gurevitch, J.; Fady, B. Large-scale longitudinal gradients of genetic diversity: A meta-analysis across six phyla in the Mediterranean Basin. Ecol. Evol. 2012, 2, 2600–2614. [Google Scholar] [CrossRef]
- Morgante, M.; Felice, N.; Vendramin, G.G. Analysis of hypervariable chloroplast microsatellites in Pinus halepensis reveals a dramatic genetic bottleneck. In Molecular Tools for Screening Biodiversity; Springer: Dordrecht, The Netherlands, 1998; pp. 407–412. [Google Scholar]
- Bucci, G.; Anzidei, M.; Madaghiele, A.; Vendramin, G.G. Detection of haplotypic variation and natural hybridization in halepensis -complex pine species using chloroplast simple sequence repeat (SSR) markers. Mol. Ecol. 1998, 7, 1633–1643. [Google Scholar] [CrossRef]
- Grivet, D.; Sebastiani, F.; González-Martínez, S.C.; Vendramin, G.G. Patterns of polymorphism resulting from long-range colonization in the Mediterranean conifer Aleppo pine. New Phytol. 2009, 184, 1016–1028. [Google Scholar] [CrossRef] [PubMed]
- Serra-Varela, M.J.; Alía, R.; Daniels, R.R.; Zimmermann, N.E.; Gonzalo-Jiménez, J.; Grivet, D. Assessing vulnerability of two Mediterranean conifers to support genetic conservation management in the face of climate change. Divers. Distrib. 2017, 23, 507–516. [Google Scholar] [CrossRef]
- Atzmon, N.; Moshe, Y.; Schiller, G. Ecophysiological response to severe drought in Pinus halepensis Mill. trees of two provenances. Plant Ecol. 2004, 171, 15–22. [Google Scholar] [CrossRef]
- Sathyan, P.; Newton, R.J.; Loopstra, C.A. Genes induced by WDS are differentially expressed in two populations of aleppo pine (Pinus halepensis). Tree Genet. Genomes 2005, 1, 166–173. [Google Scholar] [CrossRef]
- Voltas, J.; Chambel, M.R.; Prada, M.A.; Ferrio, J.P. Climate-related variability in carbon and oxygen stable isotopes among populations of Aleppo pine grown in common-garden tests. Trees 2008, 22, 759–769. [Google Scholar] [CrossRef]
- Santos-del-Blanco, L.; Bonser, S.P.; Valladares, F.; Chambel, M.R.; Climent, J. Plasticity in reproduction and growth among 52 range-wide populations of a Mediterranean conifer: Adaptive responses to environmental stress. J. Evol. Biol. 2013, 26, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Vitasse, Y.; Porté, A.J.; Kremer, A.; Michalet, R.; Delzon, S. Responses of canopy duration to temperature changes in four temperate tree species: Relative contributions of spring and autumn leaf phenology. Oecologia 2009, 161, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bresson, C.C.; Vitasse, Y.; Kremer, A.; Delzon, S. To what extent is altitudinal variation of functional traits driven by genetic adaptation in European oak and beech? Tree Physiol. 2011, 31, 1164–1174. [Google Scholar] [CrossRef]
- Grivet, D.; Avia, K.; Vaattovaara, A.; Eckert, A.J.; Neale, D.B.; Savolainen, O.; González-Martínez, S.C. High rate of adaptive evolution in two widespread European pines. Mol. Ecol. 2017, 26, 6857–6870. [Google Scholar] [CrossRef]
- Luu, K.; Bazin, E.; Blum, M.G.B. Pcadapt: An R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 2017, 17, 67–77. [Google Scholar] [CrossRef]
- Coop, G.; Witonsky, D.; Di Rienzo, A.; Pritchard, J.K. Using environmental correlations to identify loci underlying local adaptation. Genetics 2010, 185, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Coop, G. Robust identification of local adaptation from allele frequencies. Genetics 2013, 195, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M. Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 2015, 201, 1555–1579. [Google Scholar] [CrossRef] [PubMed]
- De Mita, S.; Thuillet, A.C.; Gay, L.; Ahmadi, N.; Manel, S.; Ronfort, J.; Vigouroux, Y. Detecting selection along environmental gradients: Analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol. Ecol. 2013, 22, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Blair, L.M.; Granka, J.M.; Feldman, M.W. On the stability of the Bayenv method in assessing human SNP-environment associations. Hum. Genom. 2014, 8, 1. [Google Scholar] [CrossRef]
- Lotterhos, K.E.; Whitlock, M.C. The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol. Ecol. 2015, 24, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces from global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Gonzalo, J. Phytoclimatic analysis of the Spanish Peninsula. Update and geostatistical analysis. Ph.D. Thesis, Universidad de Valladolid, Valladolid, Spain, 2007. [Google Scholar]
- Körner, C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 2007, 22, 569–574. [Google Scholar] [CrossRef]
- Pinosio, S.; González-Martínez, S.C.; Bagnoli, F.; Cattonaro, F.; Grivet, D.; Marroni, F.; Lorenzo, Z.; Pausas, J.G.; Verdú, M.; Vendramin, G.G. First insights into the transcriptome and development of new genomic tools of a widespread circum-Mediterranean tree species, Pinus halepensis Mill. Mol. Ecol. Resour. 2014, 14, 846–856. [Google Scholar] [CrossRef]
- Ruiz Daniels, R.; Taylor, R.S.; González-Martínez, S.C.; Vendramin, G.G.; Fady, B.; Oddou-Muratorio, S.; Piotti, A.; Simioni, G.; Grivet, D.; Beaumont, M.A. Looking for local adaptation: convergent microevolution in Aleppo pine (Pinus halepensis). Zenodo 2018. [Google Scholar] [CrossRef]
- Budde, K.B.; Heuertz, M.; Hernández-Serrano, A.; Pausas, J.G.; Vendramin, G.G.; Verdú, M.; González-Martínez, S.C. In situ genetic association for serotiny, a fire-related trait, in Mediterranean maritime pine (Pinus pinaster). New Phytol. 2014, 201, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Eckert, A.J.; Wegrzyn, J.L.; Liechty, J.D.; Lee, J.M.; Cumbie, W.P.; Davis, J.M.; Goldfarb, B.; Loopstra, C.A.; Palle, S.R.; Quesada, T.; et al. The evolutionary genetics of the genes underlying phenotypic associations for Loblolly pine (Pinus taeda, Pinaceae). Genetics 2013, 195, 1353–1372. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.geneious.com/ (accessed on 3 September 2019).
- François, O.; Martins, H.; Caye, K.; Schoville, S.D. Controlling false discoveries in genome scans for selection. Mol. Ecol. 2016, 25, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- McVean, G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009, 5, e1000686. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.B.; Lee, S.H.; Zhu, Z.X.; Benyamin, B.; Robinson, M.R. EigenGWAS: Finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Heredity 2016, 117, 51–61. [Google Scholar] [CrossRef]
- Duforet-Frebourg, N.; Luu, K.; Laval, G.; Bazin, E.; Blum, M.G.B. Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Mol. Biol. Evol. 2016, 33, 1082–1093. [Google Scholar] [CrossRef]
- Galinsky, K.J.; Bhatia, G.; Loh, P.R.; Georgiev, S.; Mukherjee, S.; Patterson, N.J.; Price, A.L. Fast Principal-Component Analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016, 98, 456–472. [Google Scholar] [CrossRef]
- Hao, W.; Song, M.; Storey, J.D. Probabilistic models of genetic variation in structured populations applied to global human studies. Bioinformatics 2016, 32, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Lotterhos, K.E.; Whitlock, M.C. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol. Ecol. 2014, 23, 2178–2192. [Google Scholar] [CrossRef] [PubMed]
- Kass, R.E.; Raftery, A.E. Bayes Factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, R.M.; VonHoldt, B.M.; Harrigan, R.; Knowles, J.C.; Musiani, M.; Coltman, D.; Novembre, J.; Wayne, R.K. Genetic subdivision and candidate genes under selection in North American grey wolves. Mol. Ecol. 2016, 25, 380–402. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, S.; Hodgins, K.A.; Lotterhos, K.E.; Suren, H.; Nadeau, S.; Degner, J.C.; Nurkowski, K.A.; Smets, P.; Wang, T.; Gray, L.K.; et al. Convergent local adaptation to climate in distantly related conifers. Science 2016, 353, 1431–1433. [Google Scholar] [CrossRef]
- Turchin, M.C.; Chiang, C.W.K.; Palmer, C.D.; Sankararaman, S.; Reich, D.; Hirschhorn, J.N. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 2012, 44, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Daub, J.T.; Hofer, T.; Cutivet, E.; Dupanloup, I.; Quintana-Murci, L.; Robinson-Rechavi, M.; Excoffier, L. Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 2013, 30, 1544–1558. [Google Scholar] [CrossRef]
- Berg, J.J.; Coop, G. A Population genetic signal of polygenic adaptation. PLoS Genet. 2014, 10, e1004412. [Google Scholar] [CrossRef]
- Robinson, M.R.; Hemani, G.; Medina-Gomez, C.; Mezzavilla, M.; Esko, T.; Shakhbazov, K.; Powell, J.E.; Vinkhuyzen, A.; Berndt, S.I.; Gustafsson, S.; et al. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 2015, 47, 1357–1361. [Google Scholar] [CrossRef]
- Racimo, F.; Berg, J.J.; Pickrell, J.K. Detecting polygenic adaptation in admixture graphs. Genetics 2018, 208, 1565–1584. [Google Scholar] [CrossRef] [PubMed]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zažímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Camarero, J.J.; Carrer, M. Shifts of irrigation in Aleppo pine under semi-arid conditions reveal uncoupled growth and carbon storage and legacy effects on wood anatomy. Agric. For. Meteorol. 2018, 253–254, 225–232. [Google Scholar] [CrossRef]
- Tiffin, P.; Ross-Ibarra, J. Advances and limits of using population genetics to understand local adaptation. Trends Ecol. Evol. 2014, 29, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Coop, G. Distinguishing among modes of convergent adaptation using population genomic data. Genetics 2017, 207, 1591–1619. [Google Scholar] [CrossRef] [PubMed]
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland; p. 151. Available online: http://www.ipcc.ch/report/ar5/syr/ (accessed on 3 September 2019).
- Ruiz-Labourdette, D.; Nogués-Bravo, D.; Ollero, H.S.; Schmitz, M.F.; Pineda, F.D. Forest composition in Mediterranean mountains is projected to shift along the entire elevational gradient under climate change. J. Biogeogr. 2012, 39, 162–176. [Google Scholar] [CrossRef]
- Benito-Garzón, M.; Fernández-Manjarrés, J.F. Testing scenarios for assisted migration of forest trees in Europe. New For. 2015, 46, 979–994. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).